The Molecular Biodiversity of Protein Targeting and Protein Transport Related to the Endoplasmic Reticulum
Abstract
1. Introduction
2. Two Prominent Types of Hydrophobic Targeting Signals
2.1. The Tripartite Nature of Cleavable Signal Peptides
The Correlation between Signal Peptides and Downstream Processes
2.2. Transmembrane Helices Are Efficient Targeting Signals
2.2.1. The Topological Diversity of Transmembrane Helices
2.2.2. The Crosstalk between Transmembrane Helices and Trans-Acting Factors
3. Multifactorial Polypeptide Targeting Pathways: The SRP, GET, and SND Systems
3.1. The Ancient Silk Road Project, SRP—Direct Way to the ER
3.2. The Juggling GET Cascade—Safety First
Installment of a Pre-Targeting Complex with Connections to Other Chaperone Systems
3.3. The Lonely SND Player—Mysterious All-Rounder
3.4. Comparison of Signal Recognition by the Various Chaperones Involved in Targeting
3.4.1. Cargo Capture by SRP
3.4.2. Cargo Capture by GET3
3.4.3. Cargo Capture by SGTA
3.4.4. Cargo Capture by BAG6
3.4.5. Cargo Capture by a Putative hSnd1
3.4.6. Common Cargo Capture Principles Used by Some Cytosolic Targeting Factors
4. The Interplay of Targeting Factors at the Ribosomal Exit Tunnel
4.1. Shaping the Polypeptide Fate from within the Ribosomal Tunnel
4.2. Allosteric Crosstalk from within the Tunnel to the Outside
4.3. Shaping the Polypeptide Fate from the Ribosomal Surface
5. Different ER Protein Translocases Act as Membrane-Integrated Chaperones
5.1. The Sec61 Translocase—Director for SPs and the Majority of TMHs
5.1.1. Opening of the Sec61 Complex by Targeting Signals
5.1.2. The Growing Family of Sec61 Complex-Associated Factors
5.2. The GET1/2 Complex—Post-Translational Machine for Type IV TMH of TA Proteins
The GET1/2 Duality—Receptor and Insertase Function
5.3. The EMC—Emcee for Type III, Type IV, and Charge-Containing TMHs
The Intra-Membrane Handover between EMC and Sec61
6. Disease-Causing Mutations of Targeting and Translocation Components
6.1. Disease Associations of Protein Targeting Factors
6.2. Disease Associations of Receptor and Protein Translocase Subunits
7. Conclusions and Perspectives
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
List of Abbreviations
| aa | amino acid |
| BAG6 | BCL2-associated athanogene 6 |
| cyto | cytoplasmic |
| CCDC47 | coiled-coil domain containing protein 47 |
| EMC | ER membrane protein complex |
| ER | endoplasmic reticulum |
| exo | exoplasmic |
| GET | guided entry of tail-anchored proteins |
| GPI | glycosylphosphatidylinositol |
| HSP | heat shock proteins |
| MP | membrane proteins |
| NAC | nascent-polypeptide-associated complex |
| NOMO | nodal modulator protein |
| OST | oligosaccharyltransferase |
| PAT | proteins associated with the ER translocon |
| PTC | peptidyl transferase center |
| rHRE | RNA hypoxia response elements |
| RNC | Ribosome-nascent chain complex |
| SGTA | small glutamine-rich tetratricopeptide repeat-containing protein alpha |
| SND | SRP-independent targeting pathway |
| SP | signal peptide |
| SR | SRP receptor |
| SRP | signal recognition particle |
| TA | tail-anchored |
| TMCO1 | transmembrane and coiled-coil domain-containing protein 1 |
| TMEM147 | transmembrane protein 147 |
| TMH | transmembrane helix |
| TRAM1 | translocating chain-associated membrane protein 1 |
| TRAP | translocon-associated protein |
References
- Baum, D.A.; Baum, B. An inside-out origin for the eukaryotic cell. BMC Biol. 2014, 12, 76. [Google Scholar] [CrossRef]
- Embley, T.M.; Martin, W. Eukaryotic evolution, changes and challenges. Nature 2006, 440, 623–630. [Google Scholar] [CrossRef] [PubMed]
- Sammels, E.; Parys, J.B.; Missiaen, L.; De Smedt, H.; Bultynck, G. Intracellular Ca2+ storage in health and disease: A dynamic equilibrium. Cell Calcium 2010, 47, 297–314. [Google Scholar] [CrossRef] [PubMed]
- Feske, S. Calcium signalling in lymphocyte activation and disease. Nat. Rev. Immunol. 2007, 7, 690–702. [Google Scholar] [CrossRef] [PubMed]
- Valm, A.M.; Cohen, S.; Legant, W.R.; Melunis, J.; Hershberg, U.; Wait, E.; Cohen, A.R.; Davidson, M.W.; Betzig, E.; Lippincott-Schwartz, J. Applying systems-level spectral imaging and analysis to reveal the organelle interactome. Nature 2017, 546, 162–167. [Google Scholar] [CrossRef] [PubMed]
- Palade, G.E. The endoplasmic reticulum. J. Biophys. Biochem. Cytol. 1956, 2, 85–98. [Google Scholar] [CrossRef] [PubMed]
- Pick, T.; Beck, A.; Gamayun, I.; Schwarz, Y.; Schirra, C.; Jung, M.; Krause, E.; Niemeyer, B.A.; Zimmermann, R.; Lang, S.; et al. Remodelling of Ca2+ homeostasis is linked to enlarged endoplasmic reticulum in secretory cells. Cell Calcium 2021, 99, 102473. [Google Scholar] [CrossRef] [PubMed]
- Federovitch, C.M.; Ron, D.; Hampton, R.Y. The dynamic ER: Experimental approaches and current questions. Curr. Opin. Cell Biol. 2005, 17, 409–414. [Google Scholar] [CrossRef]
- Aviram, N.; Schuldiner, M. Targeting and translocation of proteins to the endoplasmic reticulum at a glance. J. Cell Sci. 2017, 130, 4079–4085. [Google Scholar] [CrossRef] [PubMed]
- Guerriero, C.J.; Brodsky, J.L. The Delicate Balance Between Secreted Protein Folding and Endoplasmic Reticulum-Associated Degradation in Human Physiology. Physiol. Rev. 2012, 92, 537–576. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, R.; Eyrisch, S.; Ahmad, M.; Helms, V. Protein translocation across the ER membrane. Biochim. Et Biophys. Acta 2011, 1808, 912–924. [Google Scholar] [CrossRef]
- Walter, P.; Ibrahimi, I.; Blobel, G. Translocation of proteins across the endoplasmic reticulum. I. Signal recognition protein (SRP) binds to in-vitro-assembled polysomes synthesizing secretory protein. J. Cell Biol. 1981, 91, 545–550. [Google Scholar] [CrossRef] [PubMed]
- Gilmore, R.; Blobel, G.; Walter, P. Protein translocation across the endoplasmic reticulum. I. Detection in the microsomal membrane of a receptor for the signal recognition particle. J. Cell Biol. 1982, 95, 463–469. [Google Scholar] [CrossRef] [PubMed]
- Panzner, S.; Dreier, L.; Hartmann, E.; Kostka, S.; Rapoport, T.A. Posttranslational protein transport in yeast reconstituted with a purified complex of Sec proteins and Kar2p. Cell 1995, 81, 561–570. [Google Scholar] [CrossRef]
- Plath, K.; Rapoport, T.A. Spontaneous Release of Cytosolic Proteins from Posttranslational Substrates before Their Transport into the Endoplasmic Reticulum. J. Cell Biol. 2000, 151, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Johnson, N.; Powis, K.; High, S. Post-translational translocation into the endoplasmic reticulum. Biochim. Et Biophys. Acta (BBA) Mol. Cell Res. 2013, 1833, 2403–2409. [Google Scholar] [CrossRef] [PubMed]
- Haßdenteufel, S.; Nguyen, D.; Helms, V.; Lang, S.; Zimmermann, R. ER import of small human presecretory proteins: Components and mechanisms. FEBS Lett. 2019, 593, 2506–2524. [Google Scholar] [CrossRef] [PubMed]
- Stefanovic, S.; Hegde, R.S. Identification of a Targeting Factor for Posttranslational Membrane Protein Insertion into the ER. Cell 2007, 128, 1147–1159. [Google Scholar] [CrossRef] [PubMed]
- Schuldiner, M.; Metz, J.; Schmid, V.; Denic, V.; Rakwalska, M.; Schmitt, H.D.; Schwappach, B.; Weissman, J.S. The GET Complex Mediates Insertion of Tail-Anchored Proteins into the ER Membrane. Cell 2008, 134, 634–645. [Google Scholar] [CrossRef]
- Johnson, N.; Vilardi, F.; Lang, S.; Leznicki, P.; Zimmermann, R.; High, S. TRC40 can deliver short secretory proteins to the Sec61 translocon. J. Cell Sci. 2012, 125, 3612–3620. [Google Scholar] [CrossRef] [PubMed]
- Aviram, N.; Ast, T.; Costa, E.A.; Arakel, E.C.; Chuartzman, S.G.; Jan, C.H.; Haßdenteufel, S.; Dudek, J.; Jung, M.; Schorr, S.; et al. The SND proteins constitute an alternative targeting route to the endoplasmic reticulum. Nature 2016, 540, 134–138. [Google Scholar] [CrossRef] [PubMed]
- Haßdenteufel, S.; Sicking, M.; Schorr, S.; Aviram, N.; Fecher-Trost, C.; Schuldiner, M.; Jung, M.; Zimmermann, R.; Lang, S. hSnd2 protein represents an alternative targeting factor to the endoplasmic reticulum in human cells. FEBS Lett. 2017, 591, 3211–3224. [Google Scholar] [CrossRef] [PubMed]
- Casson, J.; McKenna, M.; Haßdenteufel, S.; Aviram, N.; Zimmerman, R.; High, S. Multiple pathways facilitate the biogenesis of mammalian tail-anchored proteins. J. Cell Sci. 2017, 130, 3851–3861. [Google Scholar] [CrossRef] [PubMed]
- Shao, S.; Hegde, R.S. A Calmodulin-Dependent Translocation Pathway for Small Secretory Proteins. Cell 2011, 147, 1576–1588. [Google Scholar] [CrossRef]
- Haßdenteufel, S.; Schäuble, N.; Cassella, P.; Leznicki, P.; Müller, A.; High, S.; Jung, M.; Zimmermann, R. Ca2+-calmodulin inhibits tail-anchored protein insertion into the mammalian endoplasmic reticulum membrane. FEBS Lett. 2011, 585, 3485–3490. [Google Scholar] [CrossRef] [PubMed]
- Hegde, R.S.; Keenan, R.J. The mechanisms of integral membrane protein biogenesis. Nat. Rev. Mol. Cell Biol. 2021. [Google Scholar] [CrossRef] [PubMed]
- O’Keefe, S.; Pool, M.R.; High, S. Membrane protein biogenesis at the ER: The highways and byways. FEBS J. 2021, n/a. [Google Scholar] [CrossRef]
- Park, E.; Rapoport, T.A. Mechanisms of Sec61/SecY-Mediated Protein Translocation Across Membranes. Annu. Rev. Biophys. 2012, 41, 21–40. [Google Scholar] [CrossRef] [PubMed]
- McDowell, M.A.; Heimes, M.; Sinning, I. Structural and molecular mechanisms for membrane protein biogenesis by the Oxa1 superfamily. Nat. Struct. Mol. Biol. 2021, 28, 234–239. [Google Scholar] [CrossRef] [PubMed]
- Gemmer, M.; Förster, F. A clearer picture of the ER translocon complex. J. Cell Sci. 2020, 133, jcs231340. [Google Scholar] [CrossRef] [PubMed]
- Rapoport, T.A.; Li, L.; Park, E. Structural and Mechanistic Insights into Protein Translocation. Annu. Rev. Cell Dev. Biol. 2017, 33, 369–390. [Google Scholar] [CrossRef] [PubMed]
- Chitwood, P.J.; Hegde, R.S. The Role of EMC during Membrane Protein Biogenesis. Trends Cell Biol. 2019, 29, 371–384. [Google Scholar] [CrossRef] [PubMed]
- Mateja, A.; Keenan, R.J. A structural perspective on tail-anchored protein biogenesis by the GET pathway. Curr. Opin. Struct. Biol. 2018, 51, 195–202. [Google Scholar] [CrossRef] [PubMed]
- Blobel, G.; Sabatini, D.D. Ribosome-Membrane Interaction in Eukaryotic Cells. In Biomembranes: Volume 2, Manson, L.A., Ed.; Springer: Boston, MA, USA, 1971; pp. 193–195. [Google Scholar]
- Hannigan, M.M.; Hoffman, A.M.; Thompson, W.; Zheng, T.; Nicchitta, C.V. Quantitative proteomics links the LRRC59 interactome to mRNA translation on the ER membrane. Mol. Cell. Proteom. 2020, 19, 1826–1849. [Google Scholar] [CrossRef]
- Bhadra, P.; Schorr, S.; Lerner, M.; Nguyen, D.; Dudek, J.; Förster, F.; Helms, V.; Lang, S.; Zimmermann, R. Quantitative Proteomics and Differential Protein Abundance Analysis after Depletion of Putative mRNA Receptors in the ER Membrane of Human Cells Identifies Novel Aspects of mRNA Targeting to the ER. Molecules 2021, 26, 3591. [Google Scholar] [CrossRef]
- Pyhtila, B.; Zheng, T.; Lager, P.J.; Keene, J.D.; Reedy, M.C.; Nicchitta, C.V. Signal sequence- and translation-independent mRNA localization to the endoplasmic reticulum. RNA 2008, 14, 445–453. [Google Scholar] [CrossRef]
- Béthune, J.; Jansen, R.-P.; Feldbrügge, M.; Zarnack, K. Membrane-Associated RNA-Binding Proteins Orchestrate Organelle-Coupled Translation. Trends Cell Biol. 2019, 29, 178–188. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.A.; Zhang, H.; Ilan, L.; Liu, A.X.; Kharchuk, I.; Palazzo, A.F. mRNA encoding Sec61β, a tail-anchored protein, is localized on the endoplasmic reticulum. J. Cell Sci. 2015, 128, 3398–3410. [Google Scholar] [CrossRef]
- Cui, X.A.; Zhang, H.; Palazzo, A.F. p180 Promotes the Ribosome-Independent Localization of a Subset of mRNA to the Endoplasmic Reticulum. PLoS Biol. 2012, 10, e1001336. [Google Scholar] [CrossRef] [PubMed]
- Hsu, J.C.C.; Reid, D.W.; Hoffman, A.M.; Sarkar, D.; Nicchitta, C.V. Oncoprotein AEG-1 is an endoplasmic reticulum RNA-binding protein whose interactome is enriched in organelle resident protein-encoding mRNAs. RNA 2018, 24, 688–703. [Google Scholar] [CrossRef] [PubMed]
- Hegde, R.S.; Kang, S.-W. The concept of translocational regulation. J. Cell Biol. 2008, 182, 225–232. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Hirata, T.; Liu, Y.-S.; Guo, X.-Y.; Gao, X.-D.; Kinoshita, T.; Fujita, M. Human SND2 mediates ER targeting of GPI-anchored proteins with low hydrophobic GPI attachment signals. FEBS Lett. 2021, 595, 1542–1558. [Google Scholar] [CrossRef] [PubMed]
- Davis, E.M.; Kim, J.; Menasche, B.L.; Sheppard, J.; Liu, X.; Tan, A.-C.; Shen, J. Comparative Haploid Genetic Screens Reveal Divergent Pathways in the Biogenesis and Trafficking of Glycophosphatidylinositol-Anchored Proteins. Cell Rep. 2015, 11, 1727–1736. [Google Scholar] [CrossRef] [PubMed]
- Dhiman, R.; Caesar, S.; Thiam, A.R.; Schrul, B. Mechanisms of protein targeting to lipid droplets: A unified cell biological and biophysical perspective. Semin. Cell Dev. Biol. 2020, 108, 4–13. [Google Scholar] [CrossRef]
- Allen, K.N.; Entova, S.; Ray, L.C.; Imperiali, B. Monotopic Membrane Proteins Join the Fold. Trends Biochem. Sci. 2019, 44, 7–20. [Google Scholar] [CrossRef] [PubMed]
- Blobel, G.; Dobberstein, B. Transfer of proteins across membranes. I. Presence of proteolytically processed and unprocessed nascent immunoglobulin light chains on membrane-bound ribosomes of murine myeloma. J. Cell Biol. 1975, 67, 835–851. [Google Scholar] [CrossRef] [PubMed]
- Blobel, G.; Dobberstein, B. Transfer of proteins across membranes. II. Reconstitution of functional rough microsomes from heterologous components. J. Cell Biol. 1975, 67, 852–862. [Google Scholar] [CrossRef] [PubMed]
- Gilmore, R.; Blobel, G. Transient involvement of signal recognition particle and its receptor in the microsomal membrane prior to protein translocation. Cell 1983, 35, 677–685. [Google Scholar] [CrossRef]
- Gilmore, R.; Walter, P.; Blobel, G. Protein translocation across the endoplasmic reticulum. II. Isolation and characterization of the signal recognition particle receptor. J. Cell Biol. 1982, 95, 470–477. [Google Scholar] [CrossRef] [PubMed]
- Jackson, R.C.; Blobel, G. Post-translational cleavage of presecretory proteins with an extract of rough microsomes from dog pancreas containing signal peptidase activity. Proc. Natl. Acad. Sci. USA 1977, 74, 5598–5602. [Google Scholar] [CrossRef]
- Walter, P.; Blobel, G. Purification of a membrane-associated protein complex required for protein translocation across the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 1980, 77, 7112–7116. [Google Scholar] [CrossRef] [PubMed]
- Walter, P.; Blobel, G. Translocation of proteins across the endoplasmic reticulum. II. Signal recognition protein (SRP) mediates the selective binding to microsomal membranes of in-vitro-assembled polysomes synthesizing secretory protein. J. Cell Biol. 1981, 91, 551–556. [Google Scholar] [CrossRef] [PubMed]
- Walter, P.; Blobel, G. Translocation of proteins across the endoplasmic reticulum III. Signal recognition protein (SRP) causes signal sequence-dependent and site-specific arrest of chain elongation that is released by microsomal membranes. J. Cell Biol. 1981, 91, 557–561. [Google Scholar] [CrossRef] [PubMed]
- Walter, P.; Blobel, G. Signal recognition particle contains a 7S RNA essential for protein translocation across the endoplasmic reticulum. Nature 1982, 299, 691–698. [Google Scholar] [CrossRef] [PubMed]
- Hegde, R.S.; Bernstein, H.D. The surprising complexity of signal sequences. Trends Biochem. Sci. 2006, 31, 563–571. [Google Scholar] [CrossRef] [PubMed]
- von Heijne, G. Towards a comparative anatomy of N-terminal topogenic protein sequences. J. Mol. Biol. 1986, 189, 239–242. [Google Scholar] [CrossRef]
- Nilsson, I.; Lara, P.; Hessa, T.; Johnson, A.E.; von Heijne, G.; Karamyshev, A.L. The Code for Directing Proteins for Translocation across ER Membrane: SRP Cotranslationally Recognizes Specific Features of a Signal Sequence. J. Mol. Biol. 2015, 427, 1191–1201. [Google Scholar] [CrossRef] [PubMed]
- O’Neil, K.T.; DeGrado, W.F. A Thermodynamic Scale for the Helix-Forming Tendencies of the Commonly Occurring Amino Acids. Science 1990, 250, 646–651. [Google Scholar] [CrossRef]
- Lyu, P.C.; Liff, M.I.; Marky, L.A.; Kallenbach, N.R. Side Chain Contributions to the Stability of Alpha-Helical Structure in Peptides. Science 1990, 250, 669–673. [Google Scholar] [CrossRef] [PubMed]
- Lumangtad, L.A.; Bell, T.W. The signal peptide as a new target for drug design. Bioorganic Med. Chem. Lett. 2020, 30, 127115. [Google Scholar] [CrossRef] [PubMed]
- von Heijne, G. Signal sequences: The limits of variation. J. Mol. Biol. 1985, 184, 99–105. [Google Scholar] [CrossRef]
- von Heijne, G. The signal peptide. J. Membr. Biol. 1990, 115, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Gierasch, L.M. Signal sequences. Biochemistry 1989, 28, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Käll, L.; Krogh, A.; Sonnhammer, E.L.L. A Combined Transmembrane Topology and Signal Peptide Prediction Method. J. Mol. Biol. 2004, 338, 1027–1036. [Google Scholar] [CrossRef] [PubMed]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, C.A.; Preuss, D.; Grisafi, P.; Botstein, D. Many Random Sequences Functionally Replace the Secretion Signal Sequence of Yeast Invertase. Science 1987, 235, 312–317. [Google Scholar] [CrossRef] [PubMed]
- Beck, B.B.; Trachtman, H.; Gitman, M.; Miller, I.; Sayer, J.A.; Pannes, A.; Baasner, A.; Hildebrandt, F.; Wolf, M.T.F. Autosomal Dominant Mutation in the Signal Peptide of Renin in a Kindred With Anemia, Hyperuricemia, and CKD. Am. J. Kidney Dis. 2011, 58, 821–825. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Živná, M.; Hůlková, H.; Matignon, M.; Hodaňová, K.; Vylet’al, P.; Kalbáčová, M.; Barešová, V.; Sikora, J.; Blažková, H.; Živný, J.; et al. Dominant Renin Gene Mutations Associated with Early-Onset Hyperuricemia, Anemia, and Chronic Kidney Failure. Am. J. Hum. Genet. 2009, 85, 204–213. [Google Scholar] [CrossRef]
- Clissold, R.L.; Clarke, H.C.; Spasic-Boskovic, O.; Brugger, K.; Abbs, S.; Bingham, C.; Shaw-Smith, C. Discovery of a novel dominant mutation in the REN gene after forty years of renal disease: A case report. BMC Nephrol. 2017, 18, 234. [Google Scholar] [CrossRef]
- Seppen, J.; Steenken, E.; Lindhout, D.; Bosma, P.J.; Oude Elferink, R.P.J. A mutation which disrupts the hydrophobic core of the signal peptide of bilirubin UDP-glucuronosyltransferase, an endoplasmic reticulum membrane protein, causes Crigler-Najjar type IIs. FEBS Lett. 1996, 390, 294–298. [Google Scholar] [CrossRef]
- Sneitz, N.; Bakker, C.T.; de Knegt, R.J.; Halley, D.J.J.; Finel, M.; Bosma, P.J. Crigler-Najjar syndrome in The Netherlands: Identification of four novel UGT1A1 alleles, genotype–phenotype correlation, and functional analysis of 10 missense mutants. Hum. Mutat. 2010, 31, 52–59. [Google Scholar] [CrossRef]
- Rothe, C.; Lehle, L. Sorting of invertase signal peptide mutants in yeast dependent and independent on the signal-recognition particle. Eur. J. Biochem. 1998, 252, 16–24. [Google Scholar] [CrossRef] [PubMed]
- Williams, E.J.B.; Pal, C.; Hurst, L.D. The molecular evolution of signal peptides. Gene 2000, 253, 313–322. [Google Scholar] [CrossRef]
- Rutkowski, D.T.; Lingappa, V.R.; Hegde, R.S. Substrate-specific regulation of the ribosome—Translocon junction by N-terminal signal sequences. Proc. Natl. Acad. Sci. USA 2001, 98, 7823–7828. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.-W.; Rane, N.S.; Kim, S.J.; Garrison, J.L.; Taunton, J.; Hegde, R.S. Substrate-Specific Translocational Attenuation during ER Stress Defines a Pre-Emptive Quality Control Pathway. Cell 2006, 127, 999–1013. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.J.; Mitra, D.; Salerno, J.R.; Hegde, R.S. Signal Sequences Control Gating of the Protein Translocation Channel in a Substrate-Specific Manner. Dev. Cell 2002, 2, 207–217. [Google Scholar] [CrossRef]
- Keenan, R.J.; Freymann, D.M.; Walter, P.; Stroud, R.M. Crystal Structure of the Signal Sequence Binding Subunit of the Signal Recognition Particle. Cell 1998, 94, 181–191. [Google Scholar] [CrossRef]
- Van den Berg, B.; Clemons, W.M., Jr.; Collinson, I.; Modis, Y.; Hartmann, E.; Harrison, S.C.; Rapoport, T.A. X-ray structure of a protein-conducting channel. Nature 2004, 427, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Voorhees, R.M.; Hegde, R.S. Structure of the Sec61 channel opened by a signal sequence. Science 2016, 351, 88–91. [Google Scholar] [CrossRef] [PubMed]
- Ng, D.T.; Brown, J.D.; Walter, P. Signal sequences specify the targeting route to the endoplasmic reticulum membrane. J. Cell Biol. 1996, 134, 269–278. [Google Scholar] [CrossRef] [PubMed]
- Waters, M.G.; Blobel, G. Secretory protein translocation in a yeast cell-free system can occur posttranslationally and requires ATP hydrolysis. J. Cell Biol. 1986, 102, 1543–1550. [Google Scholar] [CrossRef] [PubMed]
- Lang, S.; Benedix, J.; Fedeles, S.V.; Schorr, S.; Schirra, C.; Schäuble, N.; Jalal, C.; Greiner, M.; Haßdenteufel, S.; Tatzelt, J.; et al. Different effects of Sec61α, Sec62 and Sec63 depletion on transport of polypeptides into the endoplasmic reticulum of mammalian cells. J. Cell Sci. 2012, 125, 1958–1969. [Google Scholar] [CrossRef] [PubMed]
- Johnson, N.; Haßdenteufel, S.; Theis, M.; Paton, A.W.; Paton, J.C.; Zimmermann, R.; High, S. The Signal Sequence Influences Post-Translational ER Translocation at Distinct Stages. PLoS ONE 2013, 8, e75394. [Google Scholar] [CrossRef] [PubMed]
- Lakkaraju, A.K.K.; Thankappan, R.; Mary, C.; Garrison, J.L.; Taunton, J.; Strub, K. Efficient secretion of small proteins in mammalian cells relies on Sec62-dependent posttranslational translocation. Mol. Biol. Cell 2012, 23, 2712–2722. [Google Scholar] [CrossRef]
- Wu, X.; Cabanos, C.; Rapoport, T.A. Structure of the post-translational protein translocation machinery of the ER membrane. Nature 2019, 566, 136–139. [Google Scholar] [CrossRef] [PubMed]
- Haßdenteufel, S.; Johnson, N.; Paton, A.W.; Paton, J.C.; High, S.; Zimmermann, R. Chaperone-Mediated Sec61 Channel Gating during ER Import of Small Precursor Proteins Overcomes Sec61 Inhibitor-Reinforced Energy Barrier. Cell Rep. 2018, 23, 1373–1386. [Google Scholar] [CrossRef] [PubMed]
- Sicking, M.; Jung, M.; Lang, S. Lights, Camera, Interaction: Studying Protein–Protein Interactions of the ER Protein Translocase in Living Cells. Int. J. Mol. Sci. 2021, 22, 10358. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.; Stutz, R.; Schorr, S.; Lang, S.; Pfeffer, S.; Freeze, H.H.; Förster, F.; Helms, V.; Dudek, J.; Zimmermann, R. Proteomics reveals signal peptide features determining the client specificity in human TRAP-dependent ER protein import. Nat. Commun. 2018, 9, 3765. [Google Scholar] [CrossRef] [PubMed]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L.L. Predicting transmembrane protein topology with a hidden markov model: Application to complete genomes11Edited by F. Cohen. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [PubMed]
- Von Heijne, G. The membrane protein universe: What’s out there and why bother? J. Intern. Med. 2007, 261, 543–557. [Google Scholar] [CrossRef]
- Sharpe, H.J.; Stevens, T.J.; Munro, S. A Comprehensive Comparison of Transmembrane Domains Reveals Organelle-Specific Properties. Cell 2010, 142, 158–169. [Google Scholar] [CrossRef]
- Baker, J.A.; Wong, W.-C.; Eisenhaber, B.; Warwicker, J.; Eisenhaber, F. Charged residues next to transmembrane regions revisited: “Positive-inside rule” is complemented by the “negative inside depletion/outside enrichment rule”. BMC Biol. 2017, 15, 66. [Google Scholar] [CrossRef] [PubMed]
- Höhr, A.I.C.; Lindau, C.; Wirth, C.; Qiu, J.; Stroud, D.A.; Kutik, S.; Guiard, B.; Hunte, C.; Becker, T.; Pfanner, N.; et al. Membrane protein insertion through a mitochondrial β-barrel gate. Science 2018, 359, eaah6834. [Google Scholar] [CrossRef] [PubMed]
- Spiess, M.; Junne, T.; Janoschke, M. Membrane Protein Integration and Topogenesis at the ER. Protein J. 2019, 38, 306–316. [Google Scholar] [CrossRef] [PubMed]
- Higy, M.; Junne, T.; Spiess, M. Topogenesis of Membrane Proteins at the Endoplasmic Reticulum. Biochemistry 2004, 43, 12716–12722. [Google Scholar] [CrossRef] [PubMed]
- Devaraneni, P.K.; Conti, B.; Matsumura, Y.; Yang, Z.; Johnson, A.E.; Skach, W.R. Stepwise Insertion and Inversion of a Type II Signal Anchor Sequence in the Ribosome-Sec61 Translocon Complex. Cell 2011, 146, 134–147. [Google Scholar] [CrossRef] [PubMed]
- Goder, V.; Spiess, M. Molecular mechanism of signal sequence orientation in the endoplasmic reticulum. Embo J. 2003, 22, 3645–3653. [Google Scholar] [CrossRef] [PubMed]
- Chitwood, P.J.; Juszkiewicz, S.; Guna, A.; Shao, S.; Hegde, R.S. EMC Is Required to Initiate Accurate Membrane Protein Topogenesis. Cell 2018, 175, 1507–1519. [Google Scholar] [CrossRef]
- O’Keefe, S.; Zong, G.; Duah, K.B.; Andrews, L.E.; Shi, W.Q.; High, S. An alternative pathway for membrane protein biogenesis at the endoplasmic reticulum. Commun. Biol. 2021, 4, 828. [Google Scholar] [CrossRef] [PubMed]
- Junne, T.; Spiess, M. Integration of transmembrane domains is regulated by their downstream sequences. J. Cell Sci. 2017, 130, 372–381. [Google Scholar] [CrossRef]
- von Heijne, G. Membrane-protein topology. Nat. Rev. Mol. Cell Biol. 2006, 7, 909. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, J.; Persson, B.; von Heijne, G. Comparative analysis of amino acid distributions in integral membrane proteins from 107 genomes. Proteins Struct. Funct. Bioinform. 2005, 60, 606–616. [Google Scholar] [CrossRef] [PubMed]
- von Heijne, G. Control of topology and mode of assembly of a polytopic membrane protein by positively charged residues. Nature 1989, 341, 456–458. [Google Scholar] [CrossRef] [PubMed]
- Kida, Y.; Morimoto, F.; Mihara, K.; Sakaguchi, M. Function of Positive Charges Following Signal-Anchor Sequences during Translocation of the N-terminal Domain *. J. Biol. Chem. 2006, 281, 1152–1158. [Google Scholar] [CrossRef] [PubMed]
- Denzer, A.J.; Nabholz, C.E.; Spiess, M. Transmembrane orientation of signal-anchor proteins is affected by the folding state but not the size of the N-terminal domain. EMBO J. 1995, 14, 6311–6317. [Google Scholar] [CrossRef] [PubMed]
- Arkin, I.T.; Brunger, A.T. Statistical analysis of predicted transmembrane α-helices. Biochim. Et Biophys. Acta (BBA) Protein Struct. Mol. Enzymol. 1998, 1429, 113–128. [Google Scholar] [CrossRef]
- Dou, D.; Silva, D.V.d.; Nordholm, J.; Wang, H.; Daniels, R. Type II transmembrane domain hydrophobicity dictates the cotranslational dependence for inversion. Mol. Biol. Cell 2014, 25, 3363–3374. [Google Scholar] [CrossRef]
- Zhang, L.; Sato, Y.; Hessa, T.; von Heijne, G.; Lee, J.-K.; Kodama, I.; Sakaguchi, M.; Uozumi, N. Contribution of hydrophobic and electrostatic interactions to the membrane integration of the Shaker K+ channel voltage sensor domain. Proc. Natl. Acad. Sci. USA 2007, 104, 8263–8268. [Google Scholar] [CrossRef] [PubMed]
- Öjemalm, K.; Halling, K.K.; Nilsson, I.; von Heijne, G. Orientational Preferences of Neighboring Helices Can Drive ER Insertion of a Marginally Hydrophobic Transmembrane Helix. Mol. Cell 2012, 45, 529–540. [Google Scholar] [CrossRef] [PubMed]
- Ismail, N.; Crawshaw, S.G.; High, S. Active and passive displacement of transmembrane domains both occur during opsin biogenesis at the Sec61 translocon. J. Cell Sci. 2006, 119, 2826–2836. [Google Scholar] [CrossRef] [PubMed]
- Ismail, N.; Crawshaw, S.G.; Cross, B.C.S.; Haagsma, A.C.; High, S. Specific transmembrane segments are selectively delayed at the ER translocon during opsin biogenesis. Biochem. J. 2008, 411, 495–506. [Google Scholar] [CrossRef] [PubMed]
- Sato, M.; Hresko, R.; Mueckler, M. Testing the Charge Difference Hypothesis for the Assembly of a Eucaryotic Multispanning Membrane Protein. J. Biol. Chem. 1998, 273, 25203–25208. [Google Scholar] [CrossRef] [PubMed]
- Sato, M.; Mueckler, M. A Conserved Amino Acid Motif (R-X-G-R-R) in the Glut1 Glucose Transporter Is an Important Determinant of Membrane Topology. J. Biol. Chem. 1999, 274, 24721–24725. [Google Scholar] [CrossRef] [PubMed]
- Coelho, J.P.L.; Stahl, M.; Bloemeke, N.; Meighen-Berger, K.; Alvira, C.P.; Zhang, Z.-R.; Sieber, S.A.; Feige, M.J. A network of chaperones prevents and detects failures in membrane protein lipid bilayer integration. Nat. Commun. 2019, 10, 672. [Google Scholar] [CrossRef] [PubMed]
- Doyle, D.A.; Cabral, J.M.; Pfuetzner, R.A.; Kuo, A.; Gulbis, J.M.; Cohen, S.L.; Chait, B.T.; MacKinnon, R. The Structure of the Potassium Channel: Molecular Basis of K+ Conduction and Selectivity. Science 1998, 280, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Brambillasca, S.; Yabal, M.; Makarow, M.; Borgese, N. Unassisted translocation of large polypeptide domains across phospholipid bilayers. J. Cell Biol. 2006, 175, 767–777. [Google Scholar] [CrossRef]
- Brambillasca, S.; Yabal, M.; Soffientini, P.; Stefanovic, S.; Makarow, M.; Hegde, R.S.; Borgese, N. Transmembrane topogenesis of a tail-anchored protein is modulated by membrane lipid composition. EMBO J. 2005, 24, 2533–2542. [Google Scholar] [CrossRef]
- Dowhan, W.; Bogdanov, M. Lipid-Dependent Membrane Protein Topogenesis. Annu. Rev. Biochem. 2009, 78, 515–540. [Google Scholar] [CrossRef] [PubMed]
- Ast, T.; Schuldiner, M. All roads lead to Rome (but some may be harder to travel): SRP-independent translocation into the endoplasmic reticulum. Crit. Rev. Biochem. Mol. Biol. 2013, 48, 273–288. [Google Scholar] [CrossRef] [PubMed]
- Ast, T.; Cohen, G.; Schuldiner, M. A Network of Cytosolic Factors Targets SRP-Independent Proteins to the Endoplasmic Reticulum. Cell 2013, 152, 1134–1145. [Google Scholar] [CrossRef] [PubMed]
- Schlenstedt, G.; Zimmermann, R. Import of frog prepropeptide GLa into microsomes requires ATP but does not involve docking protein or ribosomes. EMBO J. 1987, 6, 699–703. [Google Scholar] [CrossRef]
- Wiech, H.; Sagstetter, M.; Müller, G.; Zimmermann, R. The ATP requiring step in assembly of M13 procoat protein into microsomes is related to preservation of transport competence of the precursor protein. EMBO J. 1987, 6, 1011–1016. [Google Scholar] [CrossRef] [PubMed]
- Müller, G.; Zimmermann, R. Import of honeybee prepromelittin into the endoplasmic reticulum: Structural basis for independence of SRP and docking protein. EMBO J. 1987, 6, 2099–2107. [Google Scholar] [CrossRef] [PubMed]
- Costa, E.A.; Subramanian, K.; Nunnari, J.; Weissman, J.S. Defining the physiological role of SRP in protein-targeting efficiency and specificity. Science 2018, 359, 689–692. [Google Scholar] [CrossRef] [PubMed]
- Hann, B.C.; Walter, P. The signal recognition particle in S. cerevisiae. Cell 1991, 67, 131–144. [Google Scholar] [CrossRef]
- Mukhopadhyay, R.; Ho, Y.-S.; Swiatek, P.J.; Rosen, B.P.; Bhattacharjee, H. Targeted disruption of the mouse Asna1 gene results in embryonic lethality. FEBS Lett. 2006, 580, 3889–3894. [Google Scholar] [CrossRef]
- Daniele, L.L.; Emran, F.; Lobo, G.P.; Gaivin, R.J.; Perkins, B.D. Mutation of wrb, a Component of the Guided Entry of Tail-Anchored Protein Pathway, Disrupts Photoreceptor Synapse Structure and Function. Investig. Ophthalmol. Vis. Sci. 2016, 57, 2942–2954. [Google Scholar] [CrossRef] [PubMed]
- Rivera-Monroy, J.; Musiol, L.; Unthan-Fechner, K.; Farkas, Á.; Clancy, A.; Coy-Vergara, J.; Weill, U.; Gockel, S.; Lin, S.-Y.; Corey, D.P.; et al. Mice lacking WRB reveal differential biogenesis requirements of tail-anchored proteins in vivo. Sci. Rep. 2016, 6, 39464. [Google Scholar] [CrossRef] [PubMed]
- Vogl, C.; Panou, I.; Yamanbaeva, G.; Wichmann, C.; Mangosing, S.J.; Vilardi, F.; Indzhykulian, A.A.; Pangršič, T.; Santarelli, R.; Rodriguez-Ballesteros, M.; et al. Tryptophan-rich basic protein (WRB) mediates insertion of the tail-anchored protein otoferlin and is required for hair cell exocytosis and hearing. EMBO J. 2016, 35, 2536–2552. [Google Scholar] [CrossRef]
- Coy-Vergara, J.; Rivera-Monroy, J.; Urlaub, H.; Lenz, C.; Schwappach, B. A trap mutant reveals the physiological client spectrum of TRC40. J. Cell Sci. 2019, 132, jcs230094. [Google Scholar] [CrossRef] [PubMed]
- Hein, M.Y.; Hubner, N.C.; Poser, I.; Cox, J.; Nagaraj, N.; Toyoda, Y.; Gak, I.A.; Weisswange, I.; Mansfeld, J.; Buchholz, F.; et al. A Human Interactome in Three Quantitative Dimensions Organized by Stoichiometries and Abundances. Cell 2015, 163, 712–723. [Google Scholar] [CrossRef]
- Carvalho, H.J.F.; Del Bondio, A.; Maltecca, F.; Colombo, S.F.; Borgese, N. The WRB Subunit of the Get3 Receptor is Required for the Correct Integration of its Partner CAML into the ER. Sci. Rep. 2019, 9, 11887. [Google Scholar] [CrossRef] [PubMed]
- Colombo, S.F.; Cardani, S.; Maroli, A.; Vitiello, A.; Soffientini, P.; Crespi, A.; Bram, R.F.; Benfante, R.; Borgese, N. Tail-anchored protein biogenesis in mammals: Function and reciprocal interactions of the two subunits of the TRC40 receptor. J. Biol. Chem. 2016, 291, 15292–15306. [Google Scholar] [CrossRef] [PubMed]
- Egea, P.F.; Shan, S.O.; Napetschnig, J.; Savage, D.F.; Walter, P.; Stroud, R.M. Substrate twinning activates the signal recognition particle and its receptor. Nature 2004, 427, 215–221. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Jomaa, A.; Chung, S.; Fu, Y.-H.H.; Qian, R.; Sun, X.; Hsieh, H.-H.; Chandrasekar, S.; Bi, X.; Mattei, S.; et al. Receptor compaction and GTPase rearrangement drive SRP-mediated cotranslational protein translocation into the ER. Sci. Adv. 2021, 7, eabg0942. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, K.; Jomaa, A.; Lee, J.H.; Chandrasekar, S.; Boehringer, D.; Shan, S.-O.; Ban, N. Structure of a prehandover mammalian ribosomal SRP·SRP receptor targeting complex. Science 2018, 360, 323–327. [Google Scholar] [CrossRef] [PubMed]
- Jomaa, A.; Fu, Y.-H.H.; Boehringer, D.; Leibundgut, M.; Shan, S.-O.; Ban, N. Structure of the quaternary complex between SRP, SR, and translocon bound to the translating ribosome. Nat. Commun. 2017, 8, 15470. [Google Scholar] [CrossRef]
- Bohnsack, M.T.; Schleiff, E. The evolution of protein targeting and translocation systems. Biochim. Et Biophys. Acta (BBA) Mol. Cell Res. 2010, 1803, 1115–1130. [Google Scholar] [CrossRef] [PubMed]
- Wild, K.; Becker, M.M.M.; Kempf, G.; Sinning, I. Structure, dynamics and interactions of large SRP variants. Biol. Chem. 2020, 401, 63–80. [Google Scholar] [CrossRef]
- Halic, M.; Becker, T.; Pool, M.R.; Spahn, C.M.; Grassucci, R.A.; Frank, J.; Beckmann, R. Structure of the signal recognition particle interacting with the elongation-arrested ribosome. Nature 2004, 427, 808–814. [Google Scholar] [CrossRef] [PubMed]
- Häsler, J.; Strub, K. Alu RNP and Alu RNA regulate translation initiation in vitro. Nucleic Acids Res. 2006, 34, 2374–2385. [Google Scholar] [CrossRef] [PubMed]
- Lakkaraju, A.K.; Mary, C.; Scherrer, A.; Johnson, A.E.; Strub, K. SRP keeps polypeptides translocation-competent by slowing translation to match limiting ER-targeting sites. Cell 2008, 133, 440–451. [Google Scholar] [CrossRef] [PubMed]
- Schibich, D.; Gloge, F.; Pöhner, I.; Björkholm, P.; Wade, R.C.; von Heijne, G.; Bukau, B.; Kramer, G. Global profiling of SRP interaction with nascent polypeptides. Nature 2016, 536, 219. [Google Scholar] [CrossRef] [PubMed]
- Abell, B.M.; Pool, M.R.; Schlenker, O.; Sinning, I.; High, S. Signal recognition particle mediates post-translational targeting in eukaryotes. EMBO J. 2004, 23, 2755–2764. [Google Scholar] [CrossRef] [PubMed]
- Reid, D.W.; Nicchitta, C.V. Diversity and selectivity in mRNA translation on the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 2015, 16, 221–231. [Google Scholar] [CrossRef] [PubMed]
- Jagannathan, S.; Reid, D.W.; Cox, A.H.; Nicchitta, C.V. De novo translation initiation on membrane-bound ribosomes as a mechanism for localization of cytosolic protein mRNAs to the endoplasmic reticulum. RNA 2014, 20, 1489–1498. [Google Scholar] [CrossRef] [PubMed]
- Jan, C.H.; Williams, C.C.; Weissman, J.S. Principles of ER cotranslational translocation revealed by proximity-specific ribosome profiling. Science 2014, 346, 1257521. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Jagannathan, S.; Reid, D.W.; Zheng, T.; Nicchitta, C.V. Hierarchical regulation of mRNA partitioning between the cytoplasm and the endoplasmic reticulum of mammalian cells. Mol. Biol. Cell 2011, 22, 2646–2658. [Google Scholar] [CrossRef] [PubMed]
- Leznicki, P.; High, S. SGTA associates with nascent membrane protein precursors. EMBO Rep. 2020, 21, e48835. [Google Scholar] [CrossRef] [PubMed]
- Kutay, U.; Hartmann, E.; Rapoport, T.A. A class of membrane proteins with a C-terminal anchor. Trends Cell Biol. 1993, 3, 72–75. [Google Scholar] [CrossRef]
- Shan, S.-O. Guiding Tail-anchored Membrane Proteins to the ER In a Chaperone Cascade. J. Biol. Chem. 2019, 294, 16577–16586. [Google Scholar] [CrossRef] [PubMed]
- Borgese, N.; Coy-Vergara, J.; Colombo, S.F.; Schwappach, B. The Ways of Tails: The GET Pathway and more. Protein J. 2019, 38, 289–305. [Google Scholar] [CrossRef] [PubMed]
- Stefer, S.; Reitz, S.; Wang, F.; Wild, K.; Pang, Y.-Y.; Schwarz, D.; Bomke, J.; Hein, C.; Löhr, F.; Bernhard, F.; et al. Structural Basis for Tail-Anchored Membrane Protein Biogenesis by the Get3-Receptor Complex. Science 2011, 333, 758–762. [Google Scholar] [CrossRef]
- Rome, M.E.; Rao, M.; Clemons, W.M.; Shan, S.-O. Precise timing of ATPase activation drives targeting of tail-anchored proteins. Proc. Natl. Acad. Sci. USA 2013, 110, 7666–7671. [Google Scholar] [CrossRef] [PubMed]
- Rao, M.; Okreglak, V.; Chio, U.S.; Cho, H.; Walter, P.; Shan, S.-O. Multiple selection filters ensure accurate tail-anchored membrane protein targeting. eLife 2016, 5, e21301. [Google Scholar] [CrossRef]
- Guna, A.; Volkmar, N.; Christianson, J.C.; Hegde, R.S. The ER membrane protein complex is a transmembrane domain insertase. Science 2018, 359, 470–473. [Google Scholar] [CrossRef] [PubMed]
- Gristick, H.B.; Rome, M.E.; Chartron, J.W.; Rao, M.; Hess, S.; Shan, S.-O.; Clemons, W.M. Mechanism of Assembly of a Substrate Transfer Complex during Tail-anchored Protein Targeting. J. Biol. Chem. 2015, 290, 30006–30017. [Google Scholar] [CrossRef] [PubMed]
- Darby, J.F.; Krysztofinska, E.M.; Simpson, P.J.; Simon, A.C.; Leznicki, P.; Sriskandarajah, N.; Bishop, D.S.; Hale, L.R.; Alfano, C.; Conte, M.R.; et al. Solution Structure of the SGTA Dimerisation Domain and Investigation of Its Interactions with the Ubiquitin-Like Domains of BAG6 and UBL4A. PLoS ONE 2014, 9, e113281. [Google Scholar] [CrossRef] [PubMed]
- Mock, J.-Y.; Chartron, J.W.; Zaslaver, M.A.; Xu, Y.; Ye, Y.; Clemons, W.M. Bag6 complex contains a minimal tail-anchor–targeting module and a mock BAG domain. Proc. Natl. Acad. Sci. USA 2015, 112, 106–111. [Google Scholar] [CrossRef]
- Guna, A.; Hegde, R.S. Transmembrane Domain Recognition during Membrane Protein Biogenesis and Quality Control. Curr. Biol. 2018, 28, R498–R511. [Google Scholar] [CrossRef]
- Rodrigo-Brenni, M.C.; Gutierrez, E.; Hegde, R.S. Cytosolic Quality Control of Mislocalized Proteins Requires RNF126 Recruitment to Bag6. Mol. Cell 2014, 55, 227–237. [Google Scholar] [CrossRef] [PubMed]
- Rodrigo-Brenni, M.C.; Hegde, R.S. Design Principles of Protein Biosynthesis-Coupled Quality Control. Dev. Cell 2012, 23, 896–907. [Google Scholar] [CrossRef] [PubMed]
- Hessa, T.; Sharma, A.; Mariappan, M.; Eshleman, H.D.; Gutierrez, E.; Hegde, R.S. Protein targeting and degradation are coupled for elimination of mislocalized proteins. Nature 2011, 475, 394–397. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Brown, E.C.; Mak, G.; Zhuang, J.; Denic, V. A chaperone cascade sorts proteins for posttranslational membrane insertion into the endoplasmic reticulum. Mol. Cell 2010, 40, 159–171. [Google Scholar] [CrossRef]
- Shao, S.; Rodrigo-Brenni, M.C.; Kivlen, M.H.; Hegde, R.S. Mechanistic basis for a molecular triage reaction. Science 2017, 355, 298–302. [Google Scholar] [CrossRef] [PubMed]
- Wunderley, L.; Leznicki, P.; Payapilly, A.; High, S. SGTA regulates the cytosolic quality control of hydrophobic substrates. J. Cell Sci. 2014, 127, 4728–4739. [Google Scholar] [CrossRef] [PubMed]
- Krysztofinska, E.M.; Martínez-Lumbreras, S.; Thapaliya, A.; Evans, N.J.; High, S.; Isaacson, R.L. Structural and functional insights into the E3 ligase, RNF126. Sci. Rep. 2016, 6, 26433. [Google Scholar] [CrossRef] [PubMed]
- Leznicki, P.; Roebuck, Q.P.; Wunderley, L.; Clancy, A.; Krysztofinska, E.M.; Isaacson, R.L.; Warwicker, J.; Schwappach, B.; High, S. The Association of BAG6 with SGTA and Tail-Anchored Proteins. PLoS ONE 2013, 8, e59590. [Google Scholar] [CrossRef]
- Figueiredo Costa, B.; Cassella, P.; Colombo, S.F.; Borgese, N. Discrimination between the endoplasmic reticulum and mitochondria by spontaneously inserting tail-anchored proteins. Traffic 2018, 19, 182–197. [Google Scholar] [CrossRef] [PubMed]
- Cichocki, B.A.; Krumpe, K.; Vitali, D.G.; Rapaport, D. Pex19 is involved in importing dually targeted tail-anchored proteins to both mitochondria and peroxisomes. Traffic 2018, 19, 770–785. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.; Shim, W.J.; Liu, Y.; Shan, S.-O. J-domain proteins promote client relay from Hsp70 during tail-anchored membrane protein targeting. J. Biol. Chem. 2021, 296, 100546. [Google Scholar] [CrossRef]
- Cho, H.; Shan, S.-O. Substrate relay in an Hsp70-cochaperone cascade safeguards tail-anchored membrane protein targeting. EMBO J. 2018, 37, e99264. [Google Scholar] [CrossRef]
- Martínez-Lumbreras, S.; Krysztofinska, E.M.; Thapaliya, A.; Spilotros, A.; Matak-Vinkovic, D.; Salvadori, E.; Roboti, P.; Nyathi, Y.; Muench, J.H.; Roessler, M.M.; et al. Structural complexity of the co-chaperone SGTA: A conserved C-terminal region is implicated in dimerization and substrate quality control. BMC Biol. 2018, 16, 76. [Google Scholar] [CrossRef] [PubMed]
- Hegde, R.S.; Zavodszky, E. Recognition and Degradation of Mislocalized Proteins in Health and Disease. Cold Spring Harb. Perspect. Biol. 2019, 11, a033902. [Google Scholar] [CrossRef] [PubMed]
- Powis, K.; Schrul, B.; Tienson, H.; Gostimskaya, I.; Breker, M.; High, S.; Schuldiner, M.; Jakob, U.; Schwappach, B. Get3 is a holdase chaperone and moves to deposition sites for aggregated proteins when membrane targeting is blocked. J. Cell Sci. 2013, 126, 473–483. [Google Scholar] [CrossRef] [PubMed]
- Shing, J.C.; Lindquist, L.D.; Borgese, N.; Bram, R.J. CAML mediates survival of Myc-induced lymphoma cells independent of tail-anchored protein insertion. Cell Death Discov. 2017, 3, 16098. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Shing, J.C.; Bram, R.J. Yet another hump for CAML: Support of cell survival independent of tail-anchored protein insertion. Cell Death Dis. 2017, 8, e2960. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, Y.; Sakisaka, T. The emerging role of calcium-modulating cyclophilin ligand in posttranslational insertion of tail-anchored proteins into the endoplasmic reticulum membrane. J. Biochem. 2015, 157, 419–429. [Google Scholar] [CrossRef] [PubMed]
- Inglis, A.J.; Page, K.R.; Guna, A.; Voorhees, R.M. Differential Modes of Orphan Subunit Recognition for the WRB/CAML Complex. Cell Rep. 2020, 30, 3691–3698. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Lin, Y.; Zhang, H.; Mañas, A.; Tang, W.; Zhang, Y.; Wu, D.; Lin, A.; Xiang, J. Ubl4A is required for insulin-induced Akt plasma membrane translocation through promotion of Arp2/3-dependent actin branching. Proc. Natl. Acad. Sci. USA 2015, 112, 9644–9649. [Google Scholar] [CrossRef]
- Wang, Y.; Ning, H.; Ren, F.; Zhang, Y.; Rong, Y.; Wang, Y.; Su, F.; Cai, C.; Jin, Z.; Li, Z.; et al. GdX/UBL4A Specifically Stabilizes the TC45/STAT3 Association and Promotes Dephosphorylation of STAT3 to Repress Tumorigenesis. Mol. Cell 2014, 53, 752–765. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Li, J.; Fu, Y.; Ren, F.; Xu, J.; Zhou, M.; Li, P.; Feng, H.; Wang, Y. GdX/UBL4A null mice exhibit mild kyphosis and scoliosis accompanied by dysregulation of osteoblastogenesis and chondrogenesis. Cell Biochem. Funct. 2018, 36, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Ye, P.; Yuan, D.S.; Wang, X.; Bader, J.S.; Boeke, J.D. A DNA Integrity Network in the Yeast Saccharomyces cerevisiae. Cell 2006, 124, 1069–1081. [Google Scholar] [CrossRef]
- Talbot, B.E.; Vandorpe, D.H.; Stotter, B.R.; Alper, S.L.; Schlondorff, J.S. Transmembrane insertases and N-glycosylation critically determine synthesis, trafficking, and activity of the nonselective cation channel TRPC6. J. Biol. Chem. 2019, 294, 12655–12669. [Google Scholar] [CrossRef] [PubMed]
- Fleischer, T.C.; Weaver, C.M.; McAfee, K.J.; Jennings, J.L.; Link, A.J. Systematic identification and functional screens of uncharacterized proteins associated with eukaryotic ribosomal complexes. Genes Dev. 2006, 20, 1294–1307. [Google Scholar] [CrossRef]
- Mehlhorn, D.G.; Asseck, L.Y.; Grefen, C. Looking for a safe haven: Tail-anchored proteins and their membrane insertion pathways. Plant Physiol. 2021, 187, 1916–1928. [Google Scholar] [CrossRef]
- Borgese, N. Searching for remote homologs of CAML among eukaryotes. Traffic 2020, 21, 647–658. [Google Scholar] [CrossRef] [PubMed]
- Lütcke, H.; High, S.; Römisch, K.; Ashford, A.J.; Dobberstein, B. The methionine-rich domain of the 54 kDa subunit of signal recognition particle is sufficient for the interaction with signal sequences. EMBO J. 1992, 11, 1543–1551. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, H.D.; Poritz, M.A.; Strub, K.; Hoben, P.J.; Brenner, S.; Walter, P. Model for signal sequence recognition from amino-acid sequence of 54K subunit of signal recognition particle. Nature 1989, 340, 482–486. [Google Scholar] [CrossRef]
- Voorhees, R.M.; Hegde, R.S. Structures of the scanning and engaged states of the mammalian SRP-ribosome complex. eLife 2015, 4, e07975. [Google Scholar] [CrossRef]
- Zopf, D.; Bernstein, H.; Johnson, A.; Walter, P. The methionine-rich domain of the 54 kd protein subunit of the signal recognition particle contains an RNA binding site and can be crosslinked to a signal sequence. EMBO J. 1990, 9, 4511–4517. [Google Scholar] [CrossRef] [PubMed]
- Mateja, A.; Paduch, M.; Chang, H.-Y.; Szydlowska, A.; Kossiakoff, A.A.; Hegde, R.S.; Keenan, R.J. Structure of the Get3 targeting factor in complex with its membrane protein cargo. Science 2015, 347, 1152–1155. [Google Scholar] [CrossRef] [PubMed]
- Bozkurt, G.; Stjepanovic, G.; Vilardi, F.; Amlacher, S.; Wild, K.; Bange, G.; Favaloro, V.; Rippe, K.; Hurt, E.; Dobberstein, B.; et al. Structural insights into tail-anchored protein binding and membrane insertion by Get3. Proc. Natl. Acad. Sci. USA 2009, 106, 21131–21136. [Google Scholar] [CrossRef]
- Farkas, Á.; De Laurentiis, E.I.; Schwappach, B. The natural history of Get3-like chaperones. Traffic 2019, 20, 311–324. [Google Scholar] [CrossRef] [PubMed]
- Chio, U.S.; Chung, S.; Weiss, S.; Shan, S.-o. A Chaperone Lid Ensures Efficient and Privileged Client Transfer during Tail-Anchored Protein Targeting. Cell Rep. 2019, 26, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Favaloro, V.; Vilardi, F.; Schlecht, R.; Mayer, M.P.; Dobberstein, B. Asna1/TRC40-mediated membrane insertion of tail-anchored proteins. J. Cell Sci. 2010, 123, 1522–1530. [Google Scholar] [CrossRef] [PubMed]
- Suloway, C.J.; Rome, M.E.; Clemons, W.M., Jr. Tail-anchor targeting by a Get3 tetramer: The structure of an archaeal homologue. EMBO J. 2012, 31, 707–719. [Google Scholar] [CrossRef] [PubMed]
- Chio, U.S.; Chung, S.; Weiss, S.; Shan, S.-O. A protean clamp guides membrane targeting of tail-anchored proteins. Proc. Natl. Acad. Sci. USA 2017, 114, E8585–E8594. [Google Scholar] [CrossRef] [PubMed]
- Mateja, A.; Szlachcic, A.; Downing, M.E.; Dobosz, M.; Mariappan, M.; Hegde, R.S.; Keenan, R.J. The structural basis of tail-anchored membrane protein recognition by Get3. Nature 2009, 461, 361–366. [Google Scholar] [CrossRef] [PubMed]
- Borgese, N.; Colombo, S.; Pedrazzini, E. The tale of tail-anchored proteins: Coming from the cytosol and looking for a membrane. J. Cell Biol. 2003, 161, 1013–1019. [Google Scholar] [CrossRef] [PubMed]
- Borgese, N.; Brambillasca, S.; Colombo, S. How tails guide tail-anchored proteins to their destinations. Curr. Opin. Cell Biol. 2007, 19, 368–375. [Google Scholar] [CrossRef] [PubMed]
- Borgese, N.; Fasana, E. Targeting pathways of C-tail-anchored proteins. Biochim. Et Biophys. Acta (BBA) Biomembr. 2011, 1808, 937–946. [Google Scholar] [CrossRef]
- Vitali, D.G.; Sinzel, M.; Bulthuis, E.P.; Kolb, A.; Zabel, S.; Mehlhorn, D.G.; Figueiredo Costa, B.; Farkas, Á.; Clancy, A.; Schuldiner, M.; et al. The GET pathway can increase the risk of mitochondrial outer membrane proteins to be mistargeted to the ER. J. Cell Sci. 2018, 131, jcs211110. [Google Scholar] [CrossRef] [PubMed]
- Lin, K.-F.; Fry, M.Y.; Saladi, S.M.; Clemons, W.M. The client-binding domain of the cochaperone Sgt2 has a helical-hand structure that binds a short hydrophobic helix. bioRxiv 2020, 517573. [Google Scholar] [CrossRef]
- Fry, M.Y.; Saladi, S.M.; Cunha, A.; Clemons, W.M., Jr. Sequence-based features that are determinant for tail-anchored membrane protein sorting in eukaryotes. Traffic 2021, 22, 306–318. [Google Scholar] [CrossRef]
- Roberts, J.D.; Thapaliya, A.; Martínez-Lumbreras, S.; Krysztofinska, E.M.; Isaacson, R.L. Structural and Functional Insights into Small, Glutamine-Rich, Tetratricopeptide Repeat Protein Alpha. Front. Mol. Biosci. 2015, 2, 71. [Google Scholar] [CrossRef] [PubMed]
- Lin, K.-F.; Fry, M.Y.; Saladi, S.M.; Clemons, W.M. Molecular basis of tail-anchored integral membrane protein recognition by the cochaperone Sgt2. J. Biol. Chem. 2021, 296, 100441. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, H.; Takahashi, T.; Xie, Y.; Minami, R.; Yanagi, Y.; Hayashishita, M.; Suzuki, R.; Yokota, N.; Shimada, M.; Mizushima, T.; et al. A conserved island of BAG6/Scythe is related to ubiquitin domains and participates in short hydrophobicity recognition. FEBS J. 2016, 283, 662–677. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Lei, P.; Xiao, Y.; Li, P.; Xie, P.; Wang, H.; Huang, S.; Song, P.; Zhao, Y. hSnd2/TMEM208 is an HIF-1α-targeted gene and contains a WH2 motif. Acta Biochim. Et Biophys. Sin. 2020, 52, 328–331. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.W.; Jung, C.; Hwang, I. Cytosolic events involved in chloroplast protein targeting. Biochim. Et Biophys. Acta (BBA) Mol. Cell Res. 2013, 1833, 245–252. [Google Scholar] [CrossRef] [PubMed]
- Dederer, V.; Khmelinskii, A.; Huhn, A.G.; Okreglak, V.; Knop, M.; Lemberg, M.K. Cooperation of mitochondrial and ER factors in quality control of tail-anchored proteins. eLife 2019, 8, e45506. [Google Scholar] [CrossRef] [PubMed]
- McKenna, M.J.; Sim, S.I.; Ordureau, A.; Wei, L.; Harper, J.W.; Shao, S.; Park, E. The endoplasmic reticulum P5A-ATPase is a transmembrane helix dislocase. Science 2020, 369, eabc5809. [Google Scholar] [CrossRef] [PubMed]
- Farkas, Á.; Bohnsack, K.E. Capture and delivery of tail-anchored proteins to the endoplasmic reticulum. J. Cell Biol. 2021, 220, e202105004. [Google Scholar] [CrossRef] [PubMed]
- Koch, C.; Schuldiner, M.; Herrmann, J.M. ER-SURF: Riding the Endoplasmic Reticulum Surface to Mitochondria. Int. J. Mol. Sci. 2021, 22, 9655. [Google Scholar] [CrossRef] [PubMed]
- Deuerling, E.; Gamerdinger, M.; Kreft, S.G. Chaperone Interactions at the Ribosome. Cold Spring Harb. Perspect. Biol. 2019, 11, a033977. [Google Scholar] [CrossRef]
- Genuth, N.R.; Barna, M. The Discovery of Ribosome Heterogeneity and Its Implications for Gene Regulation and Organismal Life. Mol. Cell 2018, 71, 364–374. [Google Scholar] [CrossRef] [PubMed]
- Melanson, G.; Timpano, S.; Uniacke, J. The eIF4E2-Directed Hypoxic Cap-Dependent Translation Machinery Reveals Novel Therapeutic Potential for Cancer Treatment. Oxid. Med. Cell. Longev. 2017, 2017, 6098107. [Google Scholar] [CrossRef]
- Uniacke, J.; Holterman, C.E.; Lachance, G.; Franovic, A.; Jacob, M.D.; Fabian, M.R.; Payette, J.; Holcik, M.; Pause, A.; Lee, S. An oxygen-regulated switch in the protein synthesis machinery. Nature 2012, 486, 126–129. [Google Scholar] [CrossRef]
- James, C.C.; Smyth, J.W. Alternative mechanisms of translation initiation: An emerging dynamic regulator of the proteome in health and disease. Life Sci. 2018, 212, 138–144. [Google Scholar] [CrossRef]
- Chartron, J.W.; Hunt, K.C.L.; Frydman, J. Cotranslational signal-independent SRP preloading during membrane targeting. Nature 2016, 536, 224–228. [Google Scholar] [CrossRef]
- Sabatini, D.D.; Blobel, G. Controlled proteolysis of nascent polypeptides in rat liver cell fractions: II. Location of the Polypeptides in Rough Microsomes. J. Cell Biol. 1970, 45, 146–157. [Google Scholar] [CrossRef]
- Dao Duc, K.; Batra, S.S.; Bhattacharya, N.; Cate, J.H.D.; Song, Y.S. Differences in the path to exit the ribosome across the three domains of life. Nucleic Acids Res. 2019, 47, 4198–4210. [Google Scholar] [CrossRef] [PubMed]
- Voss, N.R.; Gerstein, M.; Steitz, T.A.; Moore, P.B. The Geometry of the Ribosomal Polypeptide Exit Tunnel. J. Mol. Biol. 2006, 360, 893–906. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Deutsch, C. Folding zones inside the ribosomal exit tunnel. Nat. Struct. Mol. Biol. 2005, 12, 1123–1129. [Google Scholar] [CrossRef]
- Wruck, F.; Tian, P.; Kudva, R.; Best, R.B.; von Heijne, G.; Tans, S.J.; Katranidis, A. The ribosome modulates folding inside the ribosomal exit tunnel. Commun. Biol. 2021, 4, 523. [Google Scholar] [CrossRef] [PubMed]
- Marino, J.; von Heijne, G.; Beckmann, R. Small protein domains fold inside the ribosome exit tunnel. FEBS Lett. 2016, 590, 655–660. [Google Scholar] [CrossRef] [PubMed]
- Liutkute, M.; Samatova, E.; Rodnina, M.V. Cotranslational Folding of Proteins on the Ribosome. Biomolecules 2020, 10, 97. [Google Scholar] [CrossRef]
- Lu, J.; Deutsch, C. Electrostatics in the Ribosomal Tunnel Modulate Chain Elongation Rates. J. Mol. Biol. 2008, 384, 73–86. [Google Scholar] [CrossRef] [PubMed]
- Farías-Rico, J.A.; Ruud Selin, F.; Myronidi, I.; Frühauf, M.; von Heijne, G. Effects of protein size, thermodynamic stability, and net charge on cotranslational folding on the ribosome. Proc. Natl. Acad. Sci. USA 2018, 115, E9280–E9287. [Google Scholar] [CrossRef] [PubMed]
- Bui, P.T.; Hoang, T.X. Protein escape at the ribosomal exit tunnel: Effect of the tunnel shape. J. Chem. Phys. 2020, 153, 045105. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.K.; O’Brien, E.P. Non-equilibrium coupling of protein structure and function to translation—Elongation kinetics. Curr. Opin. Struct. Biol. 2018, 49, 94–103. [Google Scholar] [CrossRef]
- Pechmann, S.; Chartron, J.W.; Frydman, J. Local slowdown of translation by nonoptimal codons promotes nascent-chain recognition by SRP in vivo. Nat. Struct. Mol. Biol. 2014, 21, 1100–1105. [Google Scholar] [CrossRef]
- Yanagitani, K.; Kimata, Y.; Kadokura, H.; Kohno, K. Translational Pausing Ensures Membrane Targeting and Cytoplasmic Splicing of XBP1u mRNA. Science 2011, 331, 586–589. [Google Scholar] [CrossRef] [PubMed]
- Mariappan, M.; Li, X.; Stefanovic, S.; Sharma, A.; Mateja, A.; Keenan, R.J.; Hegde, R.S. A ribosome-associating factor chaperones tail-anchored membrane proteins. Nature 2010, 466, 1120–1124. [Google Scholar] [CrossRef] [PubMed]
- Bui, P.T.; Hoang, T.X. Protein escape at the ribosomal exit tunnel: Effects of native interactions, tunnel length, and macromolecular crowding. J. Chem. Phys. 2018, 149, 045102. [Google Scholar] [CrossRef] [PubMed]
- Bui, P.T.; Hoang, T.X. Folding and escape of nascent proteins at ribosomal exit tunnel. J. Chem. Phys. 2016, 144, 095102. [Google Scholar] [CrossRef] [PubMed]
- Simsek, D.; Tiu, G.C.; Flynn, R.A.; Byeon, G.W.; Leppek, K.; Xu, A.F.; Chang, H.Y.; Barna, M. The Mammalian Ribo-interactome Reveals Ribosome Functional Diversity and Heterogeneity. Cell 2017, 169, 1051–1065. [Google Scholar] [CrossRef]
- Guzel, P.; Yildirim, H.Z.; Yuce, M.; Kurkcuoglu, O. Exploring Allosteric Signaling in the Exit Tunnel of the Bacterial Ribosome by Molecular Dynamics Simulations and Residue Network Model. Front. Mol. Biosci. 2020, 7, 276. [Google Scholar] [CrossRef] [PubMed]
- Berndt, U.; Oellerer, S.; Zhang, Y.; Johnson, A.E.; Rospert, S. A signal-anchor sequence stimulates signal recognition particle binding to ribosomes from inside the exit tunnel. Proc. Natl. Acad. Sci. USA 2009, 106, 1398–1403. [Google Scholar] [CrossRef]
- Gamerdinger, M.; Kobayashi, K.; Wallisch, A.; Kreft, S.G.; Sailer, C.; Schlömer, R.; Sachs, N.; Jomaa, A.; Stengel, F.; Ban, N.; et al. Early Scanning of Nascent Polypeptides inside the Ribosomal Tunnel by NAC. Mol. Cell 2019, 75, 996–1006. [Google Scholar] [CrossRef] [PubMed]
- Denks, K.; Sliwinski, N.; Erichsen, V.; Borodkina, B.; Origi, A.; Koch, H.-G. The signal recognition particle contacts uL23 and scans substrate translation inside the ribosomal tunnel. Nat. Microbiol. 2017, 2, 16265. [Google Scholar] [CrossRef] [PubMed]
- Raue, U.; Oellerer, S.; Rospert, S. Association of Protein Biogenesis Factors at the Yeast Ribosomal Tunnel Exit Is Affected by the Translational Status and Nascent Polypeptide Sequence. J. Biol. Chem. 2007, 282, 7809–7816. [Google Scholar] [CrossRef]
- Jungnickel, B.; Rapoport, T.A. A posttargeting signal sequence recognition event in the endoplasmic reticulum membrane. Cell 1995, 82, 261–270. [Google Scholar] [CrossRef]
- Zhang, Y.; Berndt, U.; Gölz, H.; Tais, A.; Oellerer, S.; Wölfle, T.; Fitzke, E.; Rospert, S. NAC functions as a modulator of SRP during the early steps of protein targeting to the endoplasmic reticulum. Mol. Biol. Cell 2012, 23, 3027–3040. [Google Scholar] [CrossRef] [PubMed]
- Wiedmann, B.; Sakai, H.; Davis, T.A.; Wiedmann, M. A protein complex required for signal-sequence-specific sorting and translocation. Nature 1994, 370, 434–440. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, H.-H.; Lee, J.H.; Chandrasekar, S.; Shan, S.-o. A ribosome-associated chaperone enables substrate triage in a cotranslational protein targeting complex. Nat. Commun. 2020, 11, 5840. [Google Scholar] [CrossRef] [PubMed]
- del Alamo, M.; Hogan, D.J.; Pechmann, S.; Albanese, V.; Brown, P.O.; Frydman, J. Defining the specificity of cotranslationally acting chaperones by systematic analysis of mRNAs associated with ribosome-nascent chain complexes. PLOS Biol. 2011, 9, e1001100. [Google Scholar] [CrossRef]
- Zhang, Y.; De Laurentiis, E.; Bohnsack, K.E.; Wahlig, M.; Ranjan, N.; Gruseck, S.; Hackert, P.; Wölfle, T.; Rodnina, M.V.; Schwappach, B.; et al. Ribosome-bound Get4/5 facilitates the capture of tail-anchored proteins by Sgt2 in yeast. Nat. Commun. 2021, 12, 782. [Google Scholar] [CrossRef] [PubMed]
- Chitwood, P.J.; Hegde, R.S. An intramembrane chaperone complex facilitates membrane protein biogenesis. Nature 2020, 584, 630–634. [Google Scholar] [CrossRef] [PubMed]
- McGilvray, P.T.; Anghel, S.A.; Sundaram, A.; Zhong, F.; Trnka, M.J.; Fuller, J.R.; Hu, H.; Burlingame, A.L.; Keenan, R.J. An ER translocon for multi-pass membrane protein biogenesis. eLife 2020, 9, e56889. [Google Scholar] [CrossRef] [PubMed]
- Calo, D.; Eichler, J. Crossing the membrane in Archaea, the third domain of life. Biochim. Et Biophys. Acta (BBA) Biomembr. 2011, 1808, 885–891. [Google Scholar] [CrossRef] [PubMed]
- du Plessis, D.J.F.; Nouwen, N.; Driessen, A.J.M. The Sec translocase. Biochim. Et Biophys. Acta (BBA) Biomembr. 2011, 1808, 851–865. [Google Scholar] [CrossRef] [PubMed]
- Dalal, K.; Duong, F. The SecY complex: Conducting the orchestra of protein translocation. Trends Cell Biol. 2009, 21, 506–514. [Google Scholar] [CrossRef] [PubMed]
- Deshaies, R.J.; Schekman, R. A yeast mutant defective at an early stage in import of secretory protein precursors into the endoplasmic reticulum. J. Cell Biol. 1987, 105, 633–645. [Google Scholar] [CrossRef] [PubMed]
- Schekman, R. SEC mutants and the secretory apparatus. Nat. Med. 2002, 8, 1055–1058. [Google Scholar] [CrossRef] [PubMed]
- Görlich, D.; Rapoport, T.A. Protein translocation into proteoliposomes reconstituted from purified components of the endoplasmic reticulum membrane. Cell 1993, 75, 615–630. [Google Scholar] [CrossRef]
- Hartmann, E.; Sommer, T.; Prehn, S.; Görlich, D.; Jentsch, S.; Rapoport, T.A. Evolutionary conservation of components of the protein translocation complex. Nature 1994, 367, 654–657. [Google Scholar] [CrossRef] [PubMed]
- Görlich, D.; Prehn, S.; Hartmann, E.; Kalies, K.-U.; Rapoport, T.A. A mammalian homolog of SEC61p and SECYp is associated with ribosomes and nascent polypeptides during translocation. Cell 1992, 71, 489–503. [Google Scholar] [CrossRef]
- Kalies, K.U.; Görlich, D.; Rapoport, T.A. Binding of ribosomes to the rough endoplasmic reticulum mediated by the Sec61p-complex. J. Cell Biol. 1994, 126, 925–934. [Google Scholar] [CrossRef] [PubMed]
- Egea, P.F.; Stroud, R.M. Lateral opening of a translocon upon entry of protein suggests the mechanism of insertion into membranes. Proc. Natl. Acad. Sci. USA 2010, 107, 17182–17187. [Google Scholar] [CrossRef] [PubMed]
- Becker, T.; Bhushan, S.; Jarasch, A.; Armache, J.P.; Funes, S.; Jossinet, F.; Gumbart, J.; Mielke, T.; Berninghausen, O.; Schulten, K.; et al. Structure of monomeric yeast and mammalian Sec61 complexes interacting with the translating ribosome. Science 2009, 326, 1369–1373. [Google Scholar] [CrossRef] [PubMed]
- Gogala, M.; Becker, T.; Beatrix, B.; Armache, J.-P.; Barrio-Garcia, C.; Berninghausen, O.; Beckmann, R. Structures of the Sec61 complex engaged in nascent peptide translocation or membrane insertion. Nature 2014, 506, 107–110. [Google Scholar] [CrossRef] [PubMed]
- Voorhees, R.M.; Fernández, I.S.; Scheres, S.H.W.; Hegde, R.S. Structure of the Mammalian Ribosome-Sec61 Complex to 3.4 Å Resolution. Cell 2014, 157, 1632–1643. [Google Scholar] [CrossRef] [PubMed]
- Zimmer, J.; Nam, Y.; Rapoport, T.A. Structure of a complex of the ATPase SecA and the protein-translocation channel. Nature 2008, 455, 936–943. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Sugano, Y.; Takemoto, M.; Mori, T.; Furukawa, A.; Kusakizako, T.; Kumazaki, K.; Kashima, A.; Ishitani, R.; Sugita, Y.; et al. Crystal Structures of SecYEG in Lipidic Cubic Phase Elucidate a Precise Resting and a Peptide-Bound State. Cell Rep. 2015, 13, 1561–1568. [Google Scholar] [CrossRef] [PubMed]
- Pfeffer, S.; Burbaum, L.; Unverdorben, P.; Pech, M.; Chen, Y.; Zimmermann, R.; Beckmann, R.; Förster, F. Structure of the native Sec61 protein-conducting channel. Nat. Commun. 2015, 6, 8403. [Google Scholar] [CrossRef] [PubMed]
- du Plessis, D.J.F.; Berrelkamp, G.; Nouwen, N.; Driessen, A.J.M. The Lateral Gate of SecYEG Opens during Protein Translocation. J. Biol. Chem. 2009, 284, 15805–15814. [Google Scholar] [CrossRef] [PubMed]
- Bischoff, L.; Wickles, S.; Berninghausen, O.; van der Sluis, E.O.; Beckmann, R. Visualization of a polytopic membrane protein during SecY-mediated membrane insertion. Nat. Commun. 2014, 5, 4103. [Google Scholar] [CrossRef] [PubMed]
- Paetzel, M.; Karla, A.; Strynadka, N.C.J.; Dalbey, R.E. Signal Peptidases. Chem. Rev. 2002, 102, 4549–4580. [Google Scholar] [CrossRef]
- Liaci, A.M.; Steigenberger, B.; Tamara, S.; de Souza, P.C.T.; Gröllers-Mulderij, M.; Ogrissek, P.; Marrink, S.J.; Scheltema, R.A.; Förster, F. Structure of the Human Signal Peptidase Complex Reveals the Determinants for Signal Peptide Cleavage. bioRxiv 2020, 81, 3934–3948. [Google Scholar] [CrossRef]
- Kalies, K.-U.; Rapoport, T.A.; Hartmann, E. The beta-Subunit of the Sec61 Complex Facilitates Cotranslational Protein Transport and Interacts with the Signal Peptidase during Translocation. J. Cell Biol. 1998, 141, 887–894. [Google Scholar] [CrossRef] [PubMed]
- Sadlish, H.; Pitonzo, D.; Johnson, A.E.; Skach, W.R. Sequential triage of transmembrane segments by Sec61α during biogenesis of a native multispanning membrane protein. Nat. Struct. Mol. Biol. 2005, 12, 870–878. [Google Scholar] [CrossRef] [PubMed]
- Audigier, Y.; Friedlander, M.; Blobel, G. Multiple topogenic sequences in bovine opsin. Proc. Natl. Acad. Sci. USA 1987, 84, 5783–5787. [Google Scholar] [CrossRef] [PubMed]
- Schubert, D.; Klein, M.-C.; Hassdenteufel, S.; Caballero-Oteyza, A.; Yang, L.; Proietti, M.; Bulashevska, A.; Kemming, J.; Kühn, J.; Winzer, S.; et al. Plasma cell deficiency in human subjects with heterozygous mutations in Sec61 translocon alpha 1 subunit (SEC61A1). J. Allergy Clin. Immunol. 2018, 141, 1427–1438. [Google Scholar] [CrossRef] [PubMed]
- Gumbart, J.; Schulten, K. Molecular Dynamics Studies of the Archaeal Translocon. Biophys. J. 2006, 90, 2356–2367. [Google Scholar] [CrossRef] [PubMed]
- Junne, T.; Kocik, L.; Spiess, M. The hydrophobic core of the Sec61 translocon defines the hydrophobicity threshold for membrane integration. Mol. Biol. Cell 2010, 21, 1662–1670. [Google Scholar] [CrossRef] [PubMed]
- Park, E.; Rapoport, T.A. Preserving the membrane barrier for small molecules during bacterial protein translocation. Nature 2011, 473, 239–242. [Google Scholar] [CrossRef]
- Saparov, S.M.; Erlandson, K.; Cannon, K.; Schaletzky, J.; Schulman, S.; Rapoport, T.A.; Pohl, P. Determining the Conductance of the SecY Protein Translocation Channel for Small Molecules. Mol. Cell 2007, 26, 501–509. [Google Scholar] [CrossRef] [PubMed]
- Tam, P.C.; Maillard, A.P.; Chan, K.K.; Duong, F. Investigating the SecY plug movement at the SecYEG translocation channel. EMBO J. 2005, 24, 3380–3388. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Schulman, S.; Boyd, D.; Erlandson, K.; Beckwith, J.; Rapoport, T.A. The Plug Domain of the SecY Protein Stabilizes the Closed State of the Translocation Channel and Maintains a Membrane Seal. Mol. Cell 2007, 26, 511–521. [Google Scholar] [CrossRef] [PubMed]
- Bolar, N.A.; Golzio, C.; Živná, M.; Hayot, G.; Van Hemelrijk, C.; Schepers, D.; Vandeweyer, G.; Hoischen, A.; Huyghe, J.R.; Raes, A.; et al. Heterozygous Loss-of-Function SEC61A1 Mutations Cause Autosomal-Dominant Tubulo-Interstitial and Glomerulocystic Kidney Disease with Anemia. Am. J. Hum. Genet. 2016, 99, 174–187. [Google Scholar] [CrossRef] [PubMed]
- Junne, T.; Schwede, T.; Goder, V.; Spiess, M. The Plug Domain of Yeast Sec61p Is Important for Efficient Protein Translocation, but Is Not Essential for Cell Viability. Mol. Biol. Cell 2006, 17, 4063–4068. [Google Scholar] [CrossRef]
- Haßdenteufel, S.; Klein, M.-C.; Melnyk, A.; Zimmermann, R. Protein transport into the human ER and related diseases, Sec61-channelopathies. Biochem. Cell Biol. 2014, 92, 499–509. [Google Scholar] [CrossRef] [PubMed]
- Osborne, A.R.; Rapoport, T.A.; van den Berg, B. Protein translocation by the Sec61/SecY channel. Annu. Rev. Cell Dev. Biol. 2005, 21, 529–550. [Google Scholar] [CrossRef]
- Aviram, N.; Schuldiner, M. Embracing the void—how much do we really know about targeting and translocation to the endoplasmic reticulum? Curr. Opin. Cell Biol. 2014, 29, 8–17. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, R.; Sagstetter, M.; Lewis, M.J.; Pelham, H.R.B. Seventy-kilodalton heat shock proteins and an additional component from reticulocyte lysate stimulate import of M13 procoat protein into microsomes. EMBO J. 1988, 7, 2875–2880. [Google Scholar] [CrossRef] [PubMed]
- Steinberg, R.; Koch, H.-G. The largely unexplored biology of small proteins in pro- and eukaryotes. FEBS J. 2021, 7, 7002–7004. [Google Scholar] [CrossRef] [PubMed]
- Voorhees, R.M.; Hegde, R.S. Toward a structural understanding of co-translational protein translocation. Curr. Opin. Cell Biol. 2016, 41, 91–99. [Google Scholar] [CrossRef]
- Lang, S.; Nguyen, D.; Pfeffer, S.; Förster, F.; Helms, V.; Zimmermann, R. Functions and Mechanisms of the Human Ribosome-Translocon Complex. In Macromolecular Protein Complexes II: Structure and Function; Harris, J.R., Marles-Wright, J., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 83–141. [Google Scholar]
- Li, L.; Park, E.; Ling, J.; Ingram, J.; Ploegh, H.; Rapoport, T.A. Crystal structure of a substrate-engaged SecY protein-translocation channel. Nature 2016, 531, 395–399. [Google Scholar] [CrossRef] [PubMed]
- Itskanov, S.; Park, E. Structure of the posttranslational Sec protein-translocation channel complex from yeast. Science 2019, 363, 84–87. [Google Scholar] [CrossRef] [PubMed]
- Braunger, K.; Pfeffer, S.; Shrimal, S.; Gilmore, R.; Berninghausen, O.; Mandon, E.C.; Becker, T.; Förster, F.; Beckmann, R. Structural basis for coupling protein transport and N-glycosylation at the mammalian endoplasmic reticulum. Science 2018, 360, 215–219. [Google Scholar] [CrossRef] [PubMed]
- Itskanov, S.; Kuo, K.M.; Gumbart, J.C.; Park, E. Stepwise gating of the Sec61 protein-conducting channel by Sec63 and Sec62. Nat. Struct. Mol. Biol. 2021, 28, 162–172. [Google Scholar] [CrossRef] [PubMed]
- Weng, T.-H.; Steinchen, W.; Beatrix, B.; Berninghausen, O.; Becker, T.; Bange, G.; Cheng, J.; Beckmann, R. Architecture of the active post-translational Sec translocon. EMBO J. 2021, 40, e105643. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.; Jiang, Y.; Mandon, E.C.; Gilmore, R. Identification of cytoplasmic residues of Sec61p involved in ribosome binding and cotranslational translocation. J. Cell Biol. 2005, 168, 67–77. [Google Scholar] [CrossRef] [PubMed]
- Tsukazaki, T.; Mori, H.; Fukai, S.; Ishitani, R.; Mori, T.; Dohmae, N.; Perederina, A.; Sugita, Y.; Vassylyev, D.G.; Ito, K.; et al. Conformational transition of Sec machinery inferred from bacterial SecYE structures. Nature 2008, 455, 988–991. [Google Scholar] [CrossRef] [PubMed]
- Frauenfeld, J.; Gumbart, J.; Sluis, E.O.V.D.; Funes, S.; Gartmann, M.; Beatrix, B.; Mielke, T.; Berninghausen, O.; Becker, T.; Schulten, K.; et al. Cryo-EM structure of the ribosome–SecYE complex in the membrane environment. Nat. Struct. Mol. Biol. 2011, 18, 614–621. [Google Scholar] [CrossRef]
- Elia, F.; Yadhanapudi, L.; Tretter, T.; Römisch, K. The N-terminus of Sec61p plays key roles in ER protein import and ERAD. PLoS ONE 2019, 14, e0215950. [Google Scholar] [CrossRef] [PubMed]
- Erdmann, F.; Schäuble, N.; Lang, S.; Jung, M.; Honigmann, A.; Ahmad, M.; Dudek, J.; Benedix, J.; Harsman, A.; Kopp, A.; et al. Interaction of calmodulin with Sec61alpha limits Ca2+ leakage from the endoplasmic reticulum. EMBO J. 2011, 30, 17–31. [Google Scholar] [CrossRef] [PubMed]
- Mandon, E.C.; Butova, C.; Lachapelle, A.; Gilmore, R. Conserved motifs on the cytoplasmic face of the protein translocation channel are critical for the transition between resting and active conformations. J. Biol. Chem. 2018, 293, 13662–13672. [Google Scholar] [CrossRef]
- Kapp, K.; Schrempf, S.; Lemberg, M.; Dobberstein, B. Post-Targeting Functions of Signal Peptides. In Protein Transport into the Endoplasmic Reticulum; Zimmermann, R., Ed.; Landes Bioscience: Austin, TX, USA, 2009; pp. 1–16. [Google Scholar]
- Sun, S.; Li, X.; Mariappan, M. Signal Sequences Encode Information for Protein Folding in the Endoplasmic Reticulum. bioRxiv 2020. [Google Scholar] [CrossRef]
- Martoglio, B.; Graf, R.; Dobberstein, B. Signal peptide fragments of preprolactin and HIV-1 p-gp160 interact with calmodulin. EMBO J. 1997, 16, 6636–6645. [Google Scholar] [CrossRef] [PubMed]
- Lemberg, M.K.; Martoglio, B. On the mechanism of SPP-catalysed intramembrane proteolysis; conformational control of peptide bond hydrolysis in the plane of the membrane. FEBS Lett. 2004, 564, 213–218. [Google Scholar] [CrossRef]
- Mentrup, T.; Cabrera-Cabrera, F.; Fluhrer, R.; Schröder, B. Physiological functions of SPP/SPPL intramembrane proteases. Cell Mol. Life Sci. 2020, 77, 2959–2979. [Google Scholar] [CrossRef] [PubMed]
- Braud, V.; Yvonne Jones, E.; McMichael, A. The human major histocompatibility complex class Ib molecule HLA-E binds signal sequence-derived peptides with primary anchor residues at positions 2 and 9. Eur. J. Immunol. 1997, 27, 1164–1169. [Google Scholar] [CrossRef] [PubMed]
- Wirth, A.; Jung, M.; Bies, C.; Frien, M.; Tyedmers, J.; Zimmermann, R.; Wagner, R. The Sec61p complex is a dynamic precursor activated channel. Mol. Cell 2003, 12, 261–268. [Google Scholar] [CrossRef]
- Cross, B.C.S.; High, S. Dissecting the physiological role of selective transmembrane-segment retention at the ER translocon. J. Cell Sci. 2009, 122, 1768–1777. [Google Scholar] [CrossRef] [PubMed]
- McCormick, P.J.; Miao, Y.; Shao, Y.; Lin, J.; Johnson, A.E. Cotranslational Protein Integration into the ER Membrane Is Mediated by the Binding of Nascent Chains to Translocon Proteins. Mol. Cell 2003, 12, 329–341. [Google Scholar] [CrossRef]
- Heinrich, S.U.; Mothes, W.; Brunner, J.; Rapoport, T.A. The Sec61p complex mediates the integration of a membrane protein by allowing lipid partitioning of the transmembrane domain. Cell 2000, 102, 233–244. [Google Scholar] [CrossRef]
- Mothes, W.; Heinrich, S.U.; Graf, R.; Nilsson, I.; von Heijne, G.; Brunner, J.; Rapoport, T.A. Molecular mechanism of membrane protein integration into the endoplasmic reticulum. Cell 1997, 89, 523–533. [Google Scholar] [CrossRef]
- Simon, S.M.; Blobel, G. Signal peptides open protein-conducting channels in E. coli. Cell 1992, 69, 677–684. [Google Scholar] [CrossRef]
- Simon, S.M.; Blobel, G. A protein-conducting channel in the endoplasmic reticulum. Cell 1991, 65, 371–380. [Google Scholar] [CrossRef]
- Kida, Y.; Sakaguchi, M. Interaction mapping of the Sec61 translocon identifies two Sec61α regions interacting with hydrophobic segments in translocating chains. J. Biol. Chem. 2018, 293, 17050–17060. [Google Scholar] [CrossRef] [PubMed]
- Reithinger, J.H.; Yim, C.; Kim, S.; Lee, H.; Kim, H. Structural and functional profiling of the lateral gate of the Sec61 translocon. J. Biol. Chem. 2014, 289, 15845–15855. [Google Scholar] [CrossRef] [PubMed]
- Plath, K.; Mothes, W.; Wilkinson, B.M.; Stirling, C.J.; Rapoport, T.A. Signal sequence recognition in posttranslational protein transport across the yeast ER membrane. Cell 1998, 94, 795–807. [Google Scholar] [CrossRef]
- High, S.; Martoglio, B.; Görlich, D.; Andersen, S.S.; Ashford, A.J.; Giner, A.; Hartmann, E.; Prehn, S.; Rapoport, T.A.; Dobberstein, B. Site-specific photocross-linking reveals that Sec61p and TRAM contact different regions of a membrane-inserted signal sequence. J. Biol. Chem. 1993, 268, 26745–26751. [Google Scholar] [CrossRef]
- Mothes, W.; Jungnickel, B.; Brunner, J.; Rapoport, T.A. Signal sequence recognition in cotranslational translocation by protein components of the endoplasmic reticulum membrane. J. Cell Biol. 1998, 142, 355–364. [Google Scholar] [CrossRef]
- Mothes, W.; Prehn, S.; Rapoport, T.A. Systematic probing of the environment of a translocating secretory protein during translocation through the ER membrane. EMBO J. 1994, 13, 3973–3982. [Google Scholar] [CrossRef] [PubMed]
- High, S.; Andersen, S.S.L.; Görlich, D.; Hartmann, E.; Prehn, S.; Rapoport, T.A.; Dobberstein, B. Sec61p is adjecent to nascent type I and type II signal-anchor proteins during their membrane insertion. J. Cell Biol. 1993, 121, 743–750. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Miller, T.F., III. Long-Timescale Dynamics and Regulation of Sec-Facilitated Protein Translocation. Cell Rep. 2012, 2, 927–937. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Miller, T.F. Direct Simulation of Early-Stage Sec-Facilitated Protein Translocation. J. Am. Chem. Soc. 2012, 134, 13700–13707. [Google Scholar] [CrossRef]
- Rychkova, A.; Warshel, A. Exploring the nature of the translocon-assisted protein insertion. Proc. Natl. Acad. Sci. USA 2013, 110, 495–500. [Google Scholar] [CrossRef] [PubMed]
- Phillips, B.P.; Miller, E.A. Ribosome-associated quality control of membrane proteins at the endoplasmic reticulum. J. Cell Sci. 2020, 133, jcs251983. [Google Scholar] [CrossRef] [PubMed]
- Charneski, C.A.; Hurst, L.D. Positively Charged Residues Are the Major Determinants of Ribosomal Velocity. PLoS Biol. 2013, 11, e1001508. [Google Scholar] [CrossRef]
- Elazar, A.; Weinstein, J.J.; Prilusky, J.; Fleishman, S.J. Interplay between hydrophobicity and the positive-inside rule in determining membrane-protein topology. Proc. Natl. Acad. Sci. USA 2016, 113, 10340–10345. [Google Scholar] [CrossRef] [PubMed]
- Seppälä, S.; Slusky, J.S.; Lloris-Garcerá, P.; Rapp, M.; von Heijne, G. Control of Membrane Protein Topology by a Single C-Terminal Residue. Science 2010, 328, 1698–1700. [Google Scholar] [CrossRef] [PubMed]
- von Heijne, G.; Gavel, Y. Topogenic signals in integral membrane proteins. Eur. J. Biochem. 1988, 174, 671–678. [Google Scholar] [CrossRef] [PubMed]
- Wallin, E.; Heijne, G.V. Genome-wide analysis of integral membrane proteins from eubacterial, archaean, and eukaryotic organisms. Protein Sci. 1998, 7, 1029–1038. [Google Scholar] [CrossRef] [PubMed]
- Ziska, A.; Tatzelt, J.; Dudek, J.; Paton, A.W.; Paton, J.C.; Zimmermann, R.; Haßdenteufel, S. The signal peptide plus a cluster of positive charges in prion protein dictate chaperone-mediated Sec61 channel gating. Biol. Open 2019, 8, bio040691. [Google Scholar] [CrossRef] [PubMed]
- Kriegler, T.; Lang, S.; Notari, L.; Hessa, T. Prion Protein Translocation Mechanism Revealed by Pulling Force Studies. J. Mol. Biol. 2020, 432, 4447–4465. [Google Scholar] [CrossRef] [PubMed]
- Pfeffer, S.; Dudek, J.; Schaffer, M.; Ng, B.G.; Albert, S.; Plitzko, J.M.; Baumeister, W.; Zimmermann, R.; Freeze, H.H.; Engel, B.D.; et al. Dissecting the molecular organization of the translocon-associated protein complex. Nat. Commun. 2017, 8, 14516. [Google Scholar] [CrossRef]
- Kriegler, T.; Kiburg, G.; Hessa, T. Translocon-Associated Protein Complex (TRAP) is Crucial for Co-Translational Translocation of Pre-Proinsulin. J. Mol. Biol. 2020, 432, 166694. [Google Scholar] [CrossRef]
- Fons, R.D.; Bogert, B.A.; Hegde, R.S. Substrate-specific function of the translocon-associated protein complex during translocation across the ER membrane. J. Cell Biol. 2003, 160, 529–539. [Google Scholar] [CrossRef] [PubMed]
- Ménétret, J.F.; Hegde, R.S.; Aguiar, M.; Gygi, S.P.; Park, E.; Rapoport, T.A.; Akey, C.W. Single copies of Sec61 and TRAP associate with a nontranslating mammalian ribosome. Structure 2008, 16, 1126–1137. [Google Scholar] [CrossRef] [PubMed]
- Görlich, D.; Hartmann, E.; Prehn, S.; Rapoport, T.A. A protein of the endoplasmic reticulum involved early in polypeptide translocation. Nature 1992, 357, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Voigt, S.; Jungnickel, B.; Hartmann, E.; Rapoport, T.A. Signal sequence-dependent function of the TRAM protein during early phases of protein transport across the endoplasmic reticulum membrane. J. Cell Biol. 1996, 134, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Klein, M.-C.; Lerner, M.; Nguyen, D.; Pfeffer, S.; Dudek, J.; Förster, F.; Helms, V.; Lang, S.; Zimmermann, R. TRAM1 protein may support ER protein import by modulating the phospholipid bilayer near the lateral gate of the Sec61-channel. Channels 2020, 14, 28–44. [Google Scholar] [CrossRef] [PubMed]
- Conti, B.J.; Devaraneni, P.K.; Yang, Z.; David, L.L.; Skach, W.R. Cotranslational Stabilization of Sec62/63 within the ER Sec61 Translocon Is Controlled by Distinct Substrate-Driven Translocation Events. Mol. Cell 2015, 58, 269–283. [Google Scholar] [CrossRef] [PubMed]
- Schorr, S.; Nguyen, D.; Haßdenteufel, S.; Nagaraj, N.; Cavalié, A.; Greiner, M.; Weissgerber, P.; Loi, M.; Paton, A.W.; Paton, J.C.; et al. Identification of signal peptide features for substrate specificity in human Sec62/Sec63-dependent ER protein import. FEBS J. 2020, 287, 4612–4640. [Google Scholar] [CrossRef] [PubMed]
- Meacock, S.L.; Lecomte, F.J.L.; Crawshaw, S.G.; High, S. Different Transmembrane Domains Associate with Distinct Endoplasmic Reticulum Components during Membrane Integration of a Polytopic Protein. Mol. Biol. Cell 2002, 13, 4114–4129. [Google Scholar] [CrossRef] [PubMed]
- Pfeffer, S.; Dudek, J.; Gogala, M.; Schorr, S.; Linxweiler, J.; Lang, S.; Becker, T.; Beckmann, R.; Zimmermann, R.; Förster, F. Structure of the mammalian oligosaccharyl-transferase complex in the native ER protein translocon. Nat. Commun. 2014, 5, 3072. [Google Scholar] [CrossRef] [PubMed]
- Fenech, E.J.; Ben-Dor, S.; Schuldiner, M. Double the Fun, Double the Trouble: Paralogs and Homologs Functioning in the Endoplasmic Reticulum. Annu. Rev. Biochem. 2020, 89, 637–666. [Google Scholar] [CrossRef] [PubMed]
- Shrimal, S.; Trueman, S.F.; Gilmore, R. Extreme C-terminal sites are posttranslocationally glycosylated by the STT3B isoform of the OST. J. Cell Biol. 2013, 201, 81–95. [Google Scholar] [CrossRef] [PubMed]
- Tyedmers, J.; Lerner, M.; Wiedmann, M.; Volkmer, J.; Zimmermann, R. Polypeptide-binding proteins mediate completion of co-translational protein translocation into the mammalian endoplasmic reticulum. EMBO Rep. 2003, 4, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Schäuble, N.; Lang, S.; Jung, M.; Cappel, S.; Schorr, S.; Ulucan, O.; Linxweiler, J.; Dudek, J.; Blum, R.; Helms, V.; et al. BiP-mediated closing of the Sec61 channel limits Ca2+ leakage from the ER. EMBO J. 2012, 31, 3282–3296. [Google Scholar] [CrossRef] [PubMed]
- Lang, S.; Erdmann, F.; Jung, M.; Wagner, R.; Cavalié, A.; Zimmermann, R. Sec61 complexes form ubiquitous ER Ca(2+) leak channels. Channels 2011, 5, 228–235. [Google Scholar] [CrossRef]
- Wang, Q.-C.; Zheng, Q.; Tan, H.; Zhang, B.; Li, X.; Yang, Y.; Yu, J.; Liu, Y.; Chai, H.; Wang, X.; et al. TMCO1 Is an ER Ca2+ Load-Activated Ca2+ Channel. Cell 2016, 165, 1454–1466. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Yamazaki, T.; Yazawa, M.; Treves, S.; Nishi, M.; Murai, M.; Shibata, E.; Zorzato, F.; Takeshima, H. Calumin, a novel Ca2+-binding transmembrane protein on the endoplasmic reticulum. Cell Calcium 2007, 42, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Morel, J.-D.; Paatero, A.O.; Wei, J.; Yewdell, J.W.; Guenin-Macé, L.; Van Haver, D.; Impens, F.; Pietrosemoli, N.; Paavilainen, V.O.; Demangel, C. Proteomics Reveals Scope of Mycolactone-mediated Sec61 Blockade and Distinctive Stress Signature. Mol Cell Proteom. 2018, 17, 1750–1765. [Google Scholar] [CrossRef] [PubMed]
- McKenna, M.; Simmonds, R.E.; High, S. Mycolactone reveals the substrate-driven complexity of Sec61-dependent transmembrane protein biogenesis. J. Cell Sci. 2017, 130, 1307–1320. [Google Scholar] [CrossRef] [PubMed]
- Zong, G.; Hu, Z.; O’Keefe, S.; Tranter, D.; Iannotti, M.J.; Baron, L.; Hall, B.; Corfield, K.; Paatero, A.O.; Henderson, M.J.; et al. Ipomoeassin F Binds Sec61α to Inhibit Protein Translocation. J. Am. Chem. Soc. 2019, 141, 8450–8461. [Google Scholar] [CrossRef] [PubMed]
- Anghel, S.A.; McGilvray, P.T.; Hegde, R.S.; Keenan, R.J. Identification of Oxa1 Homologs Operating in the Eukaryotic Endoplasmic Reticulum. Cell Rep. 2017, 21, 3708–3716. [Google Scholar] [CrossRef] [PubMed]
- Yabal, M.; Brambillasca, S.; Soffientini, P.; Pedrazzini, E.; Borgese, N.; Makarow, M. Translocation of the C Terminus of a Tail-anchored Protein across the Endoplasmic Reticulum Membrane in Yeast Mutants Defective in Signal Peptide-driven Translocation. J. Biol. Chem. 2003, 278, 3489–3496. [Google Scholar] [CrossRef] [PubMed]
- Vilardi, F.; Stephan, M.; Clancy, A.; Janshoff, A.; Schwappach, B. WRB and CAML Are Necessary and Sufficient to Mediate Tail-Anchored Protein Targeting to the ER Membrane. PLoS ONE 2014, 9, e85033. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Chan, C.; Weir, N.R.; Denic, V. The Get1/2 transmembrane complex is an endoplasmic-reticulum membrane protein insertase. Nature 2014, 512, 441–444. [Google Scholar] [CrossRef]
- McDowell, M.A.; Heimes, M.; Fiorentino, F.; Mehmood, S.; Farkas, Á.; Coy-Vergara, J.; Wu, D.; Bolla, J.R.; Schmid, V.; Heinze, R.; et al. Structural Basis of Tail-Anchored Membrane Protein Biogenesis by the GET Insertase Complex. Mol. Cell 2020, 80, 72–86. [Google Scholar] [CrossRef] [PubMed]
- Zalisko, B.E.; Chan, C.; Denic, V.; Rock, R.S.; Keenan, R.J. Tail-Anchored Protein Insertion by a Single Get1/2 Heterodimer. Cell Rep. 2017, 20, 2287–2293. [Google Scholar] [CrossRef] [PubMed]
- Mariappan, M.; Mateja, A.; Dobosz, M.; Bove, E.; Hegde, R.S.; Keenan, R.J. The mechanism of membrane-associated steps in tail-anchored protein insertion. Nature 2011, 477, 61–66. [Google Scholar] [CrossRef]
- Rome, M.E.; Chio, U.S.; Rao, M.; Gristick, H.; Shan, S.-O. Differential gradients of interaction affinities drive efficient targeting and recycling in the GET pathway. Proc. Natl. Acad. Sci. USA 2014, 111, E4929–E4935. [Google Scholar] [CrossRef]
- Tran, D.D.; Russell, H.R.; Sutor, S.L.; van Deursen, J.; Bram, R.J. CAML Is Required for Efficient EGF Receptor Recycling. Dev. Cell 2003, 5, 245–256. [Google Scholar] [CrossRef]
- Tran, D.D.; Edgar, C.E.; Heckman, K.L.; Sutor, S.L.; Huntoon, C.J.; van Deursen, J.; McKean, D.L.; Bram, R.J. CAML Is a p56Lck-Interacting Protein that Is Required for Thymocyte Development. Immunity 2005, 23, 139–152. [Google Scholar] [CrossRef] [PubMed]
- Norlin, S.; Parekh, V.S.; Naredi, P.; Edlund, H. Asna1/TRC40 Controls β-Cell Function and Endoplasmic Reticulum Homeostasis by Ensuring Retrograde Transport. Diabetes 2016, 65, 110–119. [Google Scholar] [CrossRef] [PubMed]
- Bryda, E.C.; Johnson, N.T.; Ohlemiller, K.K.; Besch-Williford, C.L.; Moore, E.; Bram, R.J. Conditional deletion of calcium-modulating cyclophilin ligand causes deafness in mice. Mamm. Genome 2012, 23, 270–276. [Google Scholar] [CrossRef]
- Jonikas, M.C.; Collins, S.R.; Denic, V.; Oh, E.; Quan, E.M.; Schmid, V.; Weibezahn, J.; Schwappach, B.; Walter, P.; Weissman, J.S.; et al. Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science 2009, 323, 1693–1697. [Google Scholar] [CrossRef] [PubMed]
- Satoh, T.; Ohba, A.; Liu, Z.; Inagaki, T.; Satoh, A.K. dPob/EMC is essential for biosynthesis of rhodopsin and other multi-pass membrane proteins in Drosophila photoreceptors. eLife 2015, 4, e06306. [Google Scholar] [CrossRef] [PubMed]
- Richard, M.; Boulin, T.; Robert, V.J.P.; Richmond, J.E.; Bessereau, J.-L. Biosynthesis of ionotropic acetylcholine receptors requires the evolutionarily conserved ER membrane complex. Proc. Natl. Acad. Sci. USA 2013, 110, E1055–E1063. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Snowball, J.M.; Xu, Y.; Na, C.-L.; Weaver, T.E.; Clair, G.; Kyle, J.E.; Zink, E.M.; Ansong, C.; Wei, W.; et al. EMC3 coordinates surfactant protein and lipid homeostasis required for respiration. J. Clin. Investig. 2017, 127, 4314–4325. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Wu, F.; Zhang, M.; Xiong, Z.; Yin, Q.; Ru, Y.; Shi, H.; Li, J.; Mao, S.; Li, Y.; et al. EMC10 governs male fertility via maintaining sperm ion balance. J. Mol. Cell Biol. 2018, 10, 503–514. [Google Scholar] [CrossRef]
- Shurtleff, M.J.; Itzhak, D.N.; Hussmann, J.A.; Schirle Oakdale, N.T.; Costa, E.A.; Jonikas, M.; Weibezahn, J.; Popova, K.D.; Jan, C.H.; Sinitcyn, P.; et al. The ER membrane protein complex interacts cotranslationally to enable biogenesis of multipass membrane proteins. eLife 2018, 7, e37018. [Google Scholar] [CrossRef]
- Volkmar, N.; Christianson, J.C. Squaring the EMC—How promoting membrane protein biogenesis impacts cellular functions and organismal homeostasis. J. Cell Sci. 2020, 133, jcs243519. [Google Scholar] [CrossRef]
- Tian, S.; Wu, Q.; Zhou, B.; Choi, M.Y.; Ding, B.; Yang, W.; Dong, M. Proteomic Analysis Identifies Membrane Proteins Dependent on the ER Membrane Protein Complex. Cell Rep. 2019, 28, 2517–2526. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, J.P.; Phillips, B.P.; Yagita, Y.; Juszkiewicz, S.; Wagner, A.; Malinverni, D.; Keenan, R.J.; Miller, E.A.; Hegde, R.S. The architecture of EMC reveals a path for membrane protein insertion. eLife 2020, 9, e57887. [Google Scholar] [CrossRef] [PubMed]
- Miller-Vedam, L.E.; Bräuning, B.; Popova, K.D.; Schirle Oakdale, N.T.; Bonnar, J.L.; Prabu, J.R.; Boydston, E.A.; Sevillano, N.; Shurtleff, M.J.; Stroud, R.M.; et al. Structural and mechanistic basis of the EMC-dependent biogenesis of distinct transmembrane clients. eLife 2020, 9, e62611. [Google Scholar] [CrossRef] [PubMed]
- Pleiner, T.; Tomaleri, G.P.; Januszyk, K.; Inglis, A.J.; Hazu, M.; Voorhees, R.M. Structural basis for membrane insertion by the human ER membrane protein complex. Science 2020, 369, 433–436. [Google Scholar] [CrossRef] [PubMed]
- Bai, L.; You, Q.; Feng, X.; Kovach, A.; Li, H. Structure of the ER membrane complex, a transmembrane-domain insertase. Nature 2020, 584, 475–478. [Google Scholar] [CrossRef] [PubMed]
- Dudek, J.; Pfeffer, S.; Lee, P.-H.; Jung, M.; Cavalié, A.; Helms, V.; Förster, F.; Zimmermann, R. Protein Transport into the Human Endoplasmic Reticulum. J. Mol. Biol. 2015, 427, 1159–1175. [Google Scholar] [CrossRef] [PubMed]
- Braakman, I.; Hebert, D.N. Protein folding in the endoplasmic reticulum. Cold Spring Harb. Perspect. Biol. 2013, 5, a013201. [Google Scholar] [CrossRef] [PubMed]
- Phillips, B.P.; Gomez-Navarro, N.; Miller, E.A. Protein quality control in the endoplasmic reticulum. Curr. Opin. Cell Biol. 2020, 65, 96–102. [Google Scholar] [CrossRef] [PubMed]
- Melnyk, A.; Rieger, H.; Zimmermann, R. Co-chaperones of the Mammalian Endoplasmic Reticulum. In The Networking of Chaperones by Co-Chaperones: Control of Cellular Protein Homeostasis; Blatch, G.L., Edkins, A.L., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 179–200. [Google Scholar]
- Siegel, V.; Walter, P. Binding sites of the 19-kDa and 68/72-kDa signal recognition particle (SRP) proteins on SRP RNA as determined in protein-RNA “footprinting”. Proc. Natl. Acad. Sci. USA 1988, 85, 1801–1805. [Google Scholar] [CrossRef] [PubMed]
- van Nues, R.W.; Leung, E.; McDonald, J.C.; Dantuluru, I.; Brown, J.D. Roles for Srp72p in assembly, nuclear export and function of the signal recognition particle. RNA Biol. 2008, 5, 73–83. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Iakhiaeva, E.; Yin, J.; Zwieb, C. Identification of an RNA-binding Domain in Human SRP72. J. Mol. Biol. 2005, 345, 659–666. [Google Scholar] [CrossRef]
- Kirwan, M.; Walne, A.J.; Plagnol, V.; Velangi, M.; Ho, A.; Hossain, U.; Vulliamy, T.; Dokal, I. Exome Sequencing Identifies Autosomal-Dominant SRP72 Mutations Associated with Familial Aplasia and Myelodysplasia. Am. J. Hum. Genet. 2012, 90, 888–892. [Google Scholar] [CrossRef] [PubMed]
- Pool, M.R.; Stumm, J.; Fulga, T.A.; Sinning, I.; Dobberstein, B. Distinct Modes of Signal Recognition Particle Interaction with the Ribosome. Science 2002, 297, 1345–1348. [Google Scholar] [CrossRef] [PubMed]
- Akopian, D.; Shen, K.; Zhang, X.; Shan, S.-o. Signal Recognition Particle: An Essential Protein-Targeting Machine. Annu. Rev. Biochem. 2013, 82, 693–721. [Google Scholar] [CrossRef] [PubMed]
- Gowda, K.; Chittenden, K.; Zwieb, C. Binding site of the M-domain of human protein SRP54 determined by systematic site-directed mutagenesis of signal recognition particle RNA. Nucleic Acids Res. 1997, 25, 388–394. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Carapito, R.; Konantz, M.; Paillard, C.; Miao, Z.; Pichot, A.; Leduc, M.S.; Yang, Y.; Bergstrom, K.L.; Mahoney, D.H.; Shardy, D.L.; et al. Mutations in signal recognition particle SRP54 cause syndromic neutropenia with Shwachman-Diamond–like features. J. Clin. Investig. 2017, 127, 4090–4103. [Google Scholar] [CrossRef] [PubMed]
- Allenbach, Y.; Benveniste, O.; Stenzel, W.; Boyer, O. Immune-mediated necrotizing myopathy: Clinical features and pathogenesis. Nat. Rev. Rheumatol. 2020, 16, 689–701. [Google Scholar] [CrossRef] [PubMed]
- Römisch, K.; Miller, F.W.; Dobberstein, B.; High, S. Human autoantibodies against the 54 kDa protein of the signal recognition particle block function at multiple stages. Arthritis Res. 2006, 8, R39. [Google Scholar] [CrossRef]
- Benarroch, R.; Austin, J.M.; Ahmed, F.; Isaacson, R.L. Chapter Seven—The roles of cytosolic quality control proteins, SGTA and the BAG6 complex, in disease. In Advances in Protein Chemistry and Structural Biology; Donev, R., Ed.; Academic Press: Cambridge, MA, USA, 2019; Volume 114, pp. 265–313. [Google Scholar]
- Behl, C. Breaking BAG: The Co-Chaperone BAG3 in Health and Disease. Trends Pharmacol. Sci. 2016, 37, 672–688. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zang, Y.-S.; Xiu, Q. BAT3 rs1052486 and rs3117582 polymorphisms are associated with lung cancer risk: A meta-analysis. Tumor Biol. 2014, 35, 9855–9858. [Google Scholar] [CrossRef] [PubMed]
- Etokebe, G.E.; Zienolddiny, S.; Kupanovac, Z.; Enersen, M.; Balen, S.; Flego, V.; Bulat-Kardum, L.; Radojčić-Badovinac, A.; Skaug, V.; Bakke, P.; et al. Association of the FAM46A Gene VNTRs and BAG6 rs3117582 SNP with Non Small Cell Lung Cancer (NSCLC) in Croatian and Norwegian Populations. PLoS ONE 2015, 10, e0122651. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Liang, Y.; Li, H.; Li, H.; He, Q.; Xue, Y.; Shen, C.; Zhang, C.; Xiang, J.; Ding, J.; et al. Single Nucleotide Polymorphisms and Osteoarthritis: An Overview and a Meta-Analysis. Medicine 2016, 95, e2811. [Google Scholar] [CrossRef] [PubMed]
- Xue, Q.; Lv, L.; Wan, C.; Chen, B.; Li, M.; Ni, T.; Liu, Y.; Liu, Y.; Cong, X.; Zhou, Y.; et al. Expression and clinical role of small glutamine-rich tetratricopeptide repeat (TPR)-containing protein alpha (SGTA) as a novel cell cycle protein in NSCLC. J. Cancer Res. Clin. Oncol. 2013, 139, 1539–1549. [Google Scholar] [CrossRef]
- Yang, X.; Cheng, L.; Li, M.; Shi, H.; Ren, H.; Ding, Z.; Liu, F.; Wang, Y.; Cheng, C. High Expression of SGTA in Esophageal Squamous Cell Carcinoma Correlates With Proliferation and Poor Prognosis. J. Cell. Biochem. 2014, 115, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.; Ji, Z.; Xu, C.; Peng, Z.; Gu, L.; Zhang, R.; Liu, Y. Expression and prognostic role of SGTA in human breast carcinoma correlates with tumor cell proliferation. J. Mol. Histol. 2014, 45, 665–677. [Google Scholar] [CrossRef] [PubMed]
- Goodarzi, M.O.; Xu, N.; Cui, J.; Guo, X.; Chen, Y.I.; Azziz, R. Small glutamine-rich tetratricopeptide repeat-containing protein alpha (SGTA), a candidate gene for polycystic ovary syndrome. Hum. Reprod. 2008, 23, 1214–1219. [Google Scholar] [CrossRef] [PubMed]
- Sicking, M.; Lang, S.; Bochen, F.; Roos, A.; Drenth, J.P.H.; Zakaria, M.; Zimmermann, R.; Linxweiler, M. Complexity and Specificity of Sec61-Channelopathies: Human Diseases Affecting Gating of the Sec61 Complex. Cells 2021, 10, 1036. [Google Scholar] [CrossRef] [PubMed]
- Linxweiler, M.; Schick, B.; Zimmermann, R. Let’s talk about Secs: Sec61, Sec62 and Sec63 in signal transduction, oncology and personalized medicine. Signal Transduct. Target. Ther. 2017, 2, 17002. [Google Scholar] [CrossRef] [PubMed]
- Van Nieuwenhove, E.; Barber, J.S.; Neumann, J.; Smeets, E.; Willemsen, M.; Pasciuto, E.; Prezzemolo, T.; Lagou, V.; Seldeslachts, L.; Malengier-Devlies, B.; et al. Defective Sec61α1 underlies a novel cause of autosomal dominant severe congenital neutropenia. J. Allergy Clin. Immunol. 2020, 146, 1180–1193. [Google Scholar] [CrossRef] [PubMed]
- Davila, S.; Furu, L.; Gharavi, A.G.; Tian, X.; Onoe, T.; Qian, Q.; Li, A.; Cai, Y.; Kamath, P.S.; King, B.F.; et al. Mutations in SEC63 cause autosomal dominant polycystic liver disease. Nat. Genet. 2004, 36, 575–577. [Google Scholar] [CrossRef]
- Greiner, M.; Kreutzer, B.; Jung, V.; Grobholz, R.; Hasenfus, A.; Stöhr, R.F.; Tornillo, L.; Dudek, J.; Stöckle, M.; Unteregger, G.; et al. Silencing of the SEC62 gene inhibits migratory and invasive potential of various tumor cells. Int. J. Cancer 2011, 128, 2284–2295. [Google Scholar] [CrossRef] [PubMed]
- Besse, W.; Dong, K.; Choi, J.; Punia, S.; Fedeles, S.V.; Choi, M.; Gallagher, A.-R.; Huang, E.B.; Gulati, A.; Knight, J.; et al. Isolated polycystic liver disease genes define effectors of polycystin-1 function. J. Clin. Investig. 2017, 127, 1772–1785. [Google Scholar] [CrossRef] [PubMed]
- Meng, H.; Jiang, X.; Wang, J.; Sang, Z.; Guo, L.; Yin, G.; Wang, Y. SEC61G is upregulated and required for tumor progression in human kidney cancer. Mol. Med. Rep. 2021, 23, 427. [Google Scholar] [CrossRef] [PubMed]
- Ng, B.G.; Lourenço, C.M.; Losfeld, M.-E.; Buckingham, K.J.; Kircher, M.; Nickerson, D.A.; Shendure, J.; Bamshad, M.J.; University of Washington Center for Mendelian, G.; Freeze, H.H. Mutations in the translocon-associated protein complex subunit SSR3 cause a novel congenital disorder of glycosylation. J. Inherit. Metab. Dis. 2019, 42, 993–997. [Google Scholar] [CrossRef] [PubMed]
- Dittner-Moormann, S.; Lourenço, C.M.; Reunert, J.; Nishinakamura, R.; Tanaka, S.S.; Werner, C.; Debus, V.; Zimmer, K.-P.; Wetzel, G.; Naim, H.Y.; et al. TRAPγ-CDG shows asymmetric glycosylation and an effect on processing of proteins required in higher organisms. J. Med. Genet. 2021, 58, 213–216. [Google Scholar] [CrossRef] [PubMed]
- Michael Yates, T.; Ng, O.-H.; Offiah, A.C.; Willoughby, J.; Berg, J.N.; Study, D.; Johnson, D.S. Cerebrofaciothoracic dysplasia: Four new patients with a recurrent TMCO1 pathogenic variant. Am. J. Med. Genet. Part A 2019, 179, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Sharkia, R.; Zalan, A.; Jabareen-Masri, A.; Hengel, H.; Schöls, L.; Kessel, A.; Azem, A.; Mahajnah, M. A novel biallelic loss-of-function mutation in TMCO1 gene confirming and expanding the phenotype spectrum of cerebro-facio-thoracic dysplasia. Am. J. Med. Genet. Part A 2019, 179, 1338–1345. [Google Scholar] [CrossRef] [PubMed]
- Alanay, Y.; Ergüner, B.; Utine, E.; Haçarız, O.; Kiper, P.O.S.; Taşkıran, E.Z.; Perçin, F.; Uz, E.; Sağıroğlu, M.Ş.; Yuksel, B.; et al. TMCO1 deficiency causes autosomal recessive cerebrofaciothoracic dysplasia. Am. J. Med. Genet. Part A 2014, 164, 291–304. [Google Scholar] [CrossRef] [PubMed]
- Caglayan, A.; Per, H.; Akgumus, G.; Gumus, H.; Baranoski, J.; Canpolat, M.; Calik, M.; Yikilmaz, A.; Bilguvar, K.; Kumandas, S.; et al. Whole-exome sequencing identified a patient with TMCO1 defect syndrome and expands the phenotic spectrum. Clin. Genet. 2013, 84, 394–395. [Google Scholar] [CrossRef]
- Morimoto, M.; Waller-Evans, H.; Ammous, Z.; Song, X.; Strauss, K.A.; Pehlivan, D.; Gonzaga-Jauregui, C.; Puffenberger, E.G.; Holst, C.R.; Karaca, E.; et al. Bi-allelic CCDC47 Variants Cause a Disorder Characterized by Woolly Hair, Liver Dysfunction, Dysmorphic Features, and Global Developmental Delay. Am. J. Hum. Genet. 2018, 103, 794–807. [Google Scholar] [CrossRef]
- Geetha, T.S.; Lingappa, L.; Jain, A.R.; Govindan, H.; Mandloi, N.; Murugan, S.; Gupta, R.; Vedam, R. A novel splice variant in EMC1 is associated with cerebellar atrophy, visual impairment, psychomotor retardation with epilepsy. Mol. Genet. Genom. Med. 2018, 6, 282–287. [Google Scholar] [CrossRef] [PubMed]
- Harel, T.; Yesil, G.; Bayram, Y.; Coban-Akdemir, Z.; Charng, W.-L.; Karaca, E.; Al Asmari, A.; Eldomery, M.K.; Hunter, J.V.; Jhangiani, S.N.; et al. Monoallelic and Biallelic Variants in EMC1 Identified in Individuals with Global Developmental Delay, Hypotonia, Scoliosis, and Cerebellar Atrophy. Am. J. Hum. Genet. 2016, 98, 562–570. [Google Scholar] [CrossRef]
- Shao, D.D.; Straussberg, R.; Ahmed, H.; Khan, A.; Tian, S.; Hill, R.S.; Smith, R.S.; Majmundar, A.J.; Ameziane, N.; Neil, J.E.; et al. A recurrent, homozygous EMC10 frameshift variant is associated with a syndrome of developmental delay with variable seizures and dysmorphic features. Genet. Med. 2021, 23, 1158–1162. [Google Scholar] [CrossRef] [PubMed]
- Junes-Gill, K.S.; Gallaher, T.K.; Gluzman-Poltorak, Z.; Miller, J.D.; Wheeler, C.J.; Fan, X.; Basile, L.A. hHSS1: A novel secreted factor and suppressor of glioma growth located at chromosome 19q13.33. J. Neurooncol. 2011, 102, 197–211. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Kan, S.; Hu, J.; Li, M.; Lu, G.; Zhang, M.; Zhang, S.; Hou, Y.; Chen, Y.; Bai, Y. EMC6/TMEM93 suppresses glioblastoma proliferation by modulating autophagy. Cell Death Dis. 2016, 7, e2043. [Google Scholar] [CrossRef] [PubMed]
- Junes-Gill, K.S.; Lawrence, C.E.; Wheeler, C.J.; Cordner, R.; Gill, T.G.; Mar, V.; Shiri, L.; Basile, L.A. Human hematopoietic signal peptide-containing secreted 1 (hHSS1) modulates genes and pathways in glioma: Implications for the regulation of tumorigenicity and angiogenesis. BMC Cancer 2014, 14, 920. [Google Scholar] [CrossRef]
- Asseck, L.Y.; Mehlhorn, D.G.; Monroy, J.R.; Ricardi, M.M.; Breuninger, H.; Wallmeroth, N.; Berendzen, K.W.; Nowrousian, M.; Xing, S.; Schwappach, B.; et al. Endoplasmic reticulum membrane receptors of the GET pathway are conserved throughout eukaryotes. Proc. Natl. Acad. Sci. USA 2021, 118, e2017636118. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, Y.; Sakisaka, T. Molecular Machinery for Insertion of Tail-Anchored Membrane Proteins into the Endoplasmic Reticulum Membrane in Mammalian Cells. Mol. Cell 2012, 48, 387–397. [Google Scholar] [CrossRef] [PubMed]
- Egeo, A.; Mazzocco, M.; Sotgia, F.; Arrigo, P.; Oliva, R.; Bergonòn, S.; Nizetic, D.; Rasore-Quartino, A.; Scartezzini, P. Identification and characterization of a new human cDNA from chromosome 21q22.3 encoding a basic nuclear protein. Hum. Genet. 1998, 102, 289–293. [Google Scholar] [CrossRef] [PubMed]
- Murata, K.; Degmetich, S.; Kinoshita, M.; Shimada, E. Expression of the congenital heart disease 5/tryptophan rich basic protein homologue gene during heart development in Medaka fish, Oryzias latipes. Dev. Growth Differ. 2009, 51, 95–107. [Google Scholar] [CrossRef]
- Sojka, S.; Amin, N.M.; Gibbs, D.; Christine, K.S.; Charpentier, M.S.; Conlon, F.L. Congenital heart disease protein 5 associates with CASZ1 to maintain myocardial tissue integrity. Development 2014, 141, 3040–3049. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.-H.; Kim, T.-Y.; Kim, W.-H.; Park, J.-W. CAML promotes prolactin-dependent proliferation of breast cancer cells by facilitating prolactin receptor signaling pathways. Breast Cancer Res. Treat. 2011, 130, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Long, T.; Su, J.; Tang, W.; Luo, Z.; Liu, S.; Liu, Z.; Zhou, H.; Qi, M.; Zeng, W.; Zhang, J.; et al. A novel interaction between calcium-modulating cyclophilin ligand and Basigin regulates calcium signaling and matrix metalloproteinase activities in human melanoma cells. Cancer Lett. 2013, 339, 93–101. [Google Scholar] [CrossRef]
- Available online: www.mousephenotype.org/data/genes/MGI:2136882#phenotypesTab (accessed on 22 December 2021).
- Tosal-Castano, S.; Peselj, C.; Kohler, V.; Habernig, L.; Berglund, L.L.; Ebrahimi, M.; Vögtle, F.N.; Höög, J.; Andréasson, C.; Büttner, S. Snd3 controls nucleus-vacuole junctions in response to glucose signaling. Cell Rep. 2021, 34, 108637. [Google Scholar] [CrossRef] [PubMed]
- Ricarte, F.; Menjivar, R.; Chhun, S.; Soreta, T.; Oliveira, L.; Hsueh, T.; Serranilla, M.; Gharakhanian, E. A Genome-Wide Immunodetection Screen in S. cerevisiae Uncovers Novel Genes Involved in Lysosomal Vacuole Function and Morphology. PLoS ONE 2011, 6, e23696. [Google Scholar] [CrossRef][Green Version]
- Yompakdee, C.; Ogawa, N.; Harashima, S.; Oshima, Y. A putative membrane protein, Pho88p, involved in inorganic phosphate transport inSaccharomyces cerevisiae. Mol. Gen. Genet. MGG 1996, 251, 580–590. [Google Scholar] [CrossRef] [PubMed]
- Mukerji, N.; Damodaran, T.V.; Winn, M.P. TRPC6 and FSGS: The latest TRP channelopathy. Biochim. Et Biophys. Acta (BBA) Mol. Basis Dis. 2007, 1772, 859–868. [Google Scholar] [CrossRef]
- Riehle, M.; Büscher, A.K.; Gohlke, B.-O.; Kaßmann, M.; Kolatsi-Joannou, M.; Bräsen, J.H.; Nagel, M.; Becker, J.U.; Winyard, P.; Hoyer, P.F.; et al. TRPC6 G757D Loss-of-Function Mutation Associates with FSGS. J. Am. Soc. Nephrol. 2016, 27, 2771–2783. [Google Scholar] [CrossRef] [PubMed]
- Winn, M.P.; Conlon, P.J.; Lynn, K.L.; Farrington, M.K.; Creazzo, T.; Hawkins, A.F.; Daskalakis, N.; Kwan, S.Y.; Ebersviller, S.; Burchette, J.L.; et al. A Mutation in the TRPC6 Cation Channel Causes Familial Focal Segmental Glomerulosclerosis. Science 2005, 308, 1801–1804. [Google Scholar] [CrossRef] [PubMed]
- Andolfo, I.; Russo, R.; Manna, F.; Shmukler, B.E.; Gambale, A.; Vitiello, G.; De Rosa, G.; Brugnara, C.; Alper, S.L.; Snyder, L.M.; et al. Novel Gardos channel mutations linked to dehydrated hereditary stomatocytosis (xerocytosis). Am. J. Hematol. 2015, 90, 921–926. [Google Scholar] [CrossRef] [PubMed]
- Glogowska, E.; Lezon-Geyda, K.; Maksimova, Y.; Schulz, V.P.; Gallagher, P.G. Mutations in the Gardos channel (KCNN4) are associated with hereditary xerocytosis. Blood 2015, 126, 1281–1284. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, D.J.; Wheeler, M.C.; Gekakis, N. A point mutation in Sec61alpha1 leads to diabetes and hepatosteatosis in mice. Diabetes 2010, 59, 460–470. [Google Scholar] [CrossRef] [PubMed]
- Espino-Hernández, M.; Palma Milla, C.; Vara-Martín, J.; González-Granado, L.I. De novo SEC61A1 mutation in autosomal dominant tubulo-interstitial kidney disease: Phenotype expansion and review of literature. J. Paediatr. Child Health 2021, 57, 1305–1307. [Google Scholar] [CrossRef] [PubMed]
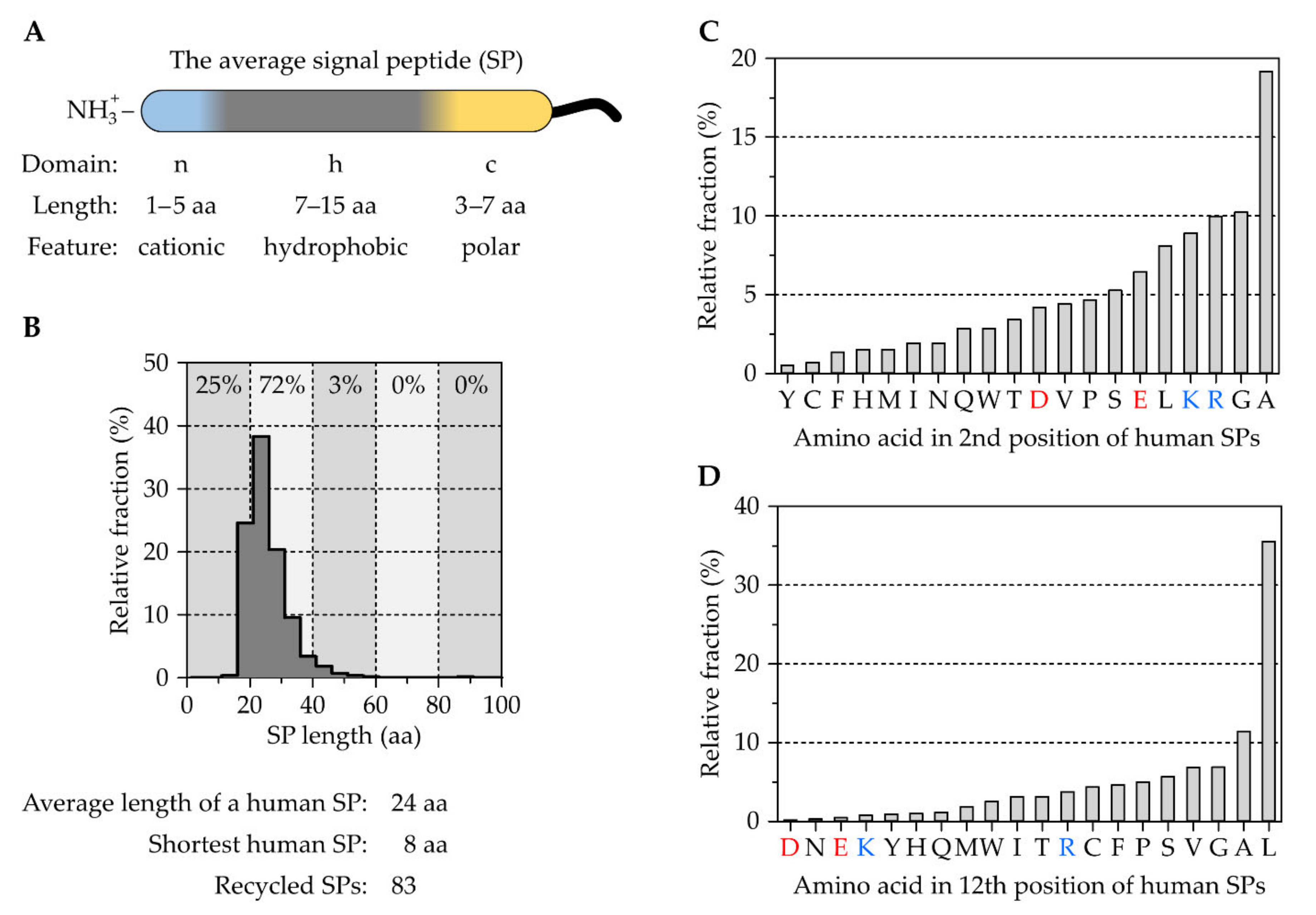

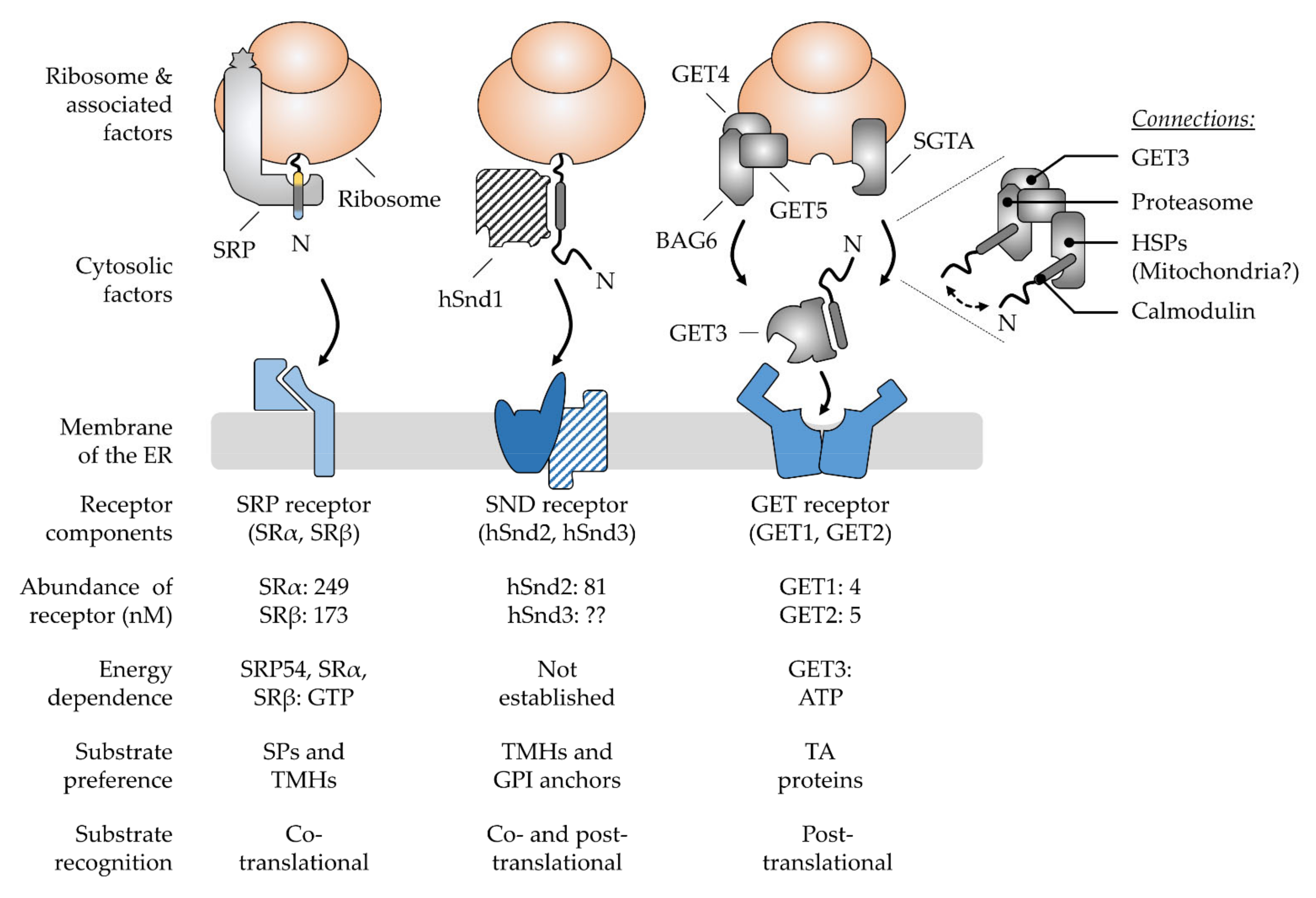
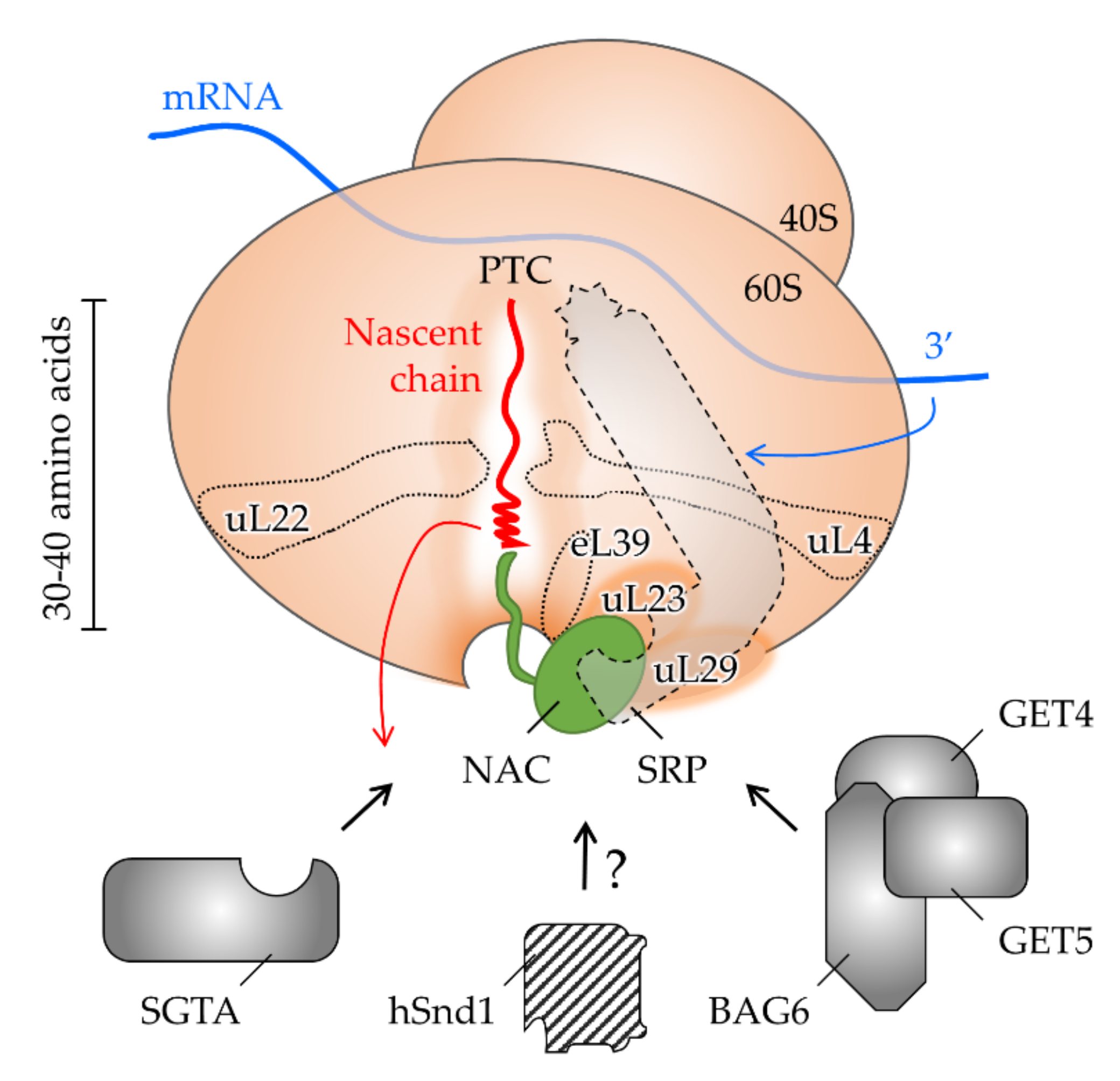
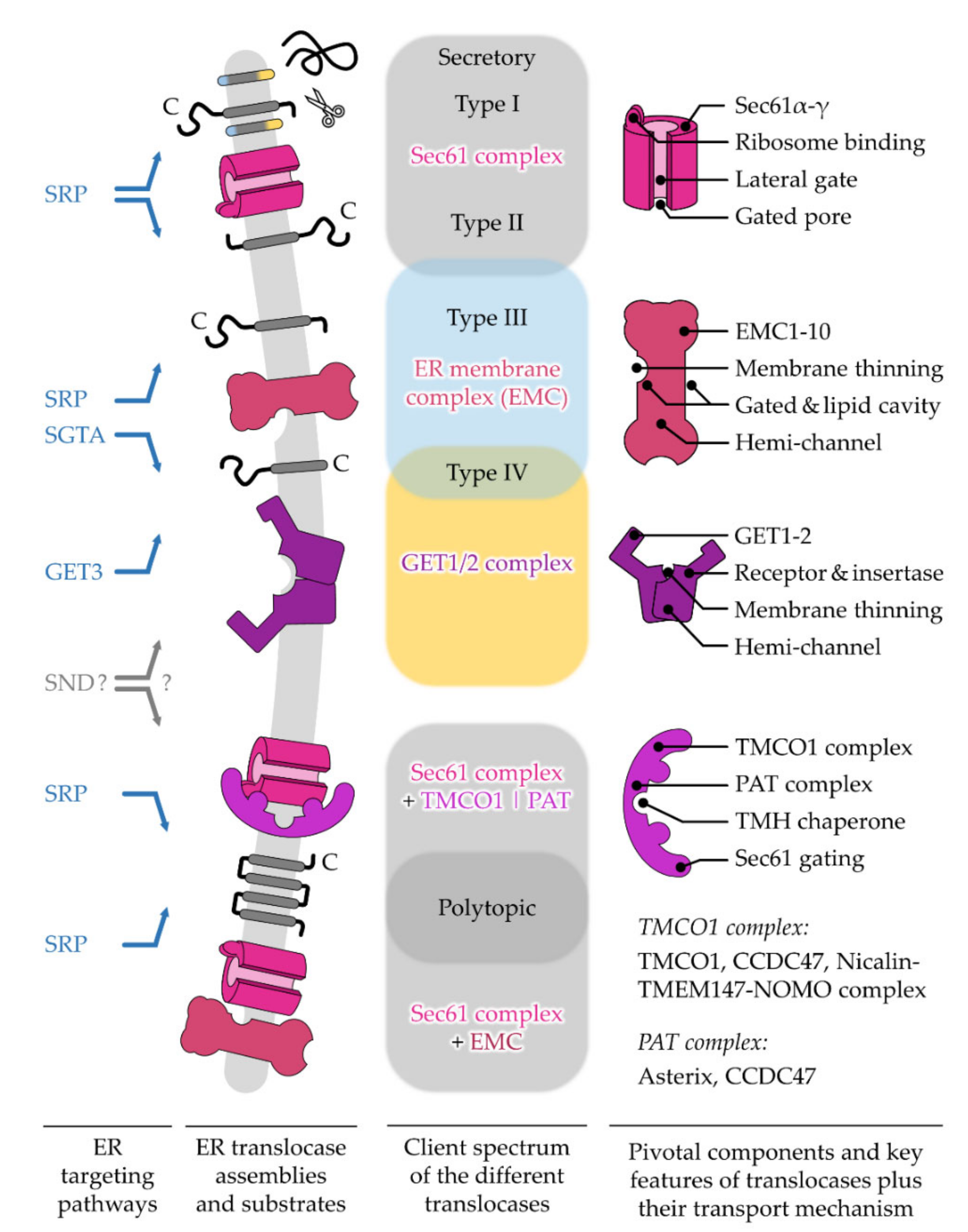
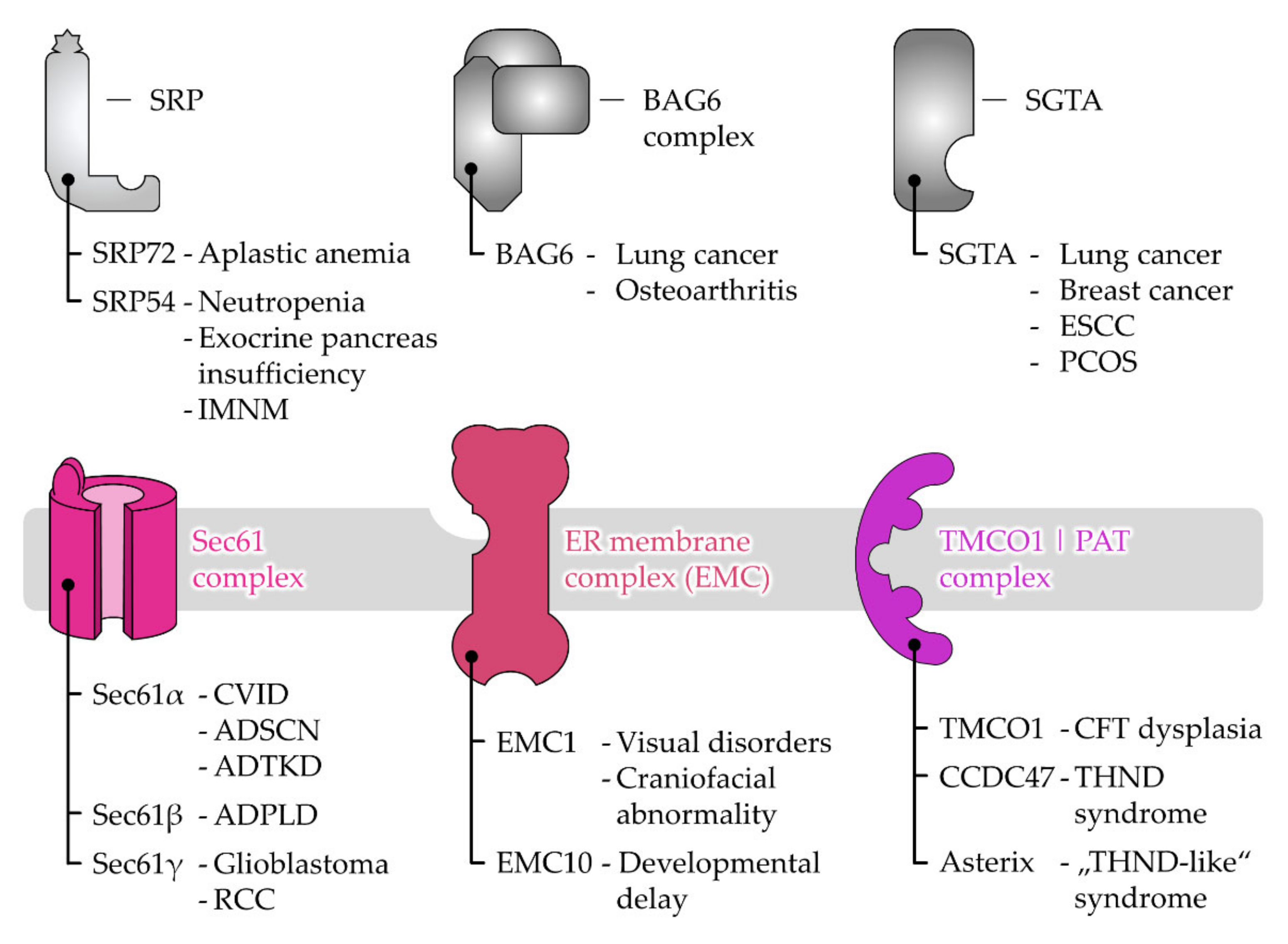
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tirincsi, A.; Sicking, M.; Hadzibeganovic, D.; Haßdenteufel, S.; Lang, S. The Molecular Biodiversity of Protein Targeting and Protein Transport Related to the Endoplasmic Reticulum. Int. J. Mol. Sci. 2022, 23, 143. https://doi.org/10.3390/ijms23010143
Tirincsi A, Sicking M, Hadzibeganovic D, Haßdenteufel S, Lang S. The Molecular Biodiversity of Protein Targeting and Protein Transport Related to the Endoplasmic Reticulum. International Journal of Molecular Sciences. 2022; 23(1):143. https://doi.org/10.3390/ijms23010143
Chicago/Turabian StyleTirincsi, Andrea, Mark Sicking, Drazena Hadzibeganovic, Sarah Haßdenteufel, and Sven Lang. 2022. "The Molecular Biodiversity of Protein Targeting and Protein Transport Related to the Endoplasmic Reticulum" International Journal of Molecular Sciences 23, no. 1: 143. https://doi.org/10.3390/ijms23010143
APA StyleTirincsi, A., Sicking, M., Hadzibeganovic, D., Haßdenteufel, S., & Lang, S. (2022). The Molecular Biodiversity of Protein Targeting and Protein Transport Related to the Endoplasmic Reticulum. International Journal of Molecular Sciences, 23(1), 143. https://doi.org/10.3390/ijms23010143





