Integrating Transcriptome and Coexpression Network Analyses to Characterize Salicylic Acid- and Jasmonic Acid-Related Genes in Tolerant Poplars Infected with Rust
Abstract
1. Introduction
2. Results
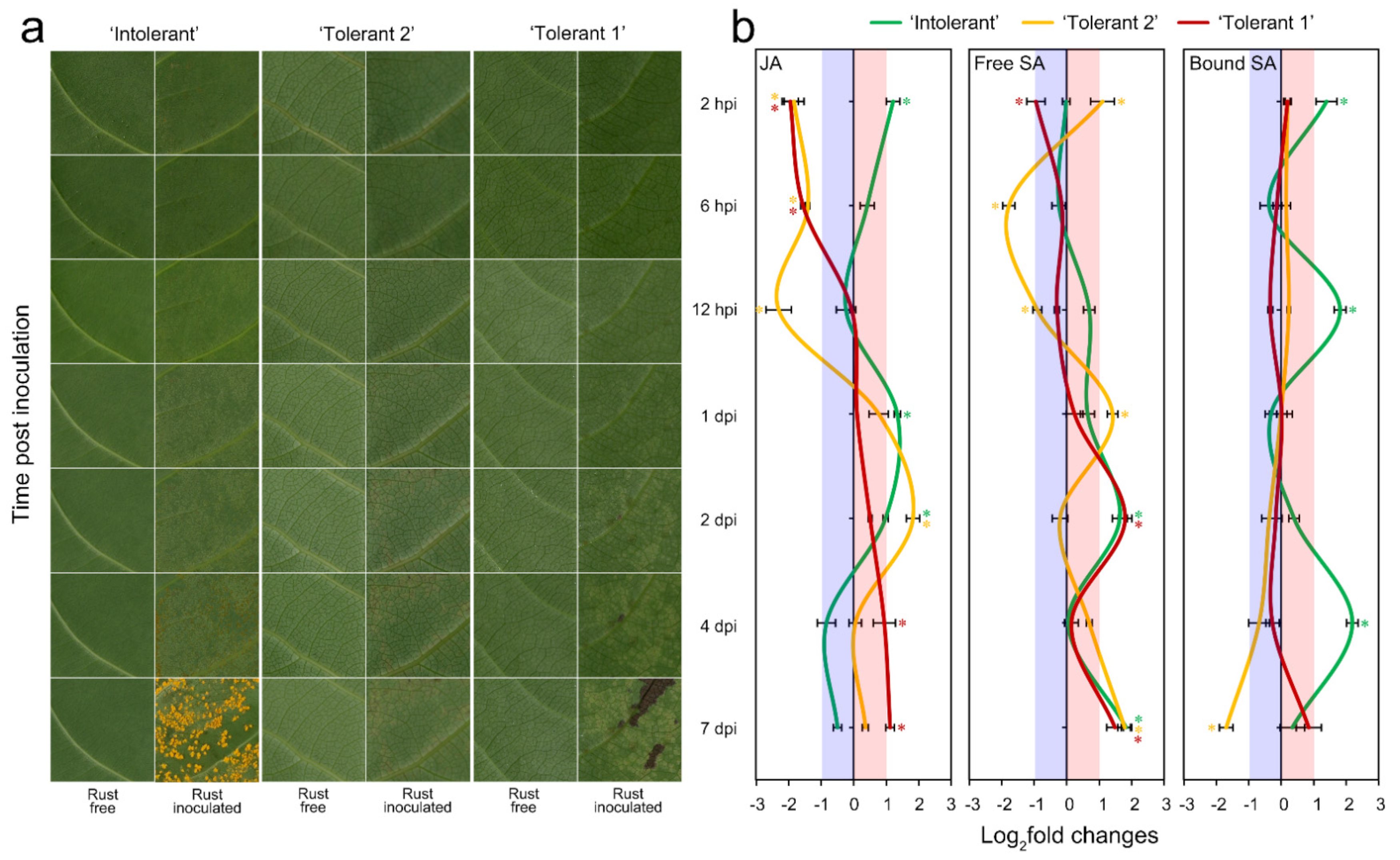
2.1. Disease Assessment
2.2. JA and SA Content Determination
2.3. Digital Gene Expression Sequencing
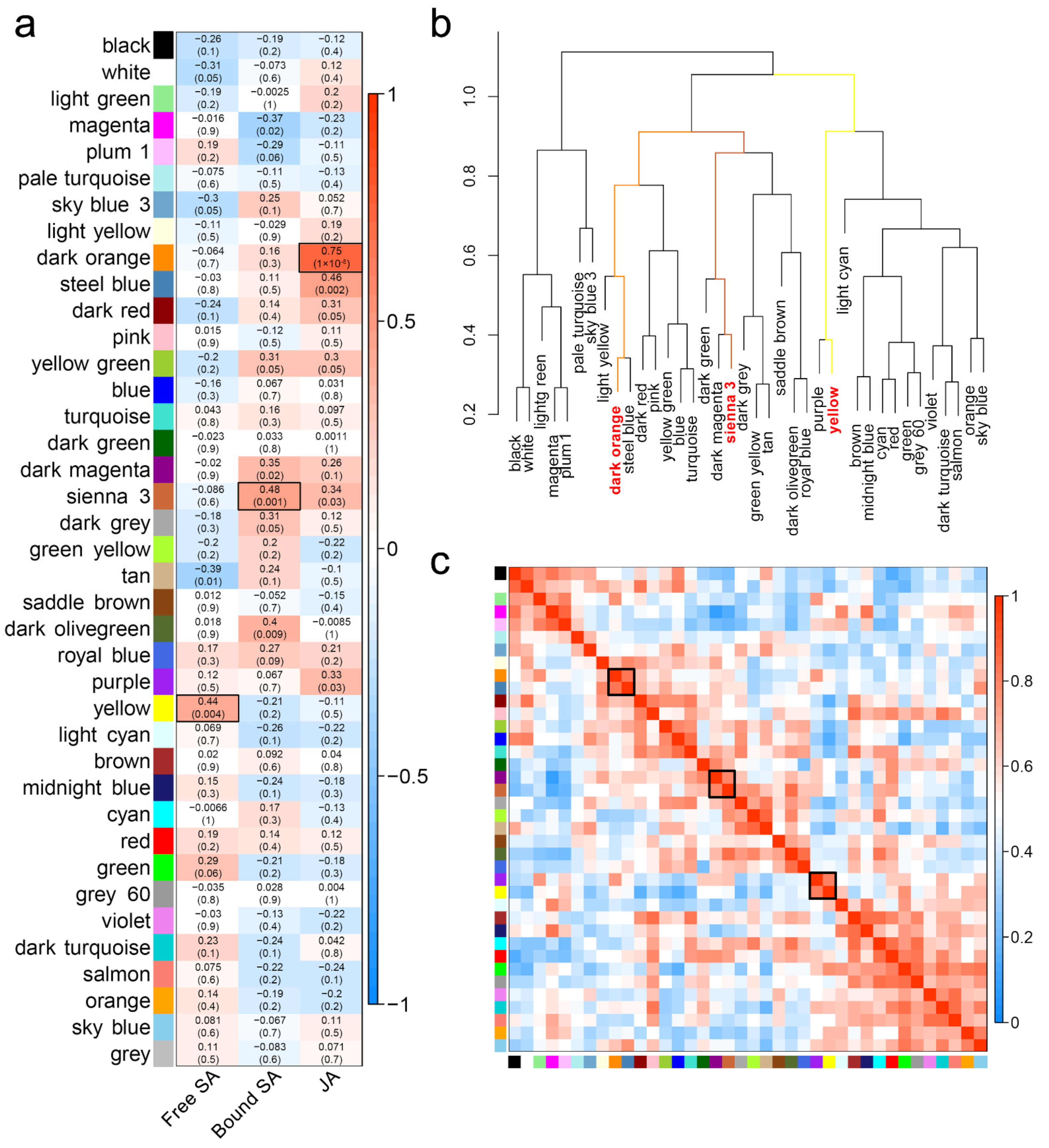
2.4. WGCNA Revealed Modules Highly Related to JA and SA
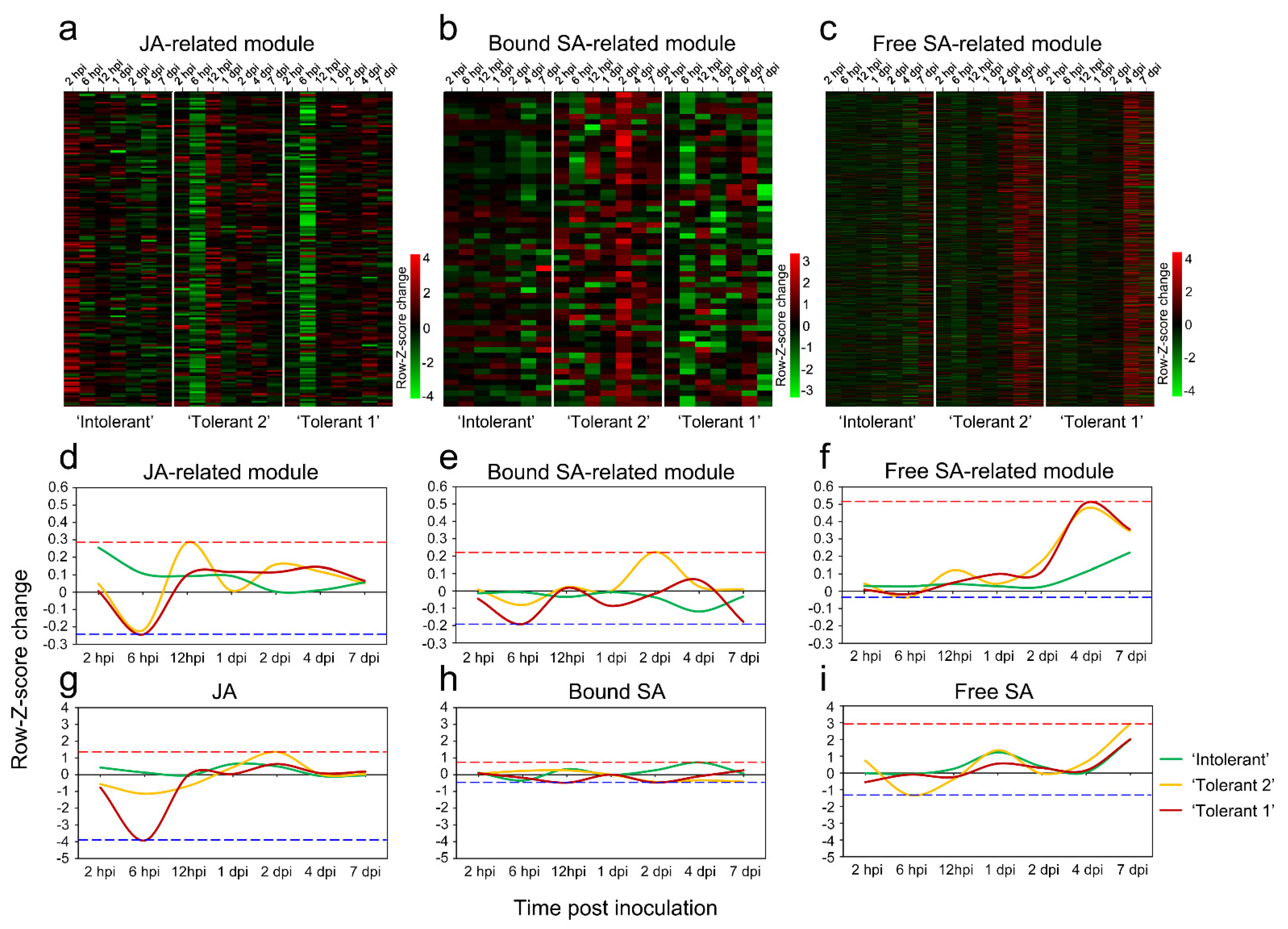
2.5. Eigengene Expression of JA- and SA-Related Modules
2.6. Expression Analysis of Plant-Pathogen Interaction-Related and Signalling-Related Genes in JA- and SA-Related Modules
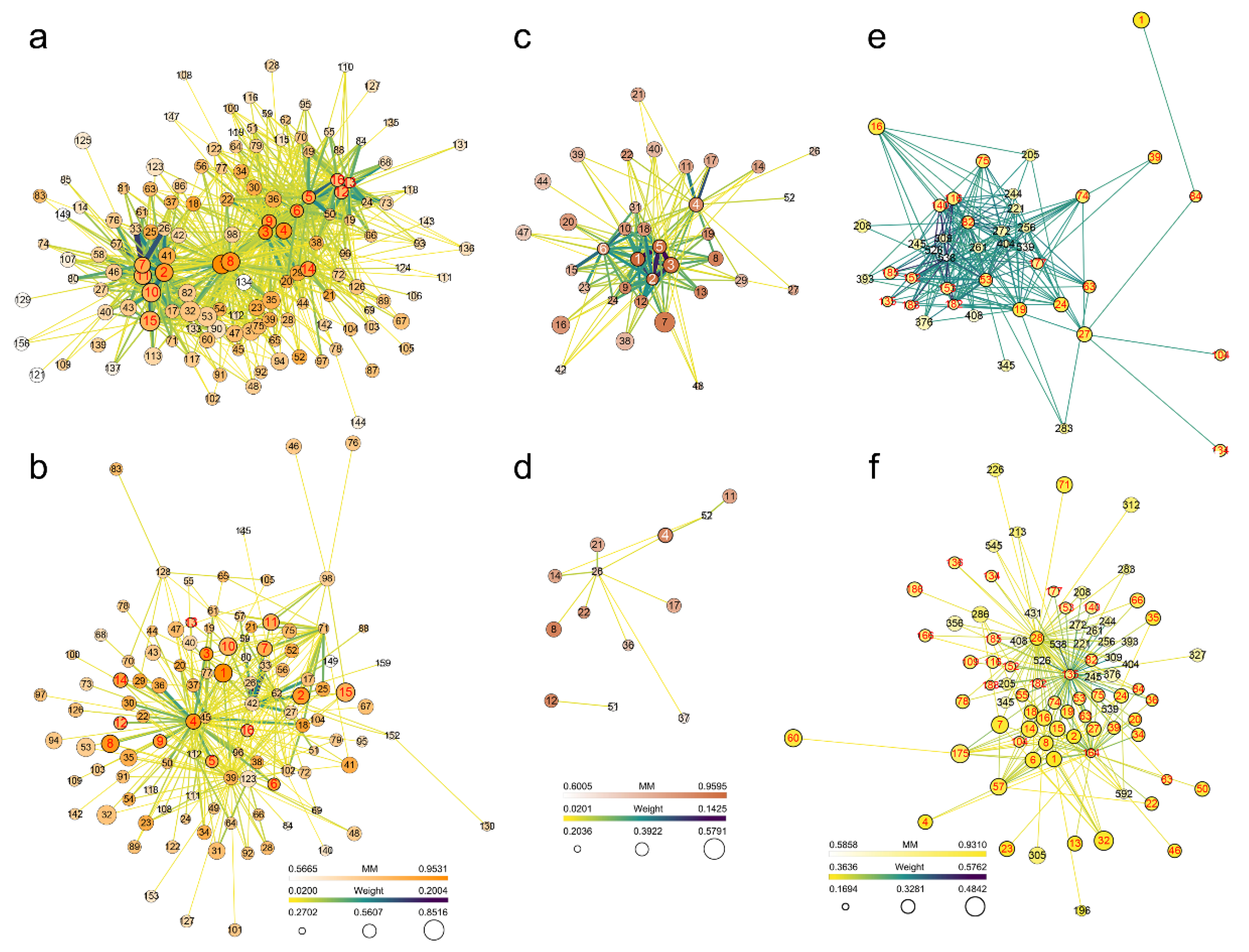
2.7. Gene Network of JA- and SA-Related Modules
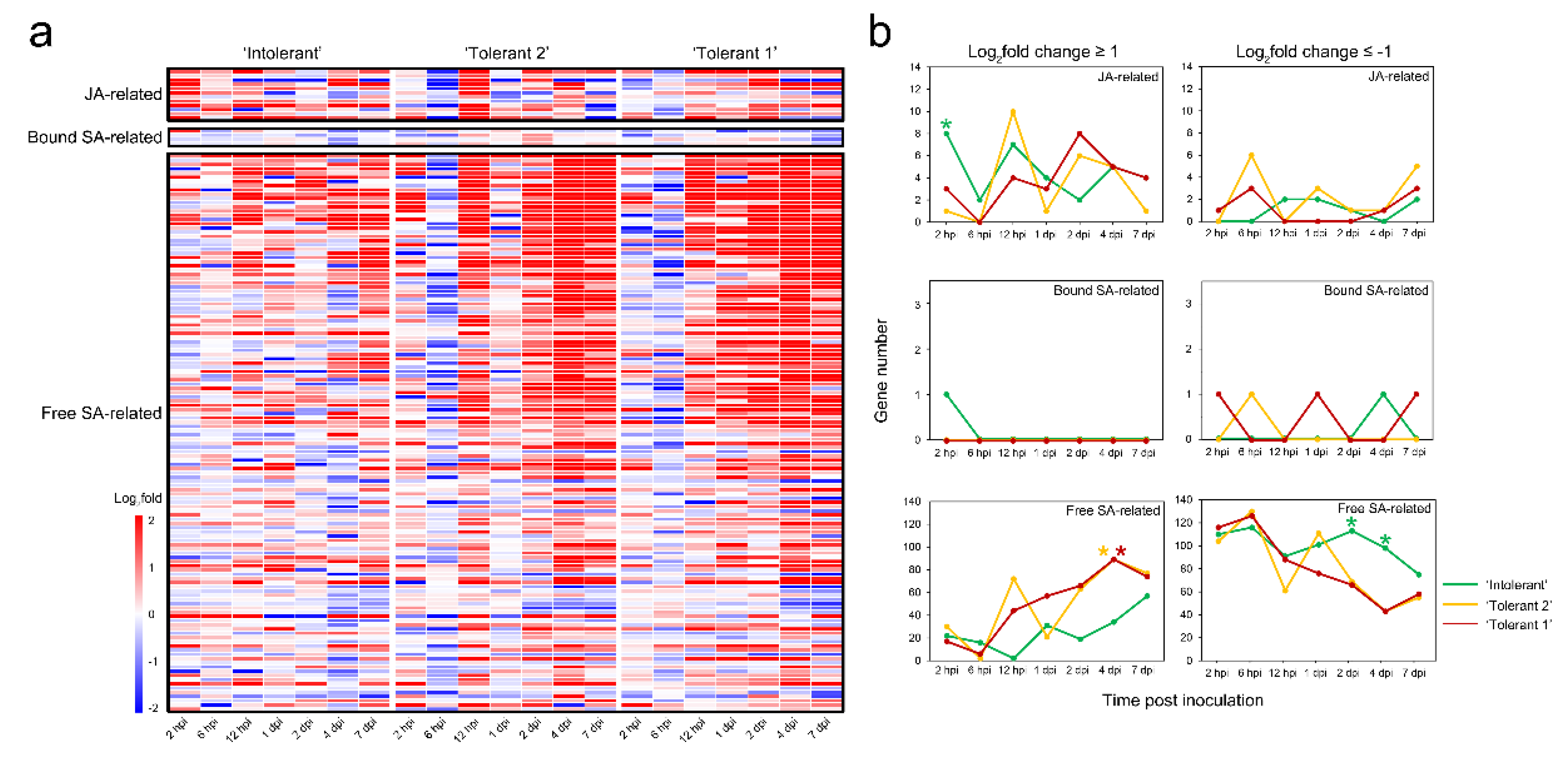
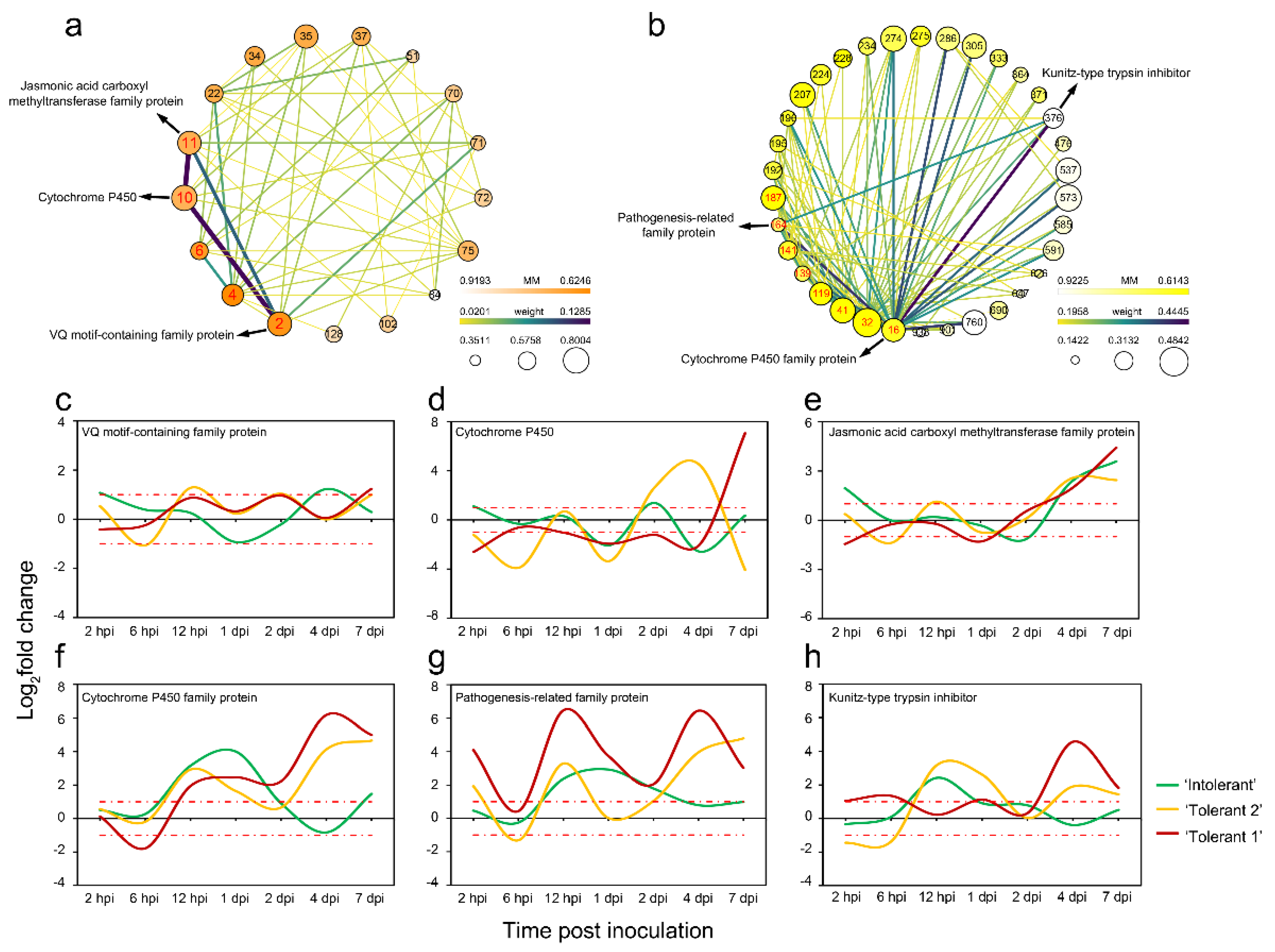
2.8. Gene Screening Based on Expression Differences
2.9. Identification of Genes Related to JA and SA Biosynthesis and Defence-Related Genes Downstream of JA and SA
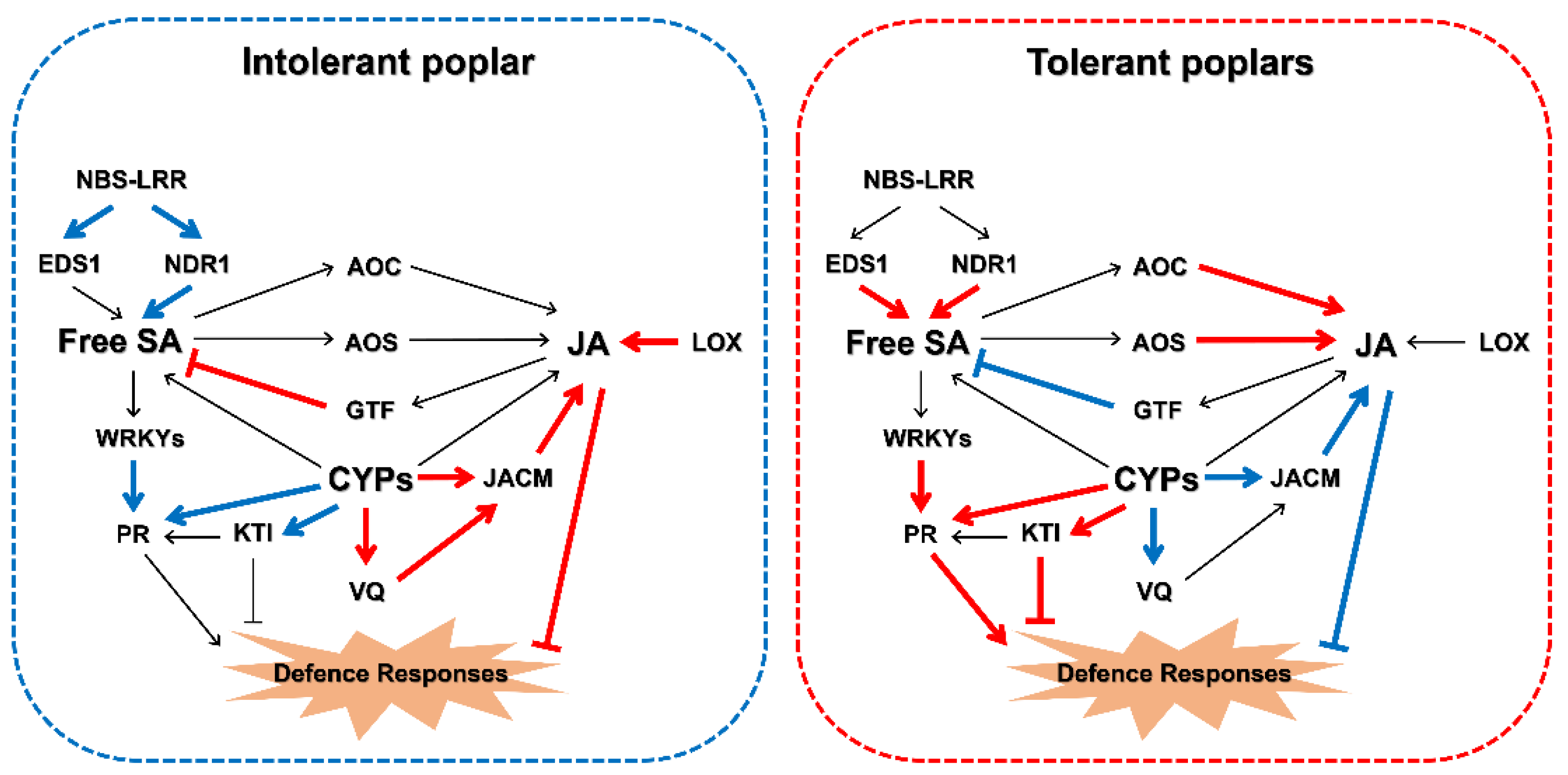
3. Discussion
4. Materials and Methods
4.1. E4 Isolates and Plant Materials
4.2. Inoculation Procedure and Total RNA Preparation
4.3. Total RNA Preparation and DGE Library Preparation and Sequencing
4.4. Quantification of SA and JA Levels
4.5. WGCNA
4.6. RT-qPCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Pei, M.; Ruiz, C.; Bayon, C.; Hunter, T.; Lonsdale, D. Pathogenic variation in poplar rust Melampsora larici-populina from England. Eur. J. Plant Pathol. 2005, 111, 147–155. [Google Scholar] [CrossRef]
- Steenackers, M. Breeding poplars for rust resistance-recent advances. Commun. Fac. Agric. Sci. State Univ. Ghent Belg. 1988, 53, 417–422. [Google Scholar]
- Steenackers, M. The phytosanity state of poplars in Belgium in 1991. Parasitica 1991, 47, 129–136. [Google Scholar]
- Steenackers, V. Nouvelle race physiologique de Melampsora larici-populina en Belgique (communication provisoire). In 22e Réunion de Travail des Maladies (6pp); FAO/CIP: Casal Monferrato, Italy, 1982; pp. 6–10. [Google Scholar]
- Pinon, J.; van Dam, B.C.; Genetet, I.; De Kam, M. Two pathogenic races of Melampsora larici-populina in northwestern Europe. Eur. J. For. Pathol. 1987, 17, 47–53. [Google Scholar] [CrossRef]
- Pinon, J.; Peulon, V. Mise en évidence d’une troisième race physiolosique de Melampsora larici-populina Kleb. En Europe. Cryptogam. Mycol. 1989, 10, 95–106. [Google Scholar]
- Steenackers, M.; Steenackers, V.; Delporte, T. A New Race of M. Larici-Populina in Belgium; International Poplar Commission: Izmit, Turkey, 1994. [Google Scholar]
- Chisholm, S.T.; Coaker, G.; Day, B.; Staskawicz, B.J. Host-microbe interactions: Shaping the evolution of the plant immune response. Cell 2006, 124, 803–814. [Google Scholar] [CrossRef]
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef]
- Somssich, I.E.; Hahlbrock, K. Pathogen defence in plants—A paradigm of biological complexity. Trends Plant Sci. 1998, 3, 86–90. [Google Scholar] [CrossRef]
- Dangl, J.L.; Jones, J.D. Plant pathogens and integrated defence responses to infection. Nature 2001, 411, 826–833. [Google Scholar] [CrossRef]
- Sanabria, N.M.; Huang, J.C.; Dubery, I.A. Self/nonself perception in plants in innate immunity and defense. Self Nonself 2010, 1, 40–54. [Google Scholar] [CrossRef]
- Boller, T.; He, S.Y. Innate immunity in plants: An arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science 2009, 324, 742–744. [Google Scholar] [CrossRef] [PubMed]
- Berens, M.L.; Berry, H.M.; Mine, A.; Argueso, C.T.; Tsuda, K. Evolution of hormone signaling networks in plant defense. Annu. Rev. Phytopathol. 2017, 55, 401–425. [Google Scholar] [CrossRef] [PubMed]
- Koornneef, A.; Pieterse, C.M. Cross talk in defense signaling. Plant Physiol. 2008, 146, 839–844. [Google Scholar] [CrossRef]
- Vlot, A.C.; Dempsey, D.A.; Klessig, D.F. Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 2009, 47, 177–206. [Google Scholar] [CrossRef] [PubMed]
- Kazan, K.; Manners, J.M. Linking development to defense: Auxin in plant–pathogen interactions. Trends Plant Sci. 2009, 14, 373–382. [Google Scholar] [CrossRef] [PubMed]
- Asselbergh, B.; De Vleesschauwer, D.; Höfte, M. Global switches and fine-tuning-ABA modulates plant pathogen defense. Mol. Plant Microbe Interact. 2008, 21, 709–719. [Google Scholar] [CrossRef]
- Denancé, N.; Sánchez-Vallet, A.; Goffner, D.; Molina, A. Disease resistance or growth: The role of plant hormones in balancing immune responses and fitness costs. Front. Plant Sci. 2013, 4, 155. [Google Scholar] [CrossRef]
- Bari, R.; Jones, J.D.G. Role of plant hormones in plant defence responses. Plant Mol. Biol. 2009, 69, 473–488. [Google Scholar] [CrossRef]
- Kazan, K.; Manners, J.M. Jasmonate signaling: Toward an integrated view. Plant Physiol. 2008, 146, 1459–1468. [Google Scholar] [CrossRef]
- Shah, J. The salicylic acid loop in plant defense. Curr. Opin. Plant Biol. 2003, 6, 365–371. [Google Scholar] [CrossRef]
- Grant, M.; Lamb, C. Systemic immunity. Curr. Opin. Plant Biol. 2006, 9, 414–420. [Google Scholar] [CrossRef] [PubMed]
- Vijayan, P.; Shockey, J.; Lévesque, C.A.; Cook, R.J.; Browse, J. A role for jasmonate in pathogen defense of Arabidopsis. Proc. Natl. Acad. Sci. USA 1998, 95, 7209–7214. [Google Scholar] [CrossRef] [PubMed]
- White, R.F. Acetylsalicylic acid (aspirin) induces resistance to tobacco mosaic virus in tobacco. Virology 1979, 99, 410–412. [Google Scholar] [CrossRef]
- Antoniw, J.F.; White, R.F. The effects of aspirin and polyacrylic acid on soluble leaf proteins and resistance to virus infection in five cultivars of tobacco. J. Phytopathol. 1980, 98, 331–341. [Google Scholar] [CrossRef]
- Raskin, I. Role of salicylic acid in plants. Annu. Rev. Plant Biol. 1992, 43, 439–463. [Google Scholar] [CrossRef]
- Klessig, D.F.; Malamy, J. The salicylic acid signal in plants. Plant Mol. Biol. 1994, 26, 1439–1458. [Google Scholar] [CrossRef]
- Vlot, A.C.; Klessig, D.F.; Park, S.W. Systemic acquired resistance: The elusive signal(s). Curr. Opin. Plant Biol. 2008, 11, 436–442. [Google Scholar] [CrossRef]
- Enyedi, A.J.; Yalpani, N.; Silverman, P.; Raskin, I. Localization, conjugation, and function of salicylic acid in tobacco during the hypersensitive reaction to tobacco mosaic virus. Proc. Natl. Acad. Sci. USA 1992, 89, 2480–2484. [Google Scholar] [CrossRef]
- Lee, H.I.; Raskin, I. Glucosylation of salicylic acid in Nicotiana tabacum Cv. Xanthi-nc. Phytopathology 1998, 88, 692–697. [Google Scholar] [CrossRef]
- Lee, H.I.; Raskin, I. Purification, cloning, and expression of a pathogen inducible UDP-glucose: Salicylic acid glucosyltransferase from tobacco. J. Biol. Chem. 1999, 274, 36637–36642. [Google Scholar] [CrossRef]
- Shulaev, V.; Silverman, P.; Raskin, I. Airborne signaling by methyl salicylate in plant pathogen resistance. Nature 1997, 385, 718–721. [Google Scholar] [CrossRef]
- Seskar, M.; Shulaev, V.; Raskin, I. Endogenous methyl salicylate in pathogen-inoculated tobacco plants. Plant Physiol. 1998, 116, 387–392. [Google Scholar] [CrossRef]
- Silverman, P.; Seskar, M.; Kanter, D.; Schweizer, P.; Metraux, J.P.; Raskin, I. Salicylic acid in rice (biosynthesis, conjugation, and possible role). Plant Physiol. 1995, 108, 633–639. [Google Scholar] [CrossRef]
- Dean, J.V.; Mohammed, L.A.; Fitzpatrick, T. The formation, vacuolar localization, and tonoplast transport of salicylic acid glucose conjugates in tobacco cell suspension cultures. Planta 2005, 221, 287–296. [Google Scholar] [CrossRef]
- Dempsey, D.A.; Vlot, A.C.; Wildermuth, M.C.; Klessig, D.F. Salicylic acid biosynthesis and metabolism. Arab. Book 2011, 9, e0156. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, X. Salicylic acid: Biosynthesis, perception, and contributions to plant immunity. Curr. Opin. Plant Biol. 2019, 50, 29–36. [Google Scholar] [CrossRef]
- Maruri-López, I.; Aviles-Baltazar, N.Y.; Buchala, A.; Serrano, M. Intra and extracellular journey of the phytohormone salicylic acid. Front. Plant Sci. 2019, 10, 423. [Google Scholar] [CrossRef]
- Pieterse, C.M.; Van der Does, D.; Zamioudis, C.; Leon-Reyes, A.; Van Wees, S.C. Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 2012, 28, 489–521. [Google Scholar] [CrossRef]
- Dong, X. SA, JA, ethylene, and disease resistance in plants. Curr. Opin. Plant Biol. 1998, 1, 316–323. [Google Scholar] [CrossRef]
- Vos, I.A.; Pieterse, C.M.J.; Wees, S.C.M.V. Costs and benefits of hormone-regulated plant defences. Plant Pathol. 2013, 62, 43–55. [Google Scholar] [CrossRef]
- Vos, I.A.; Moritz, L.; Pieterse, C.M.; Van Wees, S.C. Impact of hormonal crosstalk on plant resistance and fitness under multi-attacker conditions. Front. Plant Sci. 2015, 6, 639. [Google Scholar] [CrossRef]
- Glazebrook, J.; Chen, W.; Estes, B.; Chang, H.S.; Nawrath, C.; Métraux, J.P.; Zhu, T.; Katagiri, F. Topology of the network integrating salicylate and jasmonate signal transduction derived from global expression phenotyping. Plant J. 2003, 34, 217–228. [Google Scholar] [CrossRef]
- van Verk, M.C.; Bol, J.F.; Linthorst, H.J. Prospecting for genes involved in transcriptional regulation of plant defenses, a bioinformatics approach. BMC Plant Biol. 2011, 11, 88. [Google Scholar] [CrossRef]
- van Wees, S.C.; Luijendijk, M.; Smoorenburg, I.; van Loon, L.C.; Pieterse, C.M. Rhizobacteria-mediated induced systemic resistance (ISR) in Arabidopsis is not associated with a direct effect on expression of known defense-related genes but stimulates the expression of the jasmonate-inducible gene Atvsp upon challenge. Plant Mol. Biol. 1999, 41, 537–549. [Google Scholar] [CrossRef] [PubMed]
- Van der Does, D.; Leon-Reyes, A.; Koornneef, A.; Van Verk, M.C.; Rodenburg, N.; Pauwels, L.; Goossens, A.; Körbes, A.P.; Memelink, J.; Ritsema, T.; et al. Salicylic acid suppresses jasmonic acid signaling downstream of SCFCOI1-JAZ by targeting GCC promoter motifs via transcription factor ORA59. Plant Cell 2013, 25, 744–761. [Google Scholar] [CrossRef] [PubMed]
- Robert-Seilaniantz, A.; Grant, M.; Jones, J.D. Hormone crosstalk in plant disease and defense: More than just jasmonate-salicylate antagonism. Annu. Rev. Phytopathol. 2011, 49, 317–343. [Google Scholar] [CrossRef]
- Broekgaarden, C.; Caarls, L.; Vos, I.A.; Pieterse, C.M.; Van Wees, S.C. Ethylene: Traffic controller on hormonal crossroads to defense. Plant Physiol. 2015, 169, 2371–2379. [Google Scholar] [CrossRef]
- Kunkel, B.N.; Brooks, D.M. Cross talk between signaling pathways in pathogen defense. Curr. Opin. Plant Biol. 2002, 5, 325–331. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 599. [Google Scholar] [CrossRef] [PubMed]
- Hacquard, S.; Petre, B.; Frey, P.; Hecker, A.; Rouhier, N.; Duplessis, S. The poplar-poplar rust interaction: Insights from genomics and transcriptomics. J. Pathog. 2011, 2011, 716041. [Google Scholar] [CrossRef] [PubMed]
- Duplessis, S.; Hacquard, S.; Delaruelle, C.; Tisserant, E.; Frey, P.; Martin, F.; Kohler, A. Melampsora larici-populina transcript profiling during germination and timecourse infection of poplar leaves reveals dynamic expression patterns associated with virulence and biotrophy. Mol. Plant Microbe Interact. 2011, 24, 808–818. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Wang, J.; Li, D.; Wang, Z.; Wang, F.; Zhang, R. Molecular characterization of the transcription factors in susceptible poplar infected with virulent Melampsora larici-populina. Int. J. Mol. Sci. 2019, 20, 4806. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Wang, Z.; Li, D.; Wang, F.; Zhang, R.; Wang, J. Molecular characterization of the ERF family in susceptible poplar infected by virulent Melampsora larici-populina. Physiol. Mol. Plant Pathol. 2019, 108, 101437. [Google Scholar] [CrossRef]
- Baker, B.; Zambryski, P.; Staskawicz, B.; Dinesh-Kumar, S.P. Signaling in plant-microbe interactions. Science 1997, 276, 726–733. [Google Scholar] [CrossRef] [PubMed]
- Aarts, N.; Metz, M.; Holub, E.; Staskawicz, B.J.; Daniels, M.J.; Parker, J.E. Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis. Proc. Natl. Acad. Sci. USA 1998, 95, 10306–10311. [Google Scholar] [CrossRef]
- Falk, A.; Feys, B.J.; Frost, L.N.; Jones, J.D.; Daniels, M.J.; Parker, J.E. EDS1, an essential component of R gene-mediated disease resistance in Arabidopsis has homology to eukaryotic lipases. Proc. Natl. Acad. Sci. USA 1999, 96, 3292–3297. [Google Scholar] [CrossRef] [PubMed]
- Feys, B.J.; Moisan, L.J.; Newman, M.A.; Parker, J.E. Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO J. 2001, 20, 5400–5411. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, A.D.; Zhang, C. The role of NDR1 in avirulence gene-directed signaling and control of programmed cell death in Arabidopsis. Plant Physiol. 2001, 127, 1089–1101. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.; Boden, E.; Arias, J. Salicylic acid and NPR1 induce the recruitment of trans-activating TGA factors to a defense gene promoter in Arabidopsis. Plant Cell 2003, 15, 1846–1858. [Google Scholar] [CrossRef]
- Tada, Y.; Spoel, S.H.; Pajerowska-Mukhtar, K.; Mou, Z.; Song, J.; Wang, C.; Zuo, J.; Dong, X. Plant immunity requires conformational changes of NPR1 via S-nitrosylation and thioredoxins. Science 2008, 321, 952–956. [Google Scholar] [CrossRef]
- Lu, H.; Zhang, C.; Albrecht, U.; Shimizu, R.; Wang, G.; Bowman, K.D. Overexpression of a citrus NDR1 ortholog increases disease resistance in Arabidopsis. Front. Plant Sci. 2013, 4, 157. [Google Scholar] [CrossRef] [PubMed]
- Chern, M.S.; Fitzgerald, H.A.; Yadav, R.C.; Canlas, P.E.; Dong, X.; Ronald, P.C. Evidence for a disease-resistance pathway in rice similar to the NPR1-mediated signaling pathway in Arabidopsis. Plant J. 2001, 27, 101–113. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, H.A.; Chern, M.S.; Navarre, R.; Ronald, P.C. Overexpression of (At)NPR1 in rice leads to a BTH- and environment-induced lesion-mimic/cell death phenotype. Mol. Plant Microbe Interact. 2004, 17, 140–151. [Google Scholar] [CrossRef]
- Sandhu, D.; Tasma, I.M.; Frasch, R.; Bhattacharyya, M.K. Systemic acquired resistance in soybean is regulated by two proteins, orthologous to Arabidopsis NPR1. BMC Plant Biol. 2009, 9, 105. [Google Scholar] [CrossRef] [PubMed]
- Malnoy, M.; Jin, Q.; Borejsza-Wysocka, E.E.; He, S.Y.; Aldwinckle, H.S. Overexpression of the apple MpNPR1 gene confers increased disease resistance in Malus x domestica. Mol. Plant Microbe Interact. 2007, 20, 1568–1580. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.C.; Lu, C.F.; Wu, J.W.; Cheng, M.L.; Lin, Y.M.; Yang, N.S.; Black, L.; Green, S.K.; Wang, J.F.; Cheng, C.P. Transgenic tomato plants expressing the Arabidopsis NPR1 gene display enhanced resistance to a spectrum of fungal and bacterial diseases. Transgenic Res. 2004, 13, 567–581. [Google Scholar] [CrossRef]
- Gao, F.; Shu, X.; Ali, M.B.; Howard, S.; Li, N.; Winterhagen, P.; Qiu, W.; Gassmann, W. A functional EDS1 ortholog is differentially regulated in powdery mildew resistant and susceptible grapevines and complements an Arabidopsis eds1 mutant. Planta 2010, 231, 1037–1047. [Google Scholar] [CrossRef]
- Spoel, S.H.; Johnson, J.S.; Dong, X. Regulation of tradeoffs between plant defenses against pathogens with different lifestyles. Proc. Natl. Acad. Sci. USA 2007, 104, 18842–18847. [Google Scholar] [CrossRef]
- Li, J.; Brader, G.; Palva, E.T. The WRKY70 transcription factor: A node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 2004, 16, 319–331. [Google Scholar] [CrossRef]
- Derksen, H.; Rampitsch, C.; Daayf, F. Signaling cross-talk in plant disease resistance. Plant Sci. 2013, 207, 79–87. [Google Scholar] [CrossRef]
- Traw, M.B.; Kim, J.; Enright, S.; Cipollini, D.F.; Bergelson, J. Negative cross-talk between salicylate- and jasmonate-mediated pathways in the Wassilewskija ecotype of Arabidopsis thaliana. Mol. Ecol. 2003, 12, 1125–1135. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Wang, X.Y.; Guo, W.Z. The cytochrome P450 superfamily: Key players in plant development and defense. J. Integr. Agric. 2015, 14, 1673–1686. [Google Scholar] [CrossRef]
- Jing, Y.; Lin, R. The VQ motif-containing protein family of plant-specific transcriptional regulators. Plant Physiol. 2015, 169, 371–378. [Google Scholar] [CrossRef] [PubMed]
- Seo, H.S.; Song, J.T.; Cheong, J.J.; Lee, Y.H.; Lee, Y.W.; Hwang, I.; Lee, J.S.; Choi, Y.D. Jasmonic acid carboxyl methyltransferase: A key enzyme for jasmonate-regulated plant responses. Proc. Natl. Acad. Sci. USA 2001, 98, 4788–4793. [Google Scholar] [CrossRef] [PubMed]
- Van Loon, L.C.; Van Strien, E.A. The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol. Mol. Plant Pathol. 1999, 55, 85–97. [Google Scholar] [CrossRef]
- Penninckx, I.A.; Eggermont, K.; Terras, F.R.; Thomma, B.P.; De Samblanx, G.W.; Buchala, A.; Métraux, J.P.; Manners, J.M.; Broekaert, W.F. Pathogen-induced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid-independent pathway. Plant Cell 1996, 8, 2309–2323. [Google Scholar] [CrossRef]
- Thomma, B.P.H.J.; Eggermont, K.; Penninckx, I.A.M.A.; Mauch-Mani, B.; Vogelsang, R.; Cammue, B.P.A.; Broekaert, W.F. Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc. Natl. Acad. Sci. USA 1998, 95, 15107–15111. [Google Scholar] [CrossRef]
- Li, J.; Brader, G.; Palva, E.T. Kunitz trypsin inhibitor: An antagonist of cell death triggered by phytopathogens and fumonisin b1 in Arabidopsis. Mol. Plant 2008, 1, 482–495. [Google Scholar] [CrossRef]
- Li, D.; Wang, F.; Wang, C.; Zou, L.; Wang, Z.; Chen, Q.; Niu, C.; Zhang, R.; Ling, Y.; Wang, B. MicroRNA-mediated susceptible poplar gene expression regulation associated with the infection of virulent Melampsora larici-populina. BMC Genom. 2016, 17, 59. [Google Scholar] [CrossRef]
- Pei, M.; Ruiz, C.; Bayon, C.; Hunter, T. Rust resistance in Salix to Melampsora larici-epitea. Plant Pathol. 2004, 53, 770–779. [Google Scholar] [CrossRef]
- Pei, M.; Lindegaard, K.; Ruiz, C.; Bayon, C. Rust resistance of some varieties and recently bred genotypes of biomass willows. Biomass Bioenergy 2008, 32, 453–459. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Tuskan, G.A.; Difazio, S.; Jansson, S.; Bohlmann, J.; Grigoriev, I.; Hellsten, U.; Putnam, N.; Ralph, S.; Rombauts, S.; Salamov, A.; et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 2006, 313, 1596–1604. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Welti, R.; Wang, X. Quantitative analysis of major plant hormones in crude plant extracts by high-performance liquid chromatography-mass spectrometry. Nat. Protoc. 2010, 5, 986–992. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Q.; Zhang, R.; Li, D.; Wang, F. Integrating Transcriptome and Coexpression Network Analyses to Characterize Salicylic Acid- and Jasmonic Acid-Related Genes in Tolerant Poplars Infected with Rust. Int. J. Mol. Sci. 2021, 22, 5001. https://doi.org/10.3390/ijms22095001
Chen Q, Zhang R, Li D, Wang F. Integrating Transcriptome and Coexpression Network Analyses to Characterize Salicylic Acid- and Jasmonic Acid-Related Genes in Tolerant Poplars Infected with Rust. International Journal of Molecular Sciences. 2021; 22(9):5001. https://doi.org/10.3390/ijms22095001
Chicago/Turabian StyleChen, Qiaoli, Ruizhi Zhang, Danlei Li, and Feng Wang. 2021. "Integrating Transcriptome and Coexpression Network Analyses to Characterize Salicylic Acid- and Jasmonic Acid-Related Genes in Tolerant Poplars Infected with Rust" International Journal of Molecular Sciences 22, no. 9: 5001. https://doi.org/10.3390/ijms22095001
APA StyleChen, Q., Zhang, R., Li, D., & Wang, F. (2021). Integrating Transcriptome and Coexpression Network Analyses to Characterize Salicylic Acid- and Jasmonic Acid-Related Genes in Tolerant Poplars Infected with Rust. International Journal of Molecular Sciences, 22(9), 5001. https://doi.org/10.3390/ijms22095001





