iTRAQ-Based Quantitative Proteomics Analysis Reveals the Mechanism of Golden-Yellow Leaf Mutant in Hybrid Paper Mulberry
Abstract
:1. Introduction
2. Results
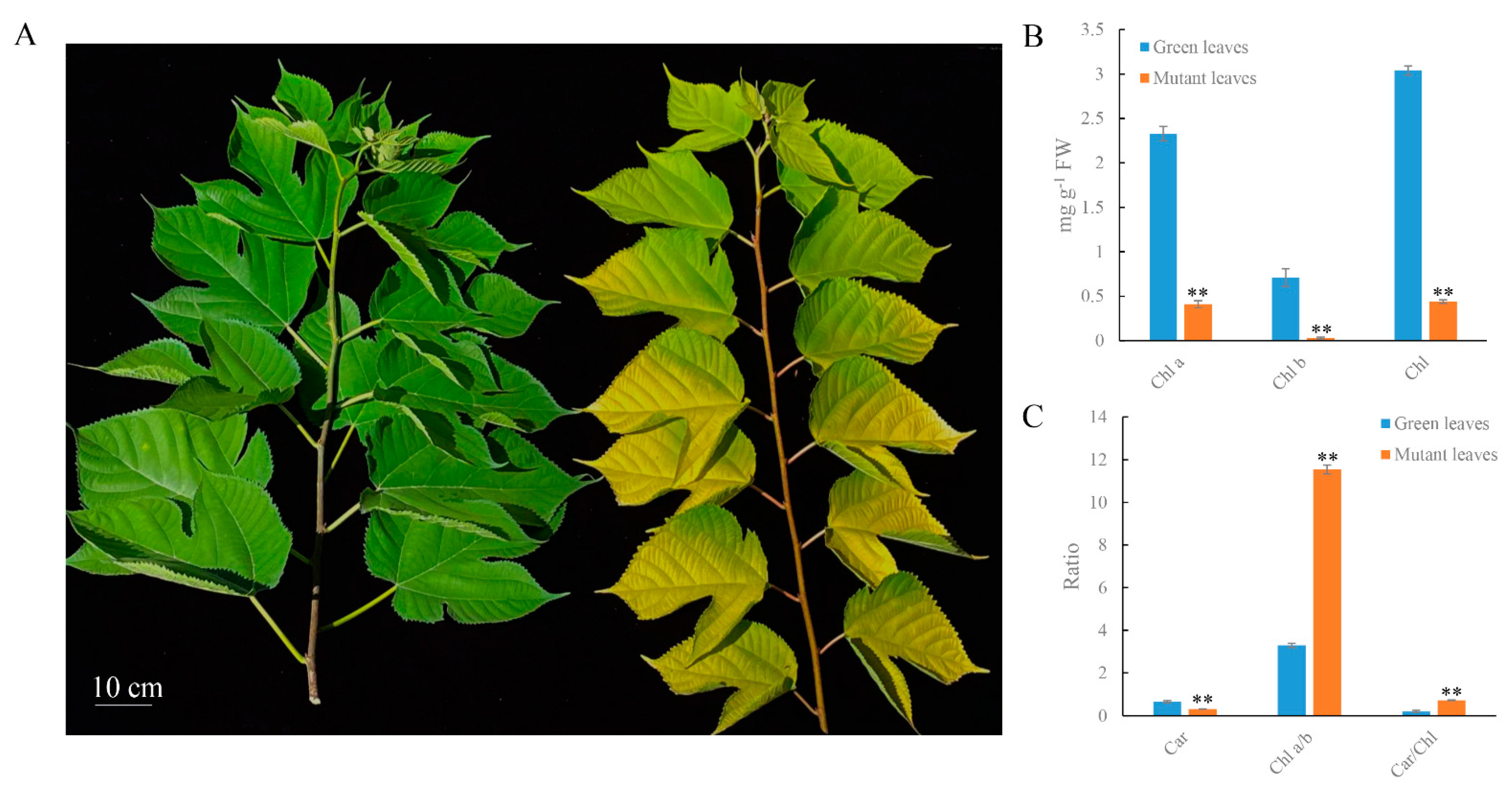
2.1. Physiological Changes in Mutant Leaves
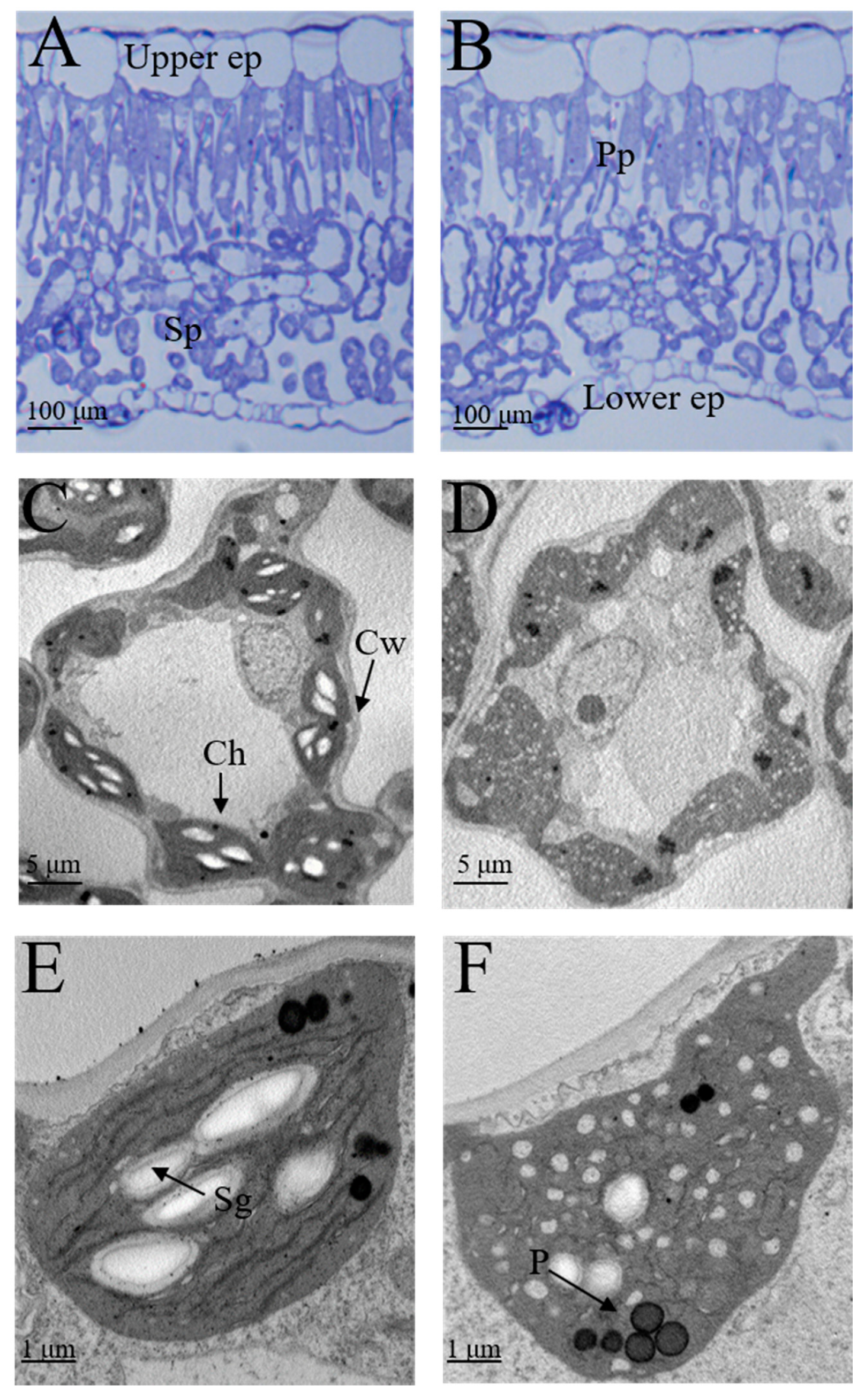
2.2. Anatomical Features and Ultrastructure of Chloroplasts in Mutant Leaves
2.3. Photosynthetic Parameter Analyses in Mutant Leaves
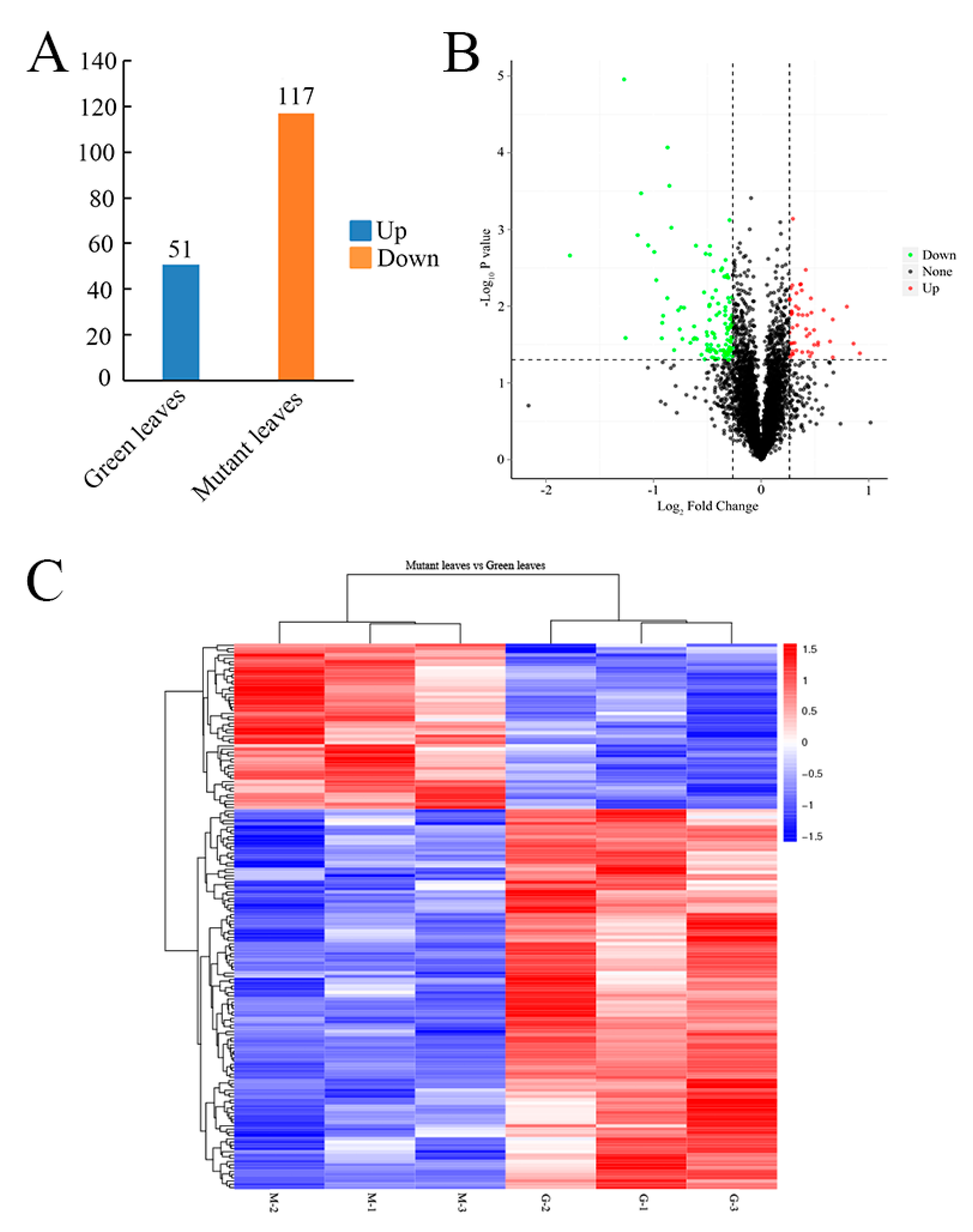
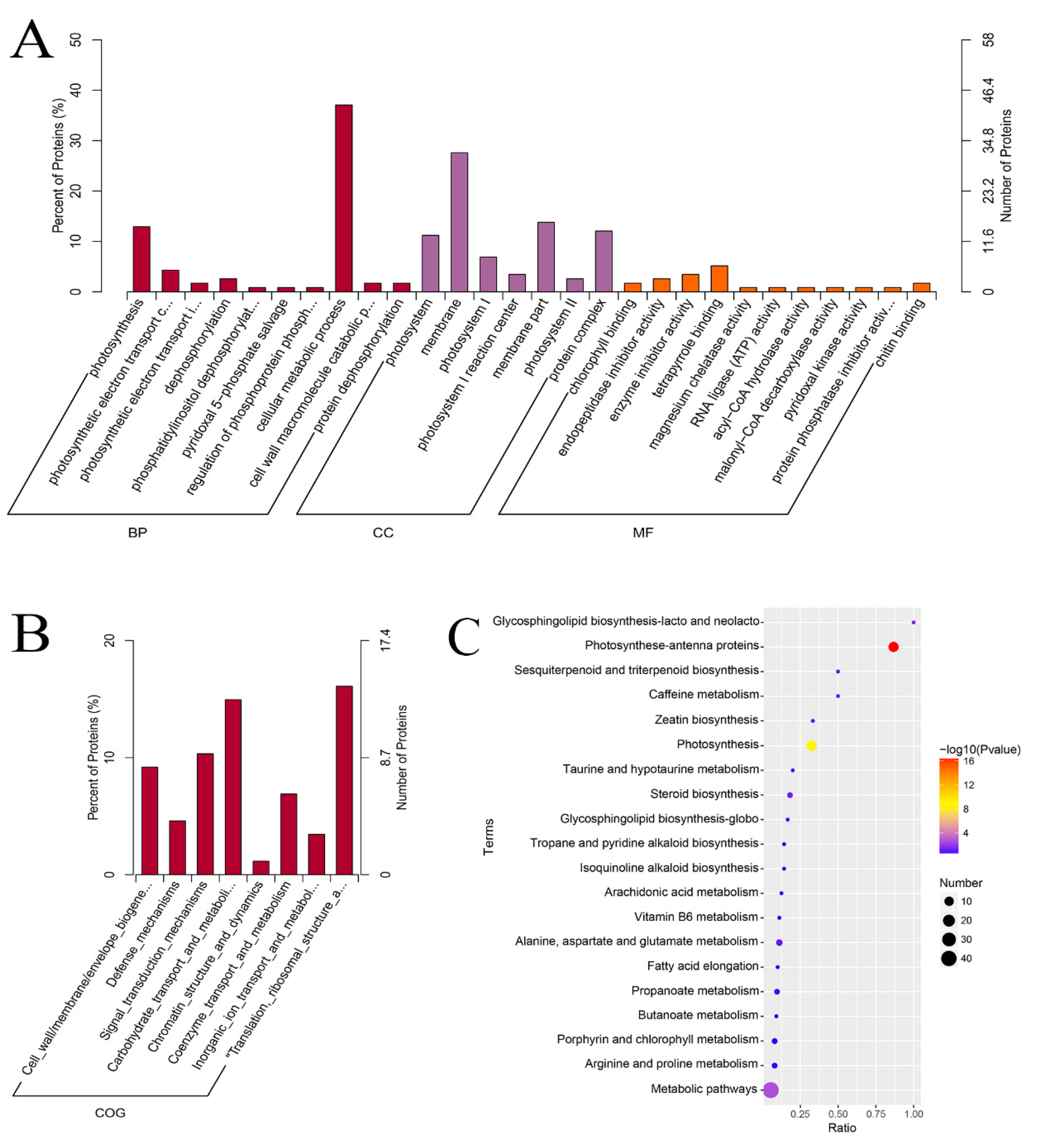
2.4. Quantitative Identification of Mutant Leaf Proteins Using iTRAQ
2.5. Protein Differences between Green Leaves and Mutant Leaves
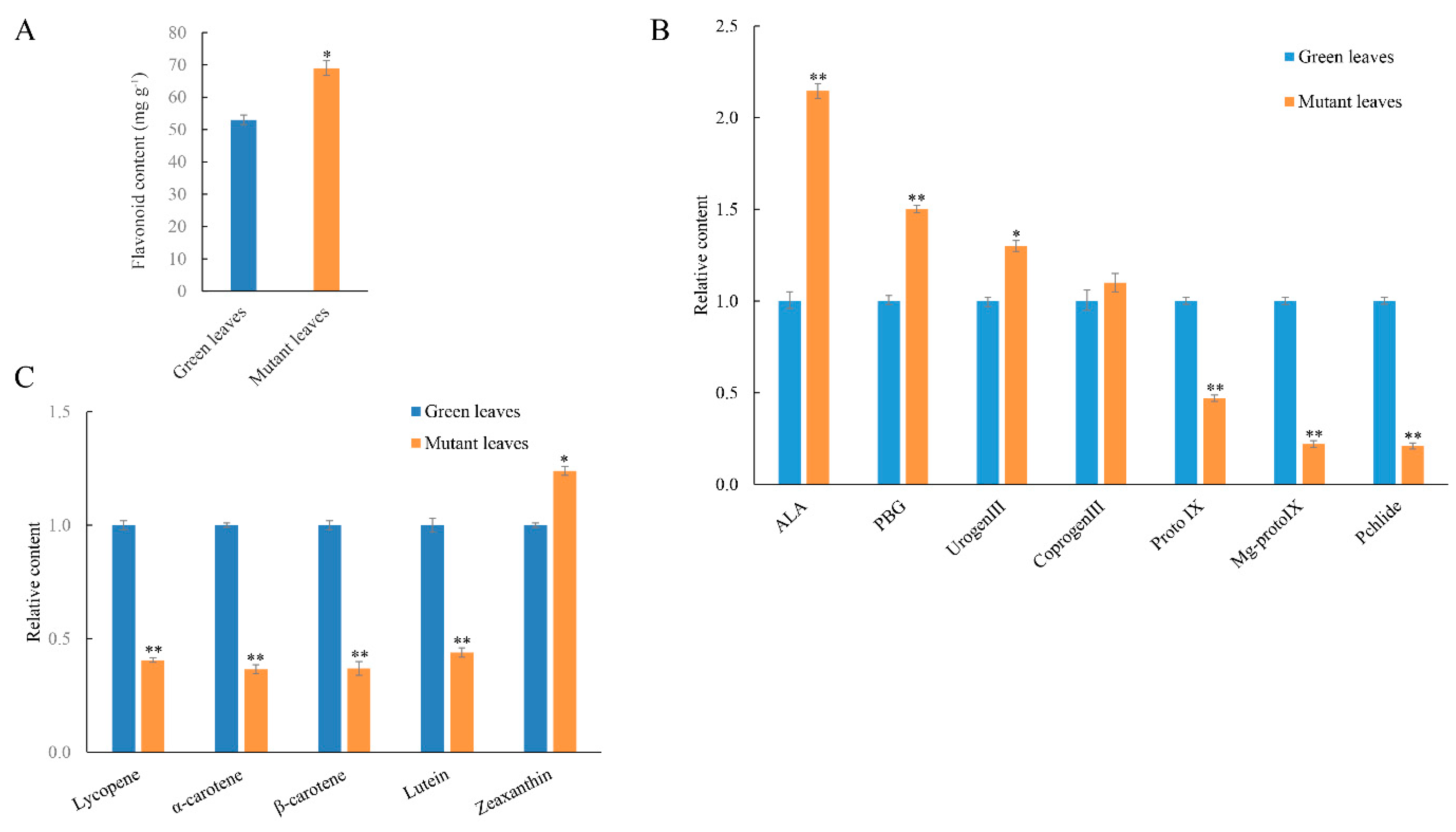
2.6. Identification of DAPs Related to Pigment Metabolism
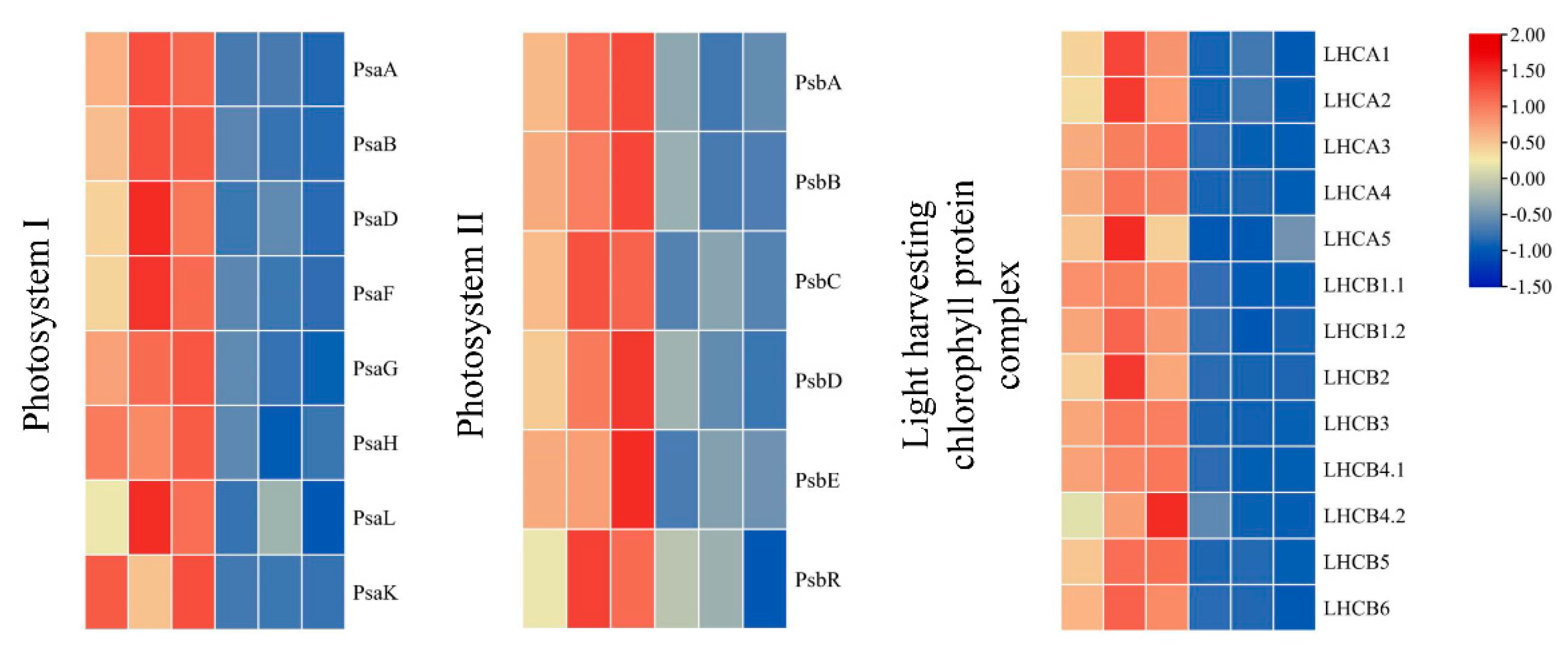
2.7. Differentially Accumulated Proteins Involved in Photosynthesis and Photosynthesis-Antenna Biosynthesis
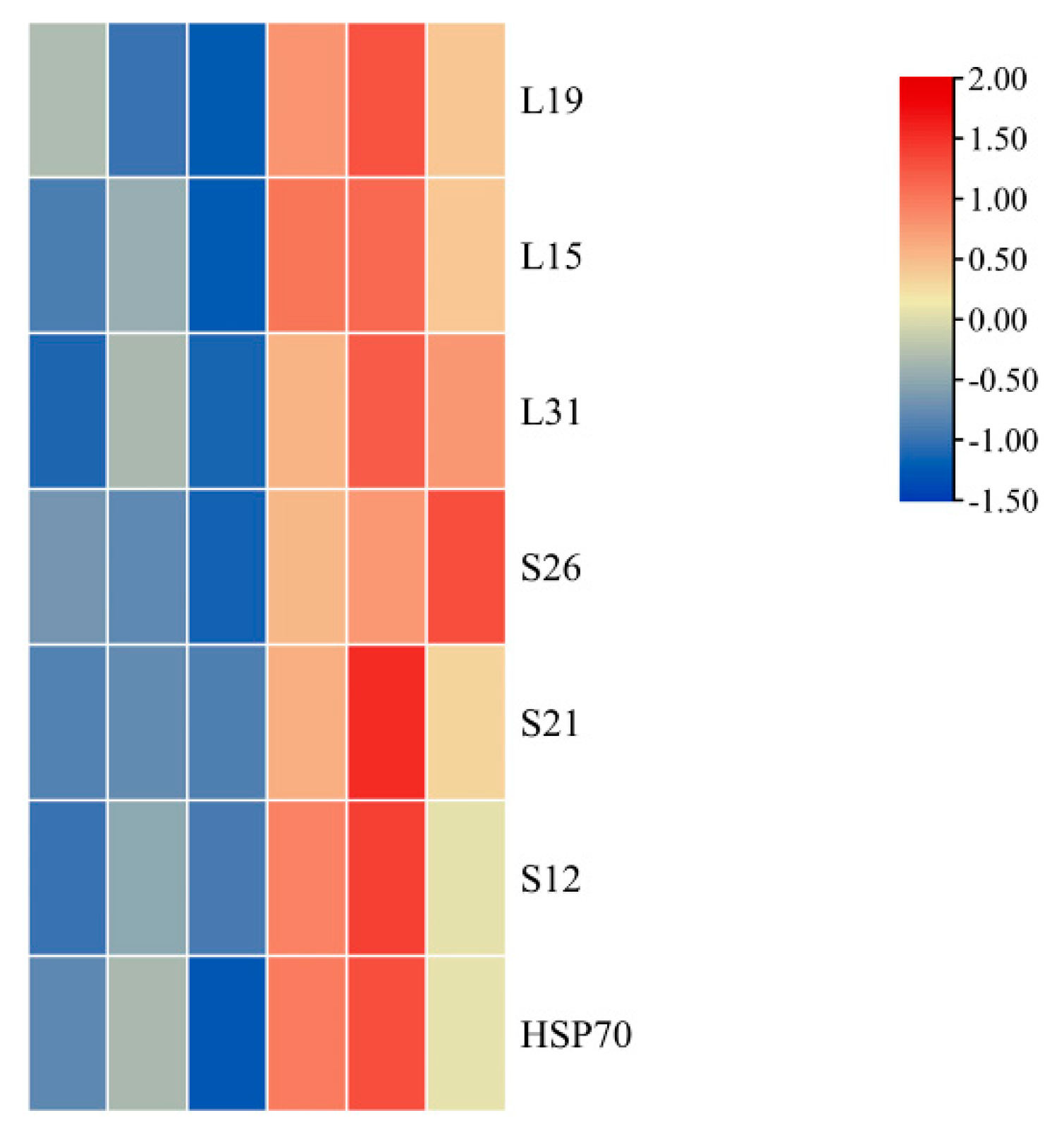
2.8. Differentially Accumulated Proteins Involved in Ribosome Pathway
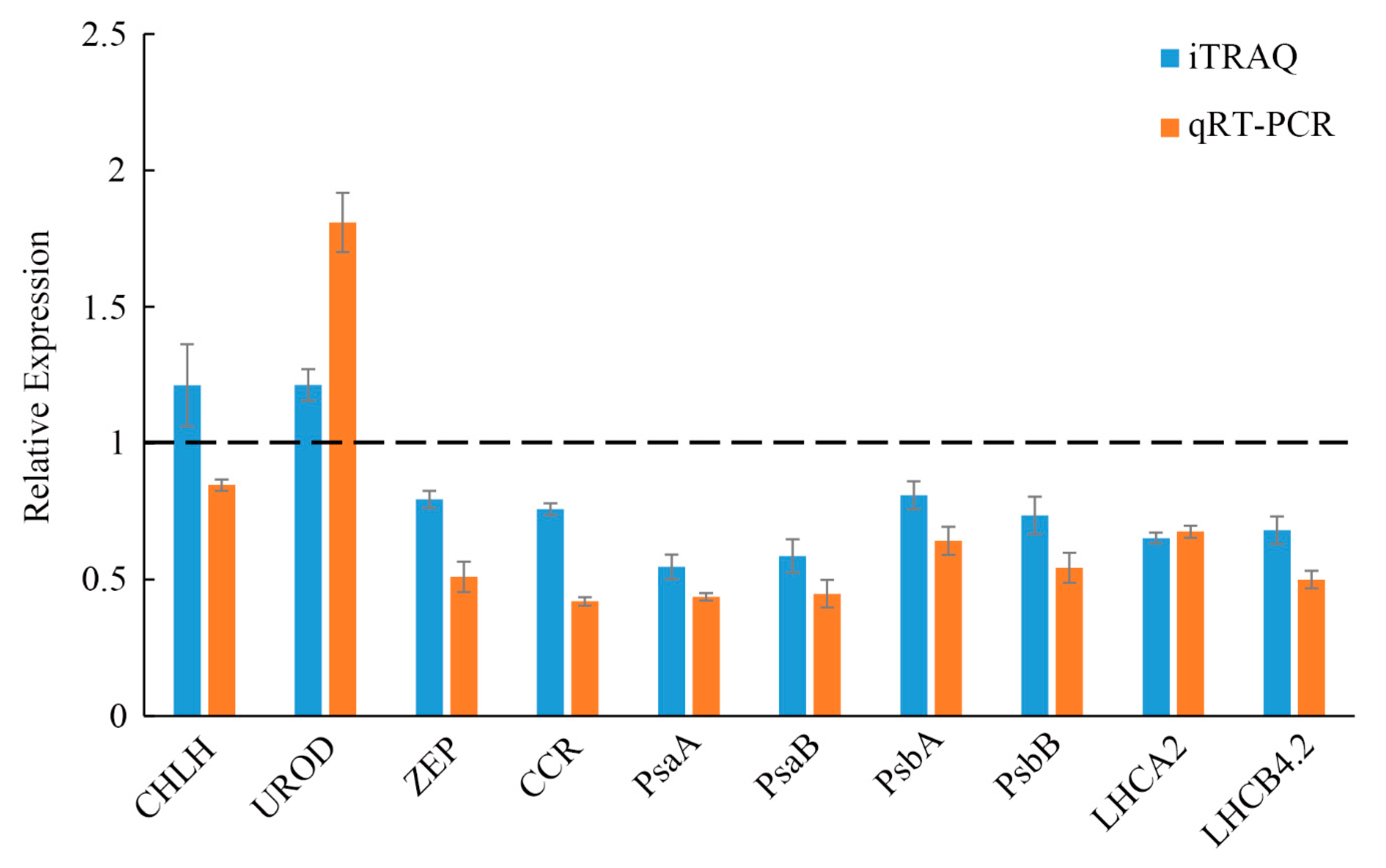
2.9. QRT-PCR Analysis of the Differentially Accumulated Proteins
3. Discussion
3.1. DAPs Involved in Chlorophyll Synthesis and Chloroplast Development
3.2. Differentially Accumulated Protein Involved in Photosynthesis Metabolism Pathways
4. Materials and Methods
4.1. Plant Materials
4.2. Determination of Chemical Composition
4.3. Transmission Electron Microscopic Analysis
4.4. Photosynthetic Parameter Measurements
4.5. Protein Extraction, Digesting, and iTRAQ Labeling
4.6. HPLC Fractionation and LC-MS/MS Analysis
4.7. Protein Identification and Quantification
4.8. Proteomic Data Analysis
4.9. RNA Extraction and qRT-PCR Analysis
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dismukes, G.C.; Klimov, V.V.; Baranov, S.V.; Kozlov, Y.N.; DasGupta, J.; Tyryshkin, A. The origin of atmospheric oxygen on Earth: The innovation of oxygenic photosynthesis. Proc. Natl. Acad. Sci. USA 2001, 98, 2170–2175. [Google Scholar] [CrossRef] [Green Version]
- Dekker, J.P.; Boekema, E.J. Supramolecular organization of thylakoid membrane proteins in green plants. Biochim. Biophys. Acta 2005, 1706, 12–39. [Google Scholar] [CrossRef] [Green Version]
- Gao, J.; Wang, H.; Yuan, Q.; Yue, F. Structure and function of the photosystem supercomplexes. Front. Plant Sci. 2018, 9, 357. [Google Scholar] [CrossRef] [PubMed]
- Zer, H.; Vink, M.; Shochat, S.; Herrmann, R.G.; Andersson, B.; Ohad, I. Light affects the accessibility of the thylakoid light harvesting complex II (LHCII) phosphorylation site to the membrane protein kinase(s). Biochemistry 2003, 42, 728–738. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, A.; Tanaka, R. Chlorophyll metabolism. Curr. Opin. Plant Biol. 2006, 9, 248–255. [Google Scholar] [CrossRef] [PubMed]
- Sandhu, D.; Atkinson, T.; Noll, A.; Johnson, C.; Espinosa, K.; Boelter, J.; Abel, S.; Dhatt, B.K.; Barta, T.; Singsaas, E.; et al. Soybean proteins GmTic110 and GmPsbP are crucial for chloroplast development and function. Plant Sci. 2016, 252, 76–87. [Google Scholar] [CrossRef] [Green Version]
- Ma, C.; Cao, J.; Li, J.; Zhou, B.; Tang, J.; Miao, A. Phenotypic, histological and proteomic analyses reveal multiple differences associated with chloroplast development in yellow and variegated variants from Camellia sinensis. Sci. Rep. 2016, 6, 33369. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Li, J.J.; Yoo, J.H.; Yoo, S.C.; Cho, S.H.; Koh, H.J.; Seo, H.S.; Paek, N.C. Rice Chlorina-1 and Chlorina-9 encode ChlD and ChlI subunits of Mg-chelatase, a key enzyme for chlorophyll synthesis and chloroplast development. Plant Mol. Biol. 2006, 62, 325–337. [Google Scholar] [CrossRef] [PubMed]
- Oda-Yamamizo, C.; Mitsuda, N.; Sakamoto, S.; Ogawa, D.; Ohme-Takagi, M.; Ohmiya, A. The NAC transcription factor ANAC046 is a positive regulator of chlorophyll degradation and senescence in Arabidopsis leaves. Sci. Rep. 2016, 6, 23609. [Google Scholar] [CrossRef] [Green Version]
- Slattery, R.A.; Andy, V.L.; Bernacchi, C.J.; Zhu, X.G.; Ort, D.R. Photosynthesis, light use efficiency, and yield of reduced-chlorophyll soybean mutants in field conditions. Front. Plant Sci. 2017, 8, 549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sang, X.C.; Fang, L.K.; Vanichpakorn, Y.; Ling, Y.H.; Du, P.; Zhao, F.M.; Yang, Z.L.; He, G.H. Physiological character and molecular mapping of leaf-color mutant wyv1 in rice (Oryza sativa L.). Genes Genom. 2010, 32, 123–128. [Google Scholar] [CrossRef]
- Zeng, Z.Q.; Lin, T.Z.; Zhao, J.Y.; Zheng, T.H.; Xu, L.F.; Wang, Y.H.; Liu, L.L.; Jiang, L.; Chen, S.H.; Wan, J.M. OsHemA gene, encoding glutamyl-tRNA reductase (GluTR) is essential for chlorophyll biosynthesis in rice (Oryza sativa). J. Integr. Agric. 2020, 19, 612–623. [Google Scholar] [CrossRef]
- Zhou, S.; Sawicki, A.; Willows, R.D.; Luo, M. C-terminal residues of oryza sativa GUN4 are required for the activation of the ChlH subunit of magnesium chelatase in chlorophyll synthesis. FEBS Lett. 2012, 586, 205–210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kong, W.Y.; Yu, X.W.; Chen, H.Y.; Liu, L.L.; Xiao, Y.J.; Wang, Y.L.; Wang, C.L.; Lin, Y.; Yu, Y.; Wang, C.M.; et al. The catalytic subunit of magnesium-protoporphyrin IX monomethyl ester cyclase forms a chloroplast complex to regulate chlorophyll biosynthesis in rice. Plant Mol. Biol. 2016, 92, 177–191. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.J.; Teng, L.H.; Wang, X.M.; Wang, Y.C.; Shen, S.H. De Novo assembly of expressed transcripts and global transcriptomic analysis from seedlings of the paper mulberry (Broussonetia kazinoki × Broussonetia papyrifera). PLoS ONE 2014, 9, e97487. [Google Scholar]
- Pi, Z.; Zhao, M.L.; Peng, X.J.; Shen, S.H. Phosphoproteomic analysis of paper mulberry reveals phosphorylation functions in chilling tolerance. J. Proteome. Res. 2017, 16, 1944–1961. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.F.; Su, Y.L.; Chen, N.Z.; Shen, S.H. Genome-wide analysis of the UGT gene family and identification of flavonoids in Broussonetia papyrifera. Molecules 2021, 26, 3449. [Google Scholar] [CrossRef]
- Si, B.; Hui, T.; Zhang, X.; Guo, J.; Diao, Q.Y. Effect of Broussonetia papyrifera L. (paper mulberry) silage on dry matter intake, milk composition, antioxidant capacity and milk fatty acid profile in dairy cows. Asian Austral. J. Anim. 2018, 31, 1259–1266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tomanek, L. Environmental proteomics: Changes in the proteome of marine organisms in response to environmental stress, pollutants, infection, symbiosis, and development. Ann. Rev. Mar. Sci. 2011, 3, 373–399. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Yu, W.; Wang, G.; Cao, F.; Cai, J.; Wang, H. Comparative proteomic and physiological analysis reveals the variation mechanisms of leaf coloration and carbon fixation in a xantha mutant of Ginkgo biloba L. Int. J. Mol. Sci. 2016, 17, 1794. [Google Scholar] [CrossRef] [Green Version]
- Dong, F.; Shi, Y.; Liu, M.; Fan, K.; Zhang, Q.; Ruan, J. iTRAQ-based quantitative proteomics analysis reveals the mechanism underlying the weakening of carbon metabolism in chlorotic tea leaves. Int. J. Mol. Sci. 2018, 19, 3943. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Cao, H.; Chen, C.; Yue, C.; Hao, X.; Yang, Y.; Wang, X. Complementary transcriptomic and proteomic analyses of a chlorophyll-deficient tea plant cultivar reveal multiple metabolic pathway changes. J. Proteom. 2016, 130, 160–169. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, R.; Kobayashi, K.; Masuda, T. Tetrapyrrole metabolism in Arabidopsis thaliana. Arab. Book 2011, 9, e0145. [Google Scholar] [CrossRef] [Green Version]
- Chu, P.; Yan, G.X.; Yang, Q.; Zhai, L.N.; Zhang, C.; Zhang, F.Q.; Guan, R.Z. iTRAQ-based quantitative proteomics analysis of Brassica napus leaves reveals pathways associated with chlorophyll deficiency. J. Proteom. 2015, 113, 244–259. [Google Scholar] [CrossRef]
- Adhikari, N.D.; Froehlich, J.E.; Strand, D.D.; Buck, S.M.; Larkin, R.M. GUN4-Porphyrin Complexes Bind the ChlH/GUN5 Subunit of Mg-Chelatase and Promote Chlorophyll Biosynthesis in Arabidopsis. Plant Cell 2011, 23, 1449–1467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cazzonelli, C.I.; Pogson, B.J. Source to sink: Regulation of carotenoid biosynthesis in plants. Trends Plant Sci. 2010, 15, 266–274. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Chen, X.; Xu, B.; Li, Y.; Ma, Y.; Wang, G. Phenotype and transcriptome analysis reveals chloroplast development and pigment biosynthesis together influenced the leaf color formation in mutants of Anthurium andraeanum ‘Sonate’. Front. Plant Sci. 2015, 6, 139. [Google Scholar] [CrossRef] [Green Version]
- Fromme, P.; Melkozernov, A.; Jordan, P.; Krauss, N. Structure and function of photosystem I: Interaction with its soluble electron carriers and external antenna systems. FEBS Lett. 2003, 555, 40–44. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, Z.; Wang, P.; Wang, S.; Ma, L.; Li, L.; Yang, R.; Ma, Y.; Wang, Q. Comprehensive transcriptome analysis discovers novel candidate genes related to leaf color in a Lagerstroemia indica yellow leaf mutant. Genes Genom. 2015, 37, 851–863. [Google Scholar] [CrossRef]
- Li, W.X.; Yang, S.B.; Lu, Z.G.; He, Z.C.; Jin, B. Cytological, physiological, and transcriptomic analyses of golden leaf coloration in Ginkgo biloba L. Hortic. Res. 2018, 5, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Wen, L.; Sha, T.; Zhang, S.; Shan, J.; Diao, X. Gene mapping and functional analysis of the novel leaf color gene SiYGL1 in foxtail millet [Setaria italica (L.) P. Beauv]. Physiol. Plant. 2016, 157, 24–37. [Google Scholar]
- Wu, Z.M.; Zhang, X.; Wang, J.L.; Wan, J.M. Leaf chloroplast ultrastructure and photosynthetic properties of a chlorophyll-deficient mutant of rice. Photosynthetica 2014, 52, 217–222. [Google Scholar] [CrossRef]
- Yoo, S.C.; Cho, S.H.; Sugimoto, H.; Li, J.; Kusumi, K.; Koh, H.-J.; Koh, I.; Chon, P.N. Rice virescent3 and stripe1 encoding the large and small subunits of ribonucleotide reductase are required for chloroplast biogenesis during early leaf development. Plant Physiol. 2009, 150, 388–401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, Y.; Lim, T.K.; Lin, Q.; Li, S. Identification of cypermethrin induced protein changes in green algae by iTRAQ quantitative proteomics. J. Proteom. 2016, 139, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Kumari, M.; Thakur, S.; Kumar, A.; Joshi, R.; Kumar, R. Regulation of color transition in purple tea (Camellia sinensis). Planta 2019, 251, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, X.; Nan, P.; Li, J.; Jin, L. De novo transcriptome sequencing of genome analysis provides insights into Solidago canadensis invasive capability via photosynthesis. J. Plant Interact. 2019, 14, 572–579. [Google Scholar] [CrossRef] [Green Version]
- Rantala, M.; Lehtimäki, N.; Aro, E.; Suorsa, M. Downregulation of TAP38/PPH1 enables LHCII hyperphosphorylation in Arabidopsis mutant lacking LHCII docking site in PSI. FEBS Lett. 2016, 590, 787–794. [Google Scholar] [CrossRef] [Green Version]
- Tiller, N.; Bock, R. The translational apparatus of plastids and its role in plant development. Plant Mol. Biol. 2014, 7, 1105–1120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Min, O.; Li, Q.; Zou, M.; Guo, J.; Ma, J.; Lu, C.; Zhang, L. The Arabidopsis chloroplast ribosome recycling factor is essential for embryogenesis and chloroplast biogenesis. Plant Mol. Biol. 2010, 74, 47–59. [Google Scholar] [CrossRef]
- Bionda, T.; Gross, L.E.; Becker, T.; Papasotiriou, D.G. Eukaryotic Hsp70 chaperones in the intermembrane space of chloroplasts. Planta 2016, 243, 733–744. [Google Scholar] [CrossRef] [PubMed]
- Arnon, D.I. Copper enzymes in isolated chloroplasts. polyphenoloxidase in beta vulgaris. Plant Physiol. 1949, 24, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dei, M. Benzyladenine-induced stimulation of 5-aminolevulinic acid accumulation under various light intensities in levulinic acid-treated cotyledons of etiolated cucumber. Physiol. Plant 1985, 64, 153–160. [Google Scholar] [CrossRef]
- Bogorad, L. Porphyrin synthesis. Method. Enzymol. 1962, 5, 885–895. [Google Scholar]
- Rebeiz, C.A.; Smith, B.B.; Mattheis, J.R.; Rebeiz, C.C.; Dayton, D.F. Chloroplast biogenesis: Biosynthesis and accumulation of protochlorophyll by isolated etioplasts and developing chloroplasts. Arch. Biochem. Biophys. 1975, 167, 351–365. [Google Scholar] [CrossRef]
- Jumaah, F.; Plaza, M.; Abrahamsson, V.; Turner, C.; Sandahl, M. A fast and sensitive method for the separation of carotenoids using ultra-high performance supercritical fluid chromatography-mass spectrometry. Anal. Bioanal. Chem. 2016, 408, 5883–5894. [Google Scholar] [CrossRef]
- Wang, L.J.; Su, S.; Wu, J.; Du, H.; Li, S.S.; Huo, J.W.; Zhang, Y.; Wang, L.S. Variation of anthocyanins and flavonols in Vaccinium uliginosum berry in Lesser Khingan Mountains and its antioxidant activity. Food Chem. 2014, 160, 357–364. [Google Scholar] [CrossRef]
- Xu, S.; Chen, W.; Huang, Y.; He, X. Responses of growth, photosynthesis and VOC emissions of Pinus tabulaeformis Carr. exposure to elevated CO2 and/or elevated O3 in an Urban Area. Bull. Environ. Contam. Toxicol. 2012, 88, 443–448. [Google Scholar] [CrossRef]
| Materials | Net Photosynthetic Rate (μmol·m−2·s−1) | Stomatal Conductance (mmol·m−2·s−1) | Intercellular CO2 Concentration (μmol·mol−1) | Transpiration Rate (mol·m−2·s−1) |
|---|---|---|---|---|
| Green leaves | 23.55 ± 0.37 | 367.83 ± 2.28 | 245.5 ± 2.67 | 6.91 ± 0.79 |
| Mutant leaves | 14.81 ± 0.08 * | 303.33 ± 1.31 | 315.1 ± 1.84 * | 7.45 ± 0.14 |
| Materials | Fo | Fm | Fv | Fv/Fm |
|---|---|---|---|---|
| Green leaves | 517.83 ± 13.5 | 2594.83 ± 26.5 | 2077.00 ± 25.9 | 0.80 ± 0.02 |
| Mutant leaves | 464.63 ± 6.43 | 1243.00 ± 9.44 * | 778.38 ± 8.14 ** | 0.58 ± 0.04 * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, F.; Chen, N.; Shen, S. iTRAQ-Based Quantitative Proteomics Analysis Reveals the Mechanism of Golden-Yellow Leaf Mutant in Hybrid Paper Mulberry. Int. J. Mol. Sci. 2022, 23, 127. https://doi.org/10.3390/ijms23010127
Wang F, Chen N, Shen S. iTRAQ-Based Quantitative Proteomics Analysis Reveals the Mechanism of Golden-Yellow Leaf Mutant in Hybrid Paper Mulberry. International Journal of Molecular Sciences. 2022; 23(1):127. https://doi.org/10.3390/ijms23010127
Chicago/Turabian StyleWang, Fenfen, Naizhi Chen, and Shihua Shen. 2022. "iTRAQ-Based Quantitative Proteomics Analysis Reveals the Mechanism of Golden-Yellow Leaf Mutant in Hybrid Paper Mulberry" International Journal of Molecular Sciences 23, no. 1: 127. https://doi.org/10.3390/ijms23010127
APA StyleWang, F., Chen, N., & Shen, S. (2022). iTRAQ-Based Quantitative Proteomics Analysis Reveals the Mechanism of Golden-Yellow Leaf Mutant in Hybrid Paper Mulberry. International Journal of Molecular Sciences, 23(1), 127. https://doi.org/10.3390/ijms23010127





