Abstract
The BAG proteins are a family of multi-functional co-chaperones. In plants, BAG proteins were found to play roles both in abiotic and biotic stress tolerance. However, the function of Arabidopsis BAG2 remains largely unknown, whereas BAG6 is required for plants’ defense to pathogens, although it remains unknown whether BAG6 is involved in plants’ tolerance to abiotic stresses. Here, we show that both BAG2 and BAG6 are expressed in various tissues and are upregulated by salt, mannitol, and heat treatments and by stress-related hormones including ABA, ethylene, and SA. Germination of bag2, bag6 and bag2 bag6 seeds is less sensitive to ABA compared to the wild type (WT), whereas BAG2 and BAG6 overexpression lines are hypersensitive to ABA. bag2, bag6, and bag2 bag6 plants show higher survival rates than WT in drought treatment but display lower survival rates in heat-stress treatment. Consistently, these mutants showed differential expression of several stress- and ABA-related genes such as RD29A, RD29B, NCED3 and ABI4 compared to the WT. Furthermore, these mutants exhibit lower levels of ROS after drought and ABA treatment but higher ROS accumulation after heat treatment than the WT. These results suggest that BAG2 and BAG6 are negatively involved in drought stress but play a positive role in heat stress in Arabidopsis.
1. Introduction
Bcl-2 (B cell lymphorma 2)-associated athanogene (BAG)-family proteins were first identified by screening for Bcl-2-interacting proteins [1,2]. BAG proteins interact physically and functionally with the Bcl-2 protein to suppress cell death [1,2]. In mammals, there are six BAG proteins (BAG1–BAG6) that contain a conserved BAG domain at their C-terminal [3]. The BAG domain is approximately 110 amino acids in length and forms three α helices in three-dimensional structures [3,4]. Through the BAG domain, BAG proteins can interact with the ATPase domain of heat shock protein 70 (both constitutive Hsc70 and inducible Hsp70) and modulate the chaperone activity of these hsc70/hsp70 proteins [5]. At their N-terminal, some BAG proteins contain an ubiquitin-like domain (UBL) and/or several other motifs such as proline rich repeat (PXXP) and WW domain [3]. These domains/motifs enable the BAG proteins to interact with diverse binding partners that are involved in protein degradation, cell migration, cell proliferation, and apoptosis [3,6]. BAG family proteins also participate in Parkin-dependent mitophagy processes [7].
The BAG proteins are evolutionarily conserved with homologs present in fungi, plants and animals [8,9,10,11,12,13]. In plants, BAG proteins perform functions in apoptosis, senescence, and responses to biotic and abiotic stress [9,10,11,14,15,16,17]. Seven BAG genes were identified in the model plant Arabidopsis thaliana (Arabidopsis), ranging from BAG1 to BAG7 [9,10,11]. The AtBAG1–4 proteins contain a BAG domain and an UBL domain, whereas the AtBAG5–7 proteins contain an IQ motif, which preferentially binds to Ca2+-free calmodulin (CaM), in addition to a BAG domain [4,9,10,11,12,15]. Phylogenetic analysis by MEGA7 software clearly revealed that the plant BAG family proteins can be divided into two subgroups: Group I including the BAG1-4 proteins, and Group II including the BAG5-7 proteins (Figure S1). Arabidopsis BAG proteins have different subcellular localizations. AtBAG1–3 are mainly localized to the cytosol [18]; AtBAG4 is present in both cytosol and nucleus [18]; AtBAG5 is localized to the mitochondria [15]; AtBAG6 is localized to the nucleus [19]; and AtBAG7 resides in the endoplasmic reticulum (ER) [18,20]. These distinct subcellular localizations suggest that AtBAG proteins are functionally distinct.
AtBAG1 was found to be upregulated in plastid protein import2 (ppi2) mutant plants, implying that it has a role in proteasomal degradation of unimported plastid proteins [18]. Further research showed that indeed, AtBAG1 interacts with the cytosolic Hsc70-4 chaperone through its BAG domain to cooperatively mediate proteasomal degradation of misfolded cytosolic proteins and unimported plastid proteins [18]. Physiologically, AtBAG1-overexpressing transgenic seedlings were more sensitive to salt treatment, suggesting that AtBAG1 might be involved in plants’ response to abiotic stress [18]. It has been reported that under normal growth conditions the atbag2 mutant exhibited slightly better growth than the WT, implying that AtBAG2 might participate in plants PCD or response to environmental stresses [4]. AtBAG3 expression was upregulated by salt stress and by the stress-related hormones ABA and ACC, but downregulated by cold stress [15]. However, so far the physiological functions of AtBAG2 and AtBAG3 remain largely unknown.
Among the plant BAG genes, BAG4 is one of the most studied. Overexpression of AtBAG4 in tobacco plants confers tolerance to UVB irradiation and cold, oxidant and salt treatments [10]. In contrast, the atbag4 mutant is more sensitive to salt treatment [11]. These results clearly indicate that AtBAG4 plays a role in plant response to abiotic stress. In rice, overexpression of OsBAG4 enhances resistance to broad-spectrum disease, indicating that OsBAG4 functions as a positive regulator of disease resistance [21]. Furthermore, a rice E3 ubiquitin ligase EBR1 (enhanced blight and blast resistance 1) targets OsBAG4 for proteasome degradation; hence EBR1 and OsBAG4 form a module to balance growth and immune response [21]. Recently, AtBAG4 was found to interact with the guard cell potassium influx channel protein KAT1 to regulate stomatal movement [22]. Rice OsHKT1;5 (HIGH-AFFINITY POTASSIUM TRANSPORTER 1;5) is a Na+-selective transporter that maintains cellular Na+/K+ homeostasis under salinity stress [23]. Recent research shows that OsBAG4 functions as a bridge between OsMYB106 (a MYB transcription factor) and OsSUVH7 (a DNA methylation reader) to facilitate OsMYB106 binding to the OsHKT1;5 promoter, thereby collaboratively regulating OsHKT1;5 expression [24]. This research provides mechanisms for BAG4 function during plant responses to biotic and abiotic stress.
AtBAG5 contains both an IQ motif that can bind calmodulin (CaM) and a BAG domain capable of binding Hsc70/Hsp70 [15]. Our previous structural and biochemical studies revealed that Ca2+-free CaM (exhibit a closed conformation) and Hsc70 bind AtBAG5 independently, whereas Ca2+–saturated CaM (adopts an open conformation) and Hsc70 bind AtBAG5 with negative cooperativity [15]. We further demonstrated that AtBAG5 physiologically acts as a hub linking calcium signaling and the Hsc70 chaperone to regulate leaf senescence [15,25].
AtBAG6 also contains an IQ motif that has higher binding affinity for Ca2+-free CaM than the Ca2+–saturated CaM [26]. AtBAG6 expression is upregulated by plant PCD inducing agent and overexpression of AtBAG6 activates cell death [26]. Moreover, both the IQ motif and the BAG domain are required for AtBAG6-activated cell death [26]. By contrast, the atbag6 mutant exhibits enhanced susceptibility to a plant fungal pathogen Botrytis cinerea [11]. These results suggest that AtBAG6 plays a role in plant response to biotic stress. Further research revealed the molecular mechanism for AtBAG6-mediated pathogen resistance [27]. Upon pathogen infection, AtBAG6 protein was cleaved by an aspartyl protease APCB1 (Aspartyl Protease Cleaving BAG) at a caspase-1 cleavage site (LATD) downstream of its BAG domain [27]. Furthermore, AtBAG6 cleavage triggers autophagy and plant defense [27]. AtBAG6 transcription is also induced by stress-related plant hormones such as SA and ABA, and by heat stress [17,19]. AtBAG6 protein level is also significantly upregulated under heat stress [28]. The atbag6 mutant is sensitive to non-acclimated severe heat stress (45 °C for 30 min) [17]. However, another report found that the atbag6 mutant exhibited mild thermotolerance to non-acclimated heat stress (45 °C for 30 min) compared to the WT plants [28]. In acclimated heat stress treatment (38 °C for 2 h, followed by 45 °C for 1-3 h), the atbag6 mutant showed no different phenotype from those of the WT [19]. However, AtBAG6 mutation could increase the thermotolerance of the fes1a mutant [19]. Fes1A is a nucleotide exchange factor (NEF) of the HSP70 chaperone, and its mutation causes degradation of the HSP70 protein and leads to reduced tolerance to heat stress [29,30].
AtBAG7 resides in the ER and binds to the ER localized Hsp70 proteins to regulate unfolded protein response (UPR) during heat and cold tolerance [20]. Further study demonstrated that AtBAG7 may play a role in ER-nucleus retrograde signaling in plant responses to heat stress [31]. Upon heat treatment, AtBAG7 is sumoylated and proteolytically cleaved at the Ile378 site of the transmembrane domain in the C-terminus, then the cleaved N-terminus of AtBAG7 is translocated from the ER to the nucleus, where it interacts with the WRKY29 transcription factor to regulate expression of heat response genes [31]. It has been reported that salt treatment downregulated expression of AtBAG6 and AtBAG7, but ACC treatment could reverse the effect of salinity [32]. Although BAG6 was found to play a significant role in biotic (pathogens) and abiotic stresses (heat), it remains unknown whether BAG6 is involved in plants’ response to other stressors or not.
Here, we have focused on characterization of the functions of BAG2 and BAG6 in Arabidopsis. We first analyzed the expression patterns of the BAG2 and BAG6 genes during Arabidopsis development and in response to various hormones and abiotic stresses. Then, we analyzed the bag2, bag6, and bag2 bag6 mutant phenotypes grown under normal or stressed conditions. Our results show that both BAG2 and BAG6 genes are expressed in various tissues in Arabidopsis. Expression of both BAG2 and BAG6 genes is induced by salt, mannitol, and heat treatments and by stress-related hormones including ABA, ACC (an ethylene precursor) and SA, while BAG6 is additionally induced by JA. Germination of the bag2 and bag6 single and bag2 bag6 double mutant seeds are less sensitive to ABA than that of the WT. Both the bag2 and bag6 single mutant and bag2 bag6 double mutant showed higher survival rates than the WT in drought treatment but displayed lower survival rates in 45 °C heat stress experiments. Consistently, these mutants displayed differential transcription levels of stress-related genes such as RD29A and RD29B, ABA biosynthesis gene NCED3, and ABA response gene ABI4 compared to the WT in response to ABA treatment. Finally, when compared to WT seedlings, the mutants exhibited higher content of reactive oxygen species (ROS) after heat stress but lower ROS levels after drought and ABA treatment.
2. Results
2.1. BAG2 and BAG6 Expression during Arabidopsis Development
To determine expression patterns of the BAG2 and BAG6 genes, we generated Arabidopsis transgenic lines expressing the GUS reporter gene under control of the promoter fragments of the BAG2 and BAG6 genes, respectively (ProBAG2:GUS and ProBAG6:GUS) and analyzed the expression of the GUS reporter gene in various tissues. Strong GUS staining was observed in most tissues throughout the ProBAG2:GUS and ProBAG6:GUS transgenic plants (Figure 1A–T). In 7-day-old seedlings, ProBAG2:GUS activity is strongly detected in cotyledons, hypocotyl, and root vascular tissues, with the strongest signal at the lower part of the hypocotyl (Figure 1A,B), while ProBAG6:GUS expression is strongly detected in all of the tissues of the whole seedling (Figure 1K,L). In 2-week-old plants and bolting plants, both ProBAG2:GUS and ProBAG6:GUS are highly expressed in rosette leaves and cauline leaves (Figure 1C–E,M–O), with ProBAG6:GUS showing additional GUS activity in stems (Figure 1P). In flowers, both ProBAG2:GUS and ProBAG6:GUS are expressed in sepals, anther filaments, pollen grains, and style (Figure 1H,Q,R), with ProBAG6:GUS showing additional expression in ovules (Figure 1R). In siliques, both ProBAG2:GUS and ProBAG6:GUS are expressed in young siliques with ProBAG6:GUS showing stronger GUS signals (Figure 1I,S), while in old siliques both genes’ expressions are very weak (Figure 1J,T).
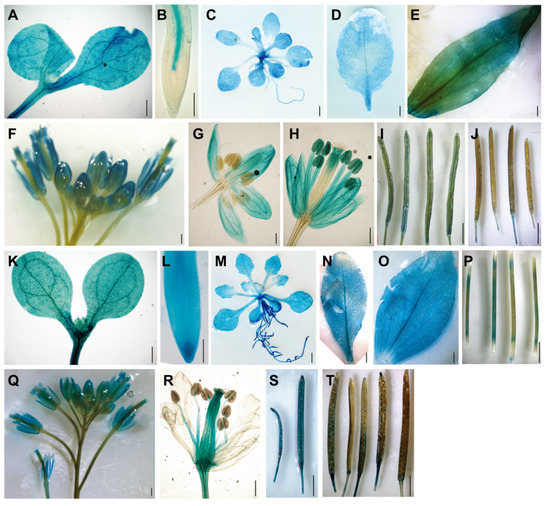
Figure 1.
Expression patterns of the Arabidopsis BAG2 and BAG6 genes. (A–J) Expression of ProBAG2:GUS in 5-day-old seedlings cotyledons and hypocotyl (A), root tip (B), 2-week-old plant (C), rosette leaf (D), cauline leaf (E), inflorescence (F), flower bud (G), opened flower (H), young siliques (I), and mature siliques (J); (K–T) Expression of ProBAG6:GUS in cotyledons and hypocotyl (K) and root tip (L) of 5-day-old seedling, 2-week-old plant (M), rosette leaf (N), cauline leaf (O), stems (P), inflorescence (Q), opened flower (R), young siliques (S), and mature siliques (T). Bars = 100 μm in (B,L), 400 μm in (A,F–H,K,Q,R) and 2 mm in (C–E,I,J,M–P,S,T).
2.2. BAG2 and BAG6 Expression Responses to Abiotic Stress and Plant Hormones
We further investigated whether BAG2 and BAG6 expression is regulated by abiotic stressors or plant hormones. The results show that ProBAG2:GUS expression is strongly increased by mannitol, salt (NaCl), heat and ABA treatments (Figure 2A,B and Figure S2A) and its expression is also slightly increased in response to other stressors and hormones such as PEG, SA and ACC, respectively. Similarly, ProBAG6:GUS showed significant enhanced GUS staining in response to JA, ABA, mannitol, salt, and heat treatment, and showed a slight increase in response to ACC, PEG and SA (Figure 2C,D and Figure S2B–D). A previous report had analyzed AtBAG genes expression response to abiotic stressors such as cold, heat, mannitol, and salt and hormones such as ABA, ACC, MeJA, and SA by reverse transcription quantitative PCR (RT-qPCR) and showed that AtBAG2 transcription is upregulated by ABA but downregulated by ACC, whereas AtBAG6 transcription is upregulated by SA and ABA [17]. As to response to abiotic stresses, AtBAG2 transcription is upregulated by salt but downregulated by cold, whereas AtBAG6 transcription is upregulated by heat and salt treatment [17]. Together, these results suggest that AtBAG2 and AtBAG6 genes are involved in Arabidopsis responses to environmental stress.
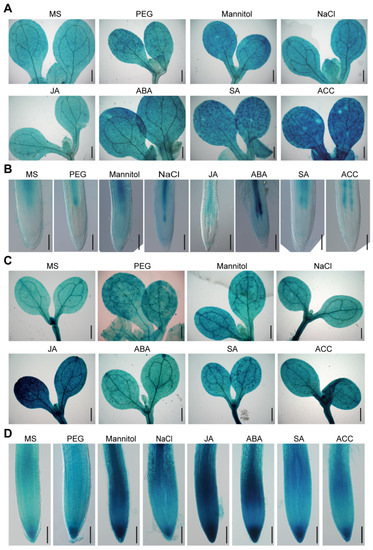
Figure 2.
Expression of ProBAG2:GUS and ProBAG6:GUS in response to various abiotic stresses and hormones. Five-day-old ProBAG2:GUS (A,B) and ProBAG6:GUS (c and d) seedlings were transferred to Murashige and Skoog (MS) media (mock) supplemented with 5% PEG, 300 mM mannitol, 200 mM NaCl, 10 μM JA, 10 μM ABA, 5 mM SA and 10 μM ACC and treated for 24 h, then were collected for GUS staining. Shown are representative images of shoots (A,C) and root tips (B,D) of three independent experiments. Bars = 400 μm in (A,C) and 100 μm in (B,D).
2.3. BAG2 and BAG6 Genes Are Involved in Arabidopsis Responses to ABA and Drought Treatment
To explore the function of the BAG2 and BAG6 genes, we analyzed localization of the genes on Arabidopsis chromosomes, and found that chromosome 5 has the highest number of genes, followed by others. AtBAG2 is located on chromosome 5 (chr.5) and AtBAG6 is located on chromosome 2 (chr.2) (Figure 3A). The transfer DNA (T-DNA) insertion mutants were isolated for BAG2 and BAG6 within the fourth exon (SALK_030295, bag2; Figure 3B) and within the first exon (SALK_004760, bag6; Figure 3D), respectively. Homozygous bag2 and bag6 mutants were crossed to generate the bag2 bag6 double mutant (Figure 3F). The bag2 and bag6 mutants had been reported previously [4,28]. RT-PCR analyses revealed that weak BAG2 transcript was detected in the homozygous bag2 and bag2 bag6 mutants (Figure 3C,G and Figure S3A,B) and no BAG6 transcript was detectable in the homozygous bag6 and bag2 bag6 mutants (Figure 3E,H and Figure S3C,D), indicating that the bag2 mutant is a knockdown allele and the bag6 mutant is a null allele. Under normal growth conditions, the bag2 and bag6 single and bag2 bag6 double mutants exhibited slightly larger rosette size than the WT (Figure S4).
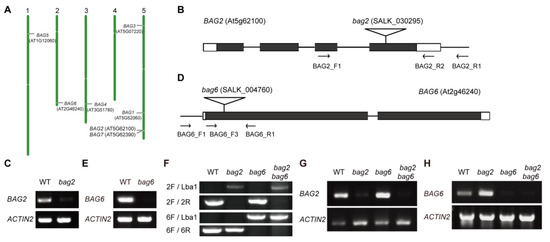
Figure 3.
Chromosomal locations of the seven BAG genes. (A) The location of the gene is marked with short lines. White mark is the centromere of the chromosome. There are 7 BAG genes that are distributed on 5 chromosomes. AtBAG2 is located on chromosome 5 and AtBAG6 is located on chromosome 2. Insertion sites and transcription analysis of the bag2 and bag6 mutants; (B,D) Structures of the AtBAG2 (B) and the AtBAG6 (D) genes with T-DNA insertion sites. White boxes at left and right borders represent UTR regions, black boxes indicate exons, lines between black boxes represent introns, flags indicate the T-DNA insertion sites, and arrows indicate the positions of primers used for PCR verification of the insertions and for RT-PCR; (C,E) RT-PCR analysis of transcription levels of AtBAG2 (C) and AtBAG6 (E) genes in WT and bag2 or bag6 mutants. The ACTIN2 gene was used as the internal control. Total RNA was extracted from 7-day-old seedlings. The presented images in (C,E) were cropped and the original, full-length gel images were provided in the additional file Figure S3; (F) Genotyping results of bag2, bag6, and bag2 bag6 mutants; (G,H) RT-PCR analysis of transcription levels of AtBAG2 and AtBAG6 genes in bag2, bag6, and bag2 bag6 mutants. The ACTIN2 gene was used Eas the internal control.
To study the response of bag2 and bag6 single and bag2 bag6 double mutants to the plant stress hormone ABA, these mutants and WT seeds were sown on Murashige and Skoog (MS) medium supplemented with different concentrations of ABA to compare the germination and greening rate between WT and mutants lines. Without ABA, the mutant seeds showed a slightly higher germination rate than that of the WT, and when ABA was applied, much higher germination and greening rates were observed in the bag2 and bag6 single and bag2 bag6 double mutants (Figure 4). When comparing germination and greening rates on 0.75 μM ABA, we found a significant difference: WT germination rate (72%) and greening rate (46%), while bag2 75% and 84%, bag6 79% and 88% and bag2 bag6 81% and 91%, respectively (Figure 4B,C). Similarly, when compared on 1 μM ABA, WT exhibited 57% of seeds germinated and after 7 days a 29% greening rate was found, but the bag2 single mutant exhibited 65% germination while a 49% greening rate was observed; bag6 also showed higher germination and greening rates than the WT (i.e., 68% and 55%, respectively) (Figure 4B,C). Interestingly, the bag2 bag6 double mutant’s germination rate was higher than each single mutant and WT, which are 75% and 71%, respectively (Figure 4B,C). Finally, when compared on 2 μM ABA, WT exhibited 37% germination and a 24% greening rate, while bag2 exhibited 50% and 36%, bag6 55% and 36% and bag2 bag6 62% and 49%, respectively (Figure 4B,C). Thus, these mutants appeared to enhance the ABA tolerance of Arabidopsis during germination and the early vegetative growth period.
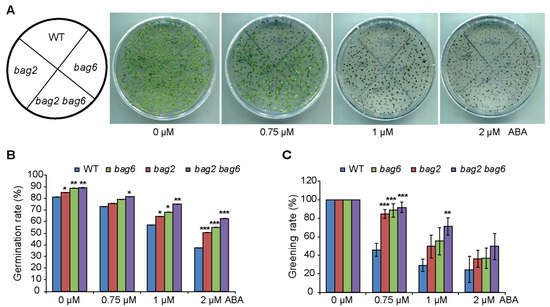
Figure 4.
bag2 and bag6 mutants are less sensitive to ABA than the WT. (A–C) Seed germination rate and greening rate of WT, bag2 and bag6 single, and bag2 bag6 double mutants. Sterilized seeds were sowed on MS medium supplemented without or with different concentrations of ABA. The germination percentage and the greening rate were scored. All the data represent the means of three independent experiments ± SD. * represent the significant differences at p < 0.05; ** represent p < 0.01; *** represent p < 0.001 (Student’s t-test).
To further analyze the role of AtBAG2 and AtBAG6 genes in Arabidopsis responses to ABA, we generated transgenic lines over expressing the AtBAG2 and the AtBAG6 gene (Figure S5A–D). Without ABA, seeds of the AtBAG2 and AtBAG6 overexpression (OE) lines showed a slightly lower germination rate and greening rate than the WT (Figure 5). When ABA was applied, much lower greening rates were observed in the AtBAG2 and AtBAG6 OE lines (Figure 5). When comparing germination and greening rates on 0.75 μM ABA, both OE lines and WT reduced significantly: WT germination rate (87%) and greening rate (74%), while AtBAG2 OE L13 (77%) and (38%), AtBAG2 OE L14 (83%) and (46%), AtBAG6 OE L20 81% and 44%, AtBAG6 OE L24 63% and 8% respectively (Figure 5B,C). Similarly, when compared on 1 μM ABA, WT germination rate and greening rate 84% and 16%, but AtBAG2 OE L13 73% and 13%, AtBAG2 OE L14 78% and 18%, AtBAG6 OE L20 77% and 10%, and AtBAG6 OE L24 59% and 4%, respectively (Figure 5B,C). Finally, when 2 μM ABA is applied, WT exhibited 82% and 0% germination and greening rates, AtBAG2 OE L13 71% and 1%, AtBAG2 OE L14 76% and 1%, AtBAG6 OE L20 74% and 1%, and AtBAG6 OE L24 57% and 0%, respectively (Figure 5B,C).
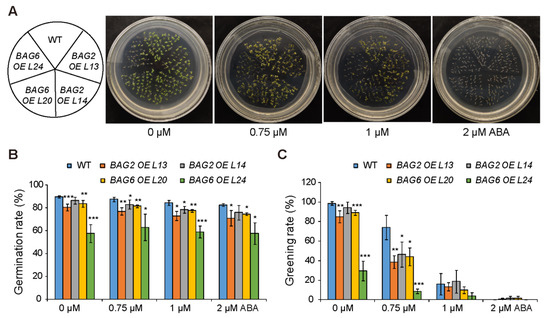
Figure 5.
AtBAG2 and AtBAG6 overexpression lines are more sensitive to ABA than the WT. (A–C) Seed germination rate and greening rate of WT, AtBAG2 and AtBAG6 overexpression lines. Sterilized seeds were sowed on MS medium supplemented without or with different concentrations of ABA. The germination percentage and the greening rate were scored. Shown are means ± SD. * represent the significant differences at p < 0.05; ** represent p < 0.01; *** represent p < 0.001 (Student’s t-test).
Collectively, these data demonstrate that mutation of AtBAG2 and AtBAG6 genes enhances the ABA tolerance of Arabidopsis during germination and the early vegetative growth period, whereas their overexpression reduced germination speed and growth capability.
To study drought stress tolerance, 3-week-old WT and mutant plants were used, and 9 plants per pot were sown and 12 pots were prepared for each genotype (108 plants in total for each genotype). After withholding water for 14 days, almost all the WT and mutant plants were wilted and near to death (Figure 6A). Then, the plants were re-watered for three days. On the third day a significant difference was noticed between WT and mutant plants (Figure 6A); bag2 showed 43%, bag6 showed 44%, and bag2 bag6 showed the highest survival rate at 73%, while the WT showed a lower survival rate at 29 % (Figure 6B). To further verify these results, we performed water loss experiments with different time intervals. Consistently, the water loss in mutant plants’ rosette leaves was slower than that of the WT (Figure 6C). These results suggest that BAG2 and BAG6 play a negative role in Arabidopsis responses to ABA and drought treatments.
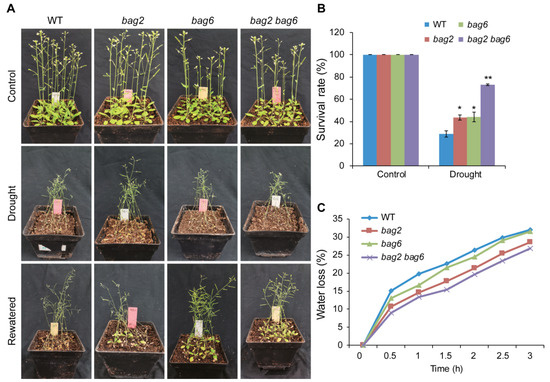
Figure 6.
bag2 and bag6 mutants are more tolerant to drought. (A) Phenotypes of WT, bag2 and bag6 single, and bag2 bag6 double mutant plants treated with drought stress. Three-week-old plants were subjected to drought stress for 14 days and re-watered for 3 days; (B) the survival rate of WT, bag2, bag6, and bag2 bag6 plants after 3 days of re-watering following the 14 days drought treatment. Values are means ± SD (n = 5 independent experiments, 108 plants for each genotype in each independent experiment were used for analysis); (C) Water loss in WT, bag2, bag6, and bag2 bag6 plants. Leaves from 3-week-old plants were collected and subjected to water loss at different time intervals. All the data represent the means ± SD of five independent experiments. * represent the significant differences at p < 0.05; ** represent p < 0.01 (Student’s t-test).
2.4. Mutation of BAG2 and BAG6 in Arabidopsis Compromises Tolerance to Heat Stress
Our ProBAG2:GUS and ProBAG6:GUS expression results and previously reported RT-PCR/RT-qPCR and proteomics results show that BAG2 and BAG6 genes are upregulated during heat stress [17,19,28]. Thus, we studied the effect of heat stress on bag2 and bag6 single and bag2 bag6 double mutants. The results showed that the bag2 and bag6 single and bag2 bag6 double mutants are hypersensitive to heat stress compared to WT seedlings (Figure 7). After being treated at 45 °C for 25 min, the survival rates of WT, bag2, bag6, and bag2 bag6 were 90%, 68%, 54%, and 70%, respectively; after being treated at 45 °C for 30 min, their survival rates were 84%, 79%, 59%, and 74%, respectively; and after being treated at 45 °C for 45 min, their survival rates were 70%, 28%, 57%, and 47%, respectively (Figure 7B). We also measured chlorophyll content in the mutants and WT on control untreated seedlings (22 °C) and 45 °C heat treated seedlings. Total chlorophyll levels decreased as a consequence of the heat treatment both in WT and mutants, but the mutants retained less chlorophyll (Figure 7C). These results indicate that mutation of BAG2 and BAG6 compromises Arabidopsis tolerance to heat stress.
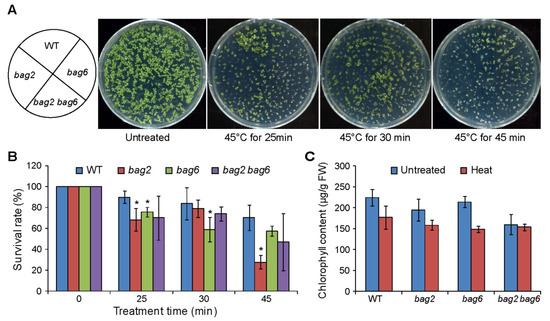
Figure 7.
bag2 and bag6 mutants are hypersensitive to heat stress. (A) Phenotype of seedlings treated with heat stress; (B) Survival rates of WT, bag2, bag6, and bag2 bag6 mutants under heat stress. Five-day-old seedlings were placed at 45 °C for 25, 30, and 45 min and then resumed growth at 22 °C; (C) Chlorophyll content measured from seedlings grown on normal growth conditions and after 45 °C heat stress. The data represent the means ± SD of two independent experiments. * represent the significant differences at p < 0.05 (Student’s t-test).
We used the AtBAG2 and AtBAG6 OE lines to further analyze the role of AtBAG2 and AtBAG6 genes in Arabidopsis responses to heat stress. After being treated at 45 °C for 25 min, the survival rates of WT, AtBAG2 OE L13 and L14, and AtBAG6 OE L20 and L24 were 78%, 68%, 91%, 89%, and 74%, respectively; and after being treated at 45 °C for 45 min, their survival rates were 62%, 38%, 59%, 54%, and 56%, respectively (Figure 8). These results indicate that AtBAG2 and AtBAG6 overexpression does not significantly enhance Arabidopsis tolerance to heat stress.
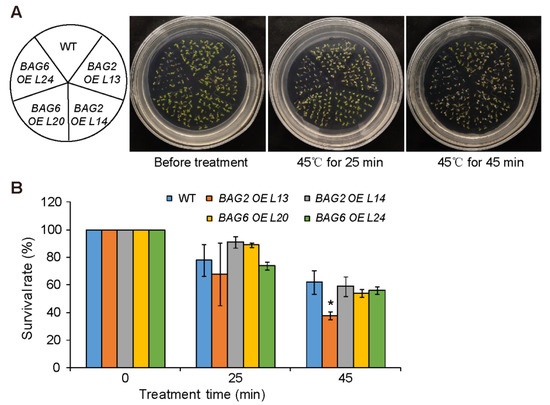
Figure 8.
AtBAG2 and AtBAG6 OE lines display no significantly enhanced tolerance to heat stress. (A) Phenotypes of seedlings treated with heat stress; (B) Survival rates of WT, AtBAG2 and AtBAG6 OE lines under heat stress. Five-day-old seedlings were placed at 45 °C for 25 and 45 min and then resumed growth at 22 °C. Shown are means ± SD. * represent the significant differences at p < 0.05 by Student’s t-test.
2.5. Stress- and ABA-Related Genes Are Differentially Regulated in the bag2, bag6, and bag2 bag6 Mutants Compared to WT
To further investigate the role of BAG2 and BAG6 in Arabidopsis responses to ABA, heat and drought treatment, we performed RT-qPCR analysis on several stress- and ABA-related genes such as RD29A and RD29B (stress related genes), NCED3 (ABA biosynthesis gene), and ABI4 (ABA response gene) [33,34,35,36,37,38]. We found that after ABA treatment the expression of RD29A and RD29B genes was upregulated in WT and bag2, bag6, and bag2 bag6 seedlings, but the mutants showed more induction compared to the WT, particularly at 6 h of ABA treatment (Figure 9A,B). Whereas expression of NCED3 and ABI4 genes was downregulated by ABA treatment in WT and bag2, bag6, and bag2 bag6 seedlings with the mutants showed more inhibition compared to the WT, particularly at 6 h of ABA treatment (Figure 9C,D). Interestingly, without ABA treatment the bag6 and bag2 bag6 seedlings already displayed lower transcription levels of the NCED3 and ABI4 genes compared to the WT (Figure 9C,D). This finding is consistent with the seed germination phenotype of these mutants. These results further support the ABA and drought phenotypic data.
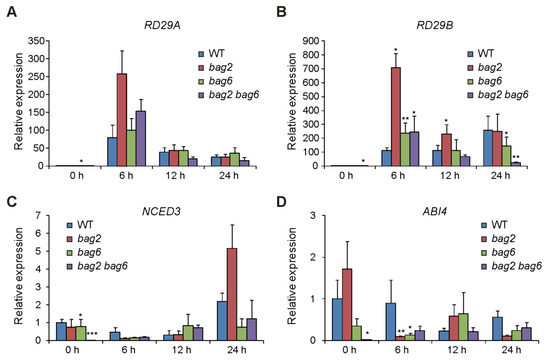
Figure 9.
The relative expression of stress- and ABA-related genes. Transcription levels of RD29A (A), RD29B (B), NCED3 (C), and ABI4 (D) genes in WT and bag2, bag6, and bag2 bag6 mutants were examined under ABA treatment for 0, 6, 12, and 24 h. Values are means ± SD of three replicates. * represent the significant differences at p < 0.05; ** represent p < 0.01; *** represent p < 0.001 (Student’s t-test).
2.6. ROS Accumulation in WT and Mutant Plants
The accumulation of H2O2 and superoxide anion (O2–) was detected using 3,3-diaminobenzidine (DAB) and nitro blue tetrazolium (NBT) staining in seedling shoots and roots. Upon 20 μM ABA treatment, the cotyledons and root of the mutant seedlings displayed lighter color than that of the WT, which indicates lower accumulation of O2– and H2O2, respectively, in the mutant seedlings (Figure 10A,B). However, upon heat stress treatment, the mutants exhibited higher accumulation of O2– than the WT (Figure 10C). Finally, DAB staining was also applied to drought stressed rosette leaves of WT and mutant plants. We obtained the same results, such that leaves of mutant lines accumulated lower H2O2 than that of the WT (Figure S6). Combined, these results indicate that mutant lines had a lower accumulation of ROS when treated with ABA and drought while having higher ROS accumulation when treated with heat stress. These results correspond well with the respective drought- and heat-tolerance phenotypes of these mutants.
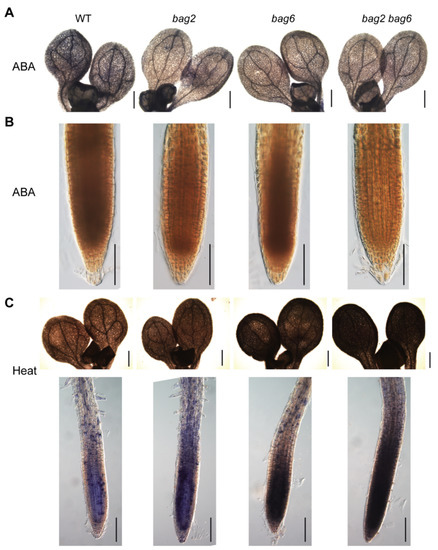
Figure 10.
BAG2 and BAG6 mutation affects the ROS levels after ABA and heat treatment in Arabidopsis seedlings. Five-day-old seedlings were transferred to 1/2 MS agar plates supplemented with 20 μM ABA for 6 h or were treated with heat stress (37 °C) for 3 h, then were stained with NBT (staining for O2–) (A,C) or DAB (staining for H2O2) (B). At least three independent experiments were performed for each analysis and 15 seedlings of each genotype were examined. Bars = 400 μm in (A) and in the shoot images in (C) and 100 μm in (B) and in the root images in (C).
3. Discussion
In plants, especially in Arabidopsis, BAG proteins are a topic of interest and they mediate multiple abiotic, biotic and developmental processes. Actually, most research on plant BAGs so far has focused on their functions in abiotic and biotic stress tolerance. The expression patterns of AtBAG genes and their roles during plant development remain largely unknown. In this study, we analyzed the expression patterns of AtBAG2 and AtBAG6 genes during development by using ProBAG2:GUS and ProBAG6:GUS transgenic lines. Interestingly, the results show that both AtBAG2 and AtBAG6 genes are expressed in various tissues with overlapping or specific expression patterns during the whole developmental process. For example, ProBAG6:GUS is expressed in all of the root tissues with stronger expression in columella root cap cells and root vascular tissue, but ProBAG2:GUS is only expressed in the root vascular tissue. The root cap surrounds and protects the root tip meristem and receives and transmits environmental signals to the growing root [39]. As the root grows, the outermost layers of the root cap cells undergo cell death [39]. The strong expression of AtBAG6 in the columella root cap cells implies that AtBAG6 might play a role in the PCD of root cap cells. In shoots, both ProBAG2:GUS and ProBAG6:GUS are strongly expressed in rosette leaves and cauline leaves with the strongest signal in the vasculature strands. The developmental process of vasculature formation involves cell differentiation and PCD [40]. The strong expression of AtBAG2 and AtBAG6 genes in the vascular tissues imply that they may have a role in the formation of vasculature strands. Detailed examination of vascular tissue development in the bag2 and bag6 mutants and AtBAG2 and AtBAG6 OE plants under normal and stressed growth conditions will provide some clue.
Mutant analysis showed that bag2 and bag6 single mutants and bag2 bag6 double mutant plants are slightly larger than the WT, and they show no developmental defect. These results suggest that the AtBAG2 and AtBAG6 genes and other AtBAG genes may have redundant and specific functions during Arabidopsis development. Analyzing the other AtBAG genes expression patterns and generating multiple atbag mutants, by crossing or by using CRISPR/Cas9 gene editing methods, will provide more information for understanding the role of the AtBAG family genes during Arabidopsis development.
Previous RT-qPCR and Northern blot analysis had shown that AtBAG2 transcript levels were significantly upregulated by ABA and salt treatment and AtBAG6 transcript levels were significantly upregulated by ABA, SA, H2O2 and heat treatment [10,17,19,28]. Our ProBAG2:GUS and ProBAG6:GUS expression results show that they both are upregulated by heat, salt and osmotic treatment and by stress-related plant hormones including ABA, SA and ACC treatment, and ProBAG6:GUS is additionally induced by JA. These expression results suggest that the AtBAG2 and AtBAG6 genes may be involved in Arabidopsis responses to multiple environmental stresses. Indeed, our stress and hormone treatments results show that the bag2 and bag6 single mutants and bag2 bag6 double mutant seeds germination are less sensitive to ABA treatment and their plants are more tolerant to drought stress than the WT, but they are more sensitive to heat stress. By contrast, AtBAG2 and AtBAG6 OE seed germination is more sensitive to ABA treatment. Interestingly, in ABA and drought treatments the bag2 bag6 double mutant shows more tolerance than either of the single mutants, suggesting an additive effect of these two genes in Arabidopsis responses to ABA or drought stress treatments. There is some discrepancy on atbag6 seedlings response to heat stress among the previously published results. Nawkar et al. (2016) showed that atbag6 (SALK_004760) seedlings were hypersensitive to heat stress treatment (45 °C for 25, 30, and 45 min) compared to the WT [17]. However, Echevarría-Zomeño et al. (2016) found that atbag6 (SALK_047959 and SALK_015968) seedlings displayed mild tolerance to heat stress (45 °C for 30 min) compared to the WT seedlings [28]. In heat-induced acclimation experiments (38 °C for 2 h, followed by 45 °C for 1–3 h), Fu et al. (2019) showed that atbag6 (SALK_047959C and SALK_073331C) seedlings exhibited the same thermotolerance phenotypes as the WT, but that they could enhance the thermotolerance of fes1a (Fes1A is a co-chaperone of the HSP70 protein) mutant [19]. Although Nawkar et al. (2016) and Echevarría-Zomeño et al. (2016) used same heat stress treatment conditions, they used a different atbag6 T-DNA insertional mutant [17,28]. We used the same atbag6 (SALK_004760) mutant and heat treatment conditions as Nawkar et al. (2016) and obtained similar results. Therefore, the discrepancy on atbag6 thermotolerance might be due to a different T-DNA insertional mutant used for the heat stress treatment. We also performed heat stress treatment on AtBAG2 and AtBAG6 OE lines (selected two OE lines for each gene). After being treated at 45 °C for 25 min, one AtBAG2 OE line and one AtBAG6 OE line exhibited a slightly higher survival rate compared to the WT, but the other line did not. Whereas after being treated at 45 °C for 45 min, both AtBAG6 OE lines and one AtBAG2 OE line showed same thermotolerance phenotypes as the WT but one AtBAG2 OE line displayed a significantly lower survival rate. In the future, more OE lines for each gene should be selected to perform heat stress treatments in order to obtain convincing conclusions.
Although our RT-qPCR analysis results showed that several stress- and ABA-related genes are differentially expressed in bag2, bag6, and bag2 bag6 seedlings compared to the WT, the underlying molecular mechanism remains to be unveiled. The molecular mechanism of how AtBAG6 mediates fungal resistance has been revealed recently [27]. Fungal infection triggers AtBAG6 cleavage by an aspartyl protease and the processed AtBAG6 activates autophagy and plant defense [27]. Whether a similar molecular mechanism could be responsible for AtBAG6-mediated abiotic stress tolerance awaits future study. atbag7 seedlings also are sensitive to heat treatment [20,31]. The underlying molecular mechanism responsible for AtBAG7 heat stress protection was revealed by in silico analysis of its amino acid sequence, by examination of its localization under normal and stress conditions, and by screening AtBAG7-interacting proteins [31]. In the future, similar experiments can be performed on AtBAG2 and AtBAG6 to provide some insight into understanding their functional mechanisms during plant responses to abiotic stress.
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
The T-DNA insertion mutants bag2 (SALK_030295) and bag6 (SALK_004760) are in Columbia-0 (Col-0) background. These mutants were obtained from the Arabidopsis Biological Resource Center (ABRC), and the double mutant was obtained by crossing bag2 and bag6. T-DNA insertion was confirmed by RT-PCR analysis. Primers used in this experiment are listed in Supplementary Table S1. Seeds were surface sterilized in 70% ethanol for 5 min and 1% Clorox bleach for 10 min, stratified for 3 days at 4 °C. The seeds were plated on solidified Murashige and Skoog (MS) medium and vertically grown at 22 °C under 16 h light/8 h dark conditions. For ABA treatment experiments, all the single, double mutant, and WT seeds were surface sterilized and, after 3 days stratification, sown on plates supplemented with or without ABA. For drought treatments, the stratified seeds were grown vertically on MS plates for seven days, and then were transferred to soil (75% soil and 25% vermiculite) for more 14 days to grow. Nine plants per pot were grown. During each independent experiment, we grew 108 plants in total for each genotype.
4.2. Vector Construction and Generation of Transgenic Plants
To create the ProBAG2:GUS construct, the 1886 bp sequence upstream of the BAG2 gene start codon was PCR-amplified. The resulting fragment was inserted upstream of the GUS reporter gene in the pGreenII0229-GUS vector via XhoI and KpnI sites [41]. Similarly, ProBAG6:GUS was also created for this study, and the 1884 bp sequence upstream of the BAG6 gene start codon was PCR-amplified. The resulting fragment was inserted upstream of the GUS reporter gene in the pGreenII0229-GUS vector via XhoI and KpnI sites [41]. Primer sequences used in this experiment are available in Supplementary Table S1. These constructs were transformed into Agrobacterium tumefaciens strain C58C1 (pMP90/pJICSa-Rep). The Arabidopsis transgenic plants were generated using floral dip method [42].
4.3. RT-PCR Analysis
RT-PCR was performed to test the transcription levels of the BAG2 and BAG6 gene in mutant or WT plants. Total RNA was extracted from 7-day-old seedlings using Trizol reagent (TransGen Biotech, Beijing, China). The first-strand cDNA was synthesized using Easy Script First-Strand cDNA Synthesis Super Mix (TransGen Biotech, Beijing, China). The ACTIN2 gene was used as an internal control. The primer sequences used in this experiment are listed in Supplementary Table S1.
4.4. GUS Histochemical Staining
For GUS staining, various tissues of ProBAG2:GUS and ProBAG6:GUS transgenic plants were incubated for 12 h at 37 °C in GUS staining buffer as previously described [43]. After staining, the tissues were cleared in chloral hydrate: distilled water: glycerol (8:3:1, w:v:v). Samples were visualized with microscopy (Olympus BX63, Tokyo, Japan).
4.5. Drought and ABA Treatments
The bag2 and bag6 single and bag2 bag6 double mutants and WT seeds were harvested at the same time and used for ABA treatments. In brief, surface-sterilized seeds were sown on MS medium plates supplemented without or with different concentrations of ABA (Sigma-Aldrich, St. Louis, MO, USA) as indicated. Plates were stratified at 4 °C in the dark for 72 h and transferred to a growth incubator for normal conditions (16 h light/8 h dark, 22 °C). For quantification of the percentage of seedlings with green cotyledons, seeds were sown on MS medium supplemented different concentrations of ABA and analyzed on the indicated days after stratification as previously described [44]. Pictures were scanned using HP LaserJet P1108 scanner.
For the drought tolerance test, 3-week-old plants grown in soil with sufficient water were treated with natural drought (water was withheld). After 14 days without watering, the drought-treated plants were re-watered and recovery was checked after 3 days. Drought experiments were repeated three times, and at least 9 plants for each individual genotype were used per experiment.
4.6. Heat Treatment and Measurement of Chlorophyll Content
For heat treatment, 5-day-old WT, bag2, bag6 and bag2 bag6 seedlings were exposed to 45 °C for 45 min. After three days of recovery at 22 °C, the seedlings’ survival rate was counted [15]. For the measurement of chlorophyll content, mesophyll cells were spectrophotometrically measured as previously reported [45].
4.7. DAB and NBT Staining
DAB and NBT staining were performed as previously described [46]. The experiment was performed three times and each time 20 seedlings of each genotype were stained. Five-day-old seedlings were used in this experiment.
4.8. Hormone Treatment
Hormonal treatment of the ProBAG2:GUS and ProBAG6:GUS seedling was performed as previously described [47].
4.9. RT-qPCR
For RT-qPCR analysis, of different stress related genes, we followed previously described protocol [48].
5. Conclusions
Results in the present study show that the AtBAG2 and AtBAG6 genes are broadly expressed in various tissues during Arabidopsis development. In addition, AtBAG2 and AtBAG6 expression is induced by abiotic stresses such as heat, salt and osmotic treatment and by stress-related plant hormones including ABA, SA, JA, and ACC. Seed germinations of bag2 and bag6 single and bag2 bag6 double mutants are more tolerant to ABA treatment and their plants are more tolerant to drought stress but sensitive to heat stress. These findings suggest that AtBAG2 and AtBAG6 play important roles in Arabidopsis response to multiple abiotic stresses and hormones.
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/article/10.3390/ijms22115856/s1.
Author Contributions
S.M. and Y.S. conceived the project. S.M. planned and designed the research. M.A. performed experiments with the help of Q.L., Z.L. and L.L., M.A. and S.M. analysed data and wrote the manuscript with contributions of all the authors. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the National Natural Science Foundation of China (Grants 31870230, 31570247, and 91417308 to S.M.).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Arabidopis mutant lines (SALK_030295, SALK_004760) were kindly provided by Arabidopsis Biological Resource Center (ABRC) and the double mutant was obtained by crossing bag2 and bag6.
Acknowledgments
We thank the Arabidopsis Biological Resource Center (ABRC) for providing mutant seeds, John Innes Centre for providing the pGREENII0229 vector.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Abbreviations
| BAG | Bcl-2 associated athanogene |
| ABA | Abscisic acid |
| ACC | 1-aminocyclopropane-1-carboxylic acid |
| JA | Jasmonate |
| SA | Salicylic acid |
| GA | Gibberellic acid |
| WT | Wild type |
| GUS | β-glucuronidase |
| PCD | programmed cell death |
| ROS | reactive oxygen species |
References
- Takayama, S.; Sato, T.; Krajewski, S.; Kochel, K.; Irie, S.; Millan, J.A.; Reed, J.C. Cloning and functional analysis of BAG-1: A novel Bcl-2-binding protein with anti-cell death activity. Cell 1995, 80, 279–284. [Google Scholar] [CrossRef]
- Antoku, K.; Maser, R.S.; Scully, W.J., Jr.; Delach, S.M.; Johnson, D.E. Isolation of Bcl-2 binding proteins that exhibit homology with BAG-1 and suppressor of death domains protein. Biochem. Biophys. Res. Commun. 2001, 286, 1003–1010. [Google Scholar] [CrossRef] [PubMed]
- Doong, H.; Vrailas, A.; Kohn, E.C. What’s in the ‘BAG’?—A functional domain analysis of the BAG-family proteins. Cancer Lett. 2002, 188, 25–32. [Google Scholar] [CrossRef]
- Fang, S.; Li, L.; Cui, B.; Men, S.; Shen, Y.; Yang, X. Structural insight into plant programmed cell death mediated by BAG proteins in Arabidopsis thaliana. Acta Crystallogr. D Biol. Crystallogr. 2013, 69, 934–945. [Google Scholar] [CrossRef]
- Takayama, S.; Bimston, D.N.; Matsuzawa, S.; Freeman, B.C.; Aime-Sempe, C.; Xie, Z.; Morimoto, R.I.; Reed, J.C. BAG-1 modulates the chaperone activity of Hsp70/Hsc70. EMBO J. 1997, 16, 4887–4896. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, H.; Minami, R.; Yokota, N. BAG6/BAT3: Emerging roles in quality control for nascent polypeptides. J. Biochem. 2013, 153, 147–160. [Google Scholar] [CrossRef] [PubMed]
- Kapadia, M.; De Snoo, M.L.; Kalia, L.V.; Kalia, S.K. Regulation of Parkin-dependent mitophagy by Bcl-2-associated athanogene (BAG) family members. Neural. Regen. Res. 2021, 16, 684–685. [Google Scholar]
- Jain, S.; Wiemann, P.; Thill, E.; Williams, B.; Keller, N.P.; Kabbage, M. A Bcl-2 associated athanogene (bagA) modulates sexual development and secondary metabolism in the filamentous fungus Aspergillus nidulans. Front. Microbiol. 2018, 9, 1316. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; He, C.; Zhang, H. The BAG-family proteins in Arabidopsis thaliana. Plant Sci. 2003, 165, 1–7. [Google Scholar] [CrossRef]
- Doukhanina, E.V.; Chen, S.; Van der Zalm, E.; Godzik, A.; Reed, J.; Dickman, M.B. Identification and functional characterization of the BAG protein family in Arabidopsis thaliana. J. Biol. Chem. 2006, 281, 18793–18801. [Google Scholar] [CrossRef]
- Kabbage, M.; Dickman, M.B. The BAG proteins: A ubiquitous family of chaperone regulators. Cell. Mol. Life Sci. 2008, 65, 1390–1402. [Google Scholar] [CrossRef] [PubMed]
- Castro, A.; Saavedra, L.; Ruibal, C.; Lascano, R.; Vidal, S. Genome-wide identification, characterization and expression analysis of the Bcl-2 associated athanogene (BAG) gene family in Physcomitrium patens. BioRxiv 2020. [Google Scholar] [CrossRef]
- Nguyen, P.; Hess, K.; Smulders, L.; Le, D.; Briseno, C.; Chavez, C.M.; Nikolaidis, N. Origin and evolution of the human Bcl2-associated athanogene-1 (BAG-1). Int. J. Mol. Sci. 2020, 21, 9701. [Google Scholar] [CrossRef] [PubMed]
- Thanthrige, N.; Jain, S.; Bhowmik, S.D.; Ferguson, B.J.; Kabbage, M.; Mundree, S.; Williams, B. Centrality of BAGs in plant PCD, stress responses, and host defense. Trends Plant Sci. 2020, 25, 1131–1140. [Google Scholar] [CrossRef]
- Li, L.; Xing, Y.; Chang, D.; Fang, S.; Cui, B.; Li, Q.; Wang, X.; Guo, S.; Yang, X.; Men, S.; et al. CaM/BAG5/Hsc70 signaling complex dynamically regulates leaf senescence. Sci. Rep. 2016, 6, 31889. [Google Scholar] [CrossRef] [PubMed]
- Ren, K.; Feng, L.; Sun, S.; Zhuang, X. Plant mitophagy in comparison to mammals: What is still missing? Int. J. Mol. Sci. 2021, 27, 1236. [Google Scholar] [CrossRef] [PubMed]
- Nawkar, G.M.; Maibam, P.; Park, J.H.; Woo, S.G.; Kim, C.Y.; Lee, S.Y.; Kang, C.H. In silico study on Arabidopsis BAG gene expression in response to environmental stresses. Protoplasma 2017, 254, 409–421. [Google Scholar] [CrossRef]
- Lee, D.W.; Kim, S.J.; Oh, Y.J.; Choi, B.; Lee, J.; Hwang, I. Arabidopsis BAG1 functions as a cofactor in Hsc70-mediated proteasomal degradation of unimported plastid proteins. Mol. Plant 2016, 9, 1428–1431. [Google Scholar] [CrossRef]
- Fu, C.; Hou, Y.; Ge, J.; Zhang, L.; Liu, X.; Huo, P.; Liu, J. Increased fes1a thermotolerance is induced by BAG6 knockout. Plant Mol. Biol. 2019, 100, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Williams, B.; Kabbage, M.; Britt, R.; Dickman, M.B. AtBAG7, an Arabidopsis Bcl-2–associated athanogene, resides in the endoplasmic reticulum and is involved in the unfolded protein response. Proc. Natl. Acad. Sci. USA 2010, 107, 6088–6093. [Google Scholar] [CrossRef]
- You, Q.; Zhai, K.; Yang, D.; Yang, W.; Wu, J.; Liu, J.; Pan, W.; Wang, J.; Zhu, X.; Jian, Y.; et al. An E3 ubiquitin ligase-BAG protein module controls plant innate immunity and broad-spectrum disease resistance. Cell Host Microbe 2016, 20, 758–769. [Google Scholar] [CrossRef] [PubMed]
- Locascio, A.; Marqués, M.C.; García-Martínez, G.; Corratgé-Faillie, C.; Andrés-Colás, N.; Rubio, L.; Fernández, J.A.; Véry, A.A.; Mulet, J.M.; Yenush, L. BCL2-ASSOCIATED ATHANOGENE4 regulates the KAT1 potassium channel and controls stomatal movement. Plant Physiol. 2019, 181, 1277–1294. [Google Scholar] [CrossRef] [PubMed]
- Ren, Z.H.; Gao, J.; Li, L.G.; Cai, X.L.; Huang, W.; Chao, D.Y.; Zhu, M.Z.; Wang, Z.Y.; Luan, S.; Lin, H.X. A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat. Genet. 2005, 37, 1141–1146. [Google Scholar] [CrossRef]
- Wang, J.; Nan, N.; Li, N.; Liu, Y.; Wang, T.J.; Hwang, I.; Liu, B.; Xu, Z.Y. A DNA methylation reader-chaperone regulator-transcription factor complex activates OsHKT1;5 expression during salinity stress. Plant Cell 2020, 32, 3535–3558. [Google Scholar] [CrossRef] [PubMed]
- Fu, S.; Li, L.; Kang, H.; Yang, X.; Men, S.; Shen, Y. Chronic mitochondrial calcium elevation suppresses leaf senescence. Biochem. Biophys. Res. Commun. 2017, 487, 672–677. [Google Scholar] [CrossRef] [PubMed]
- Kang, C.H.; Jung, W.Y.; Kang, Y.H.; Kim, J.Y.; Kim, D.G.; Jeong, J.C.; Baek, D.W.; Jin, J.B.; Lee, J.Y.; Kim, M.O.; et al. AtBAG6, a novel calmodulin-binding protein, induces programmed cell death in yeast and plants. Cell Death Differ. 2006, 13, 84–95. [Google Scholar] [CrossRef]
- Li, Y.; Kabbage, M.; Liu, W.; Dickman, M.B. Aspartyl protease-mediated cleavage of BAG6 is necessary for autophagy and fungal resistance in plants. Plant Cell 2016, 28, 233–247. [Google Scholar] [CrossRef]
- Echevarría-Zomeño, S.; Fernández-Calvino, L.; Castro-Sanz, A.B.; López, J.A.; Vázquez, J.; Castellano, M.M. Dissecting the proteome dynamics of the early heat stress response leading to plant survival or death in Arabidopsis. Plant Cell Environ. 2016, 39, 1264–1278. [Google Scholar] [CrossRef]
- Zhang, J.X.; Wang, C.; Yang, C.Y.; Wang, J.Y.; Chen, L.; Bao, X.M.; Zhao, Y.X.; Zhang, H.; Liu, J. The role of Arabidopsis AtFes1A in cytosolic Hsp70 stability and abiotic stress tolerance. Plant J. 2010, 62, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Fu, C.; Zhang, J.; Liu, X.; Yang, W.; Yu, H.; Liu, J. AtFes1A is essential for highly efficient molecular chaperone function in Arabidopsis. J. Plant Biol. 2015, 59, 366–373. [Google Scholar] [CrossRef]
- Li, Y.; Williams, B.; Dickman, M. Arabidopsis B-cell lymphoma2 (Bcl-2)-associated athanogene 7 (BAG7)-mediated heat tolerance requires translocation, sumoylation and binding to WRKY 29. New Phytol. 2017, 214, 695–705. [Google Scholar] [CrossRef]
- Pan, Y.J.; Liu, L.; Lin, Y.C.; Zu, Y.G.; Li, L.P.; Tang, Z.H. Ethylene antagonizes salt-induced growth retardation and cell death process via transcriptional controlling of ethylene-, BAG- and senescence-associated genes in Arabidopsis. Front. Plant Sci. 2016, 7, 696. [Google Scholar] [CrossRef]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Characterization of the expression of a desiccation-responsive rd29 gene of Arabidopsis thaliana and analysis of its promoter in transgenic plants. Mol. Gen. Genet. 1993, 236, 331–340. [Google Scholar] [CrossRef]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 1994, 6, 251–264. [Google Scholar]
- Narusaka, Y.; Nakashima, K.; Shinwari, Z.K.; Sakuma, Y.; Furihata, T.; Abe, H.; Narusaka, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J. 2003, 34, 137–148. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, K.; Fujita, Y.; Katsura, K.; Maruyama, K.; Narusaka, Y.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Transcriptional regulation of ABI3- and ABA-responsive genes including RD29B and RD29A in seeds, germinating embryos, and seedlings of Arabidopsis. Plant Mol. Biol. 2006, 60, 51–68. [Google Scholar] [CrossRef] [PubMed]
- Iuchi, S.; Kobayashi, M.; Taji, T.; Naramoto, M.; Seki, M.; Kato, T.; Tabata, S.; Kakubari, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J. 2001, 27, 325–333. [Google Scholar] [CrossRef] [PubMed]
- Soderman, E.M.; Brocard, I.M.; Lynch, T.J.; Finkelstein, R.R. Regulation and function of the arabidopsis ABA-insensitive4 gene in seed and abscisic acid response signaling networks. Plant Physiol. 2000, 124, 1752–1765. [Google Scholar] [CrossRef]
- Kumpf, R.P.; Nowack, M.K. The root cap: A short story of life and death. J. Exp. Bot. 2015, 66, 5651–5662. [Google Scholar] [CrossRef]
- Hellmann, E.; Ko, D.; Ruonala, R.; Helariutta, Y. Plant vascular tissues-connecting tissue comes in all shapes. Plants 2018, 7, 109. [Google Scholar] [CrossRef]
- Bao, S.; Shen, G.; Li, G.; Liu, Z.; Arif, M.; Wei, Q.; Men, S. The Arabidopsis nucleoporin NUP1 is essential for megasporogenesis and early stages of pollen development. Plant Cell Rep. 2019, 38, 59–74. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Liu, H.; Liu, B.; Chen, X.; Zhu, H.; Zou, C.; Men, S. AUX1 acts upstream of PIN2 in regulating root gravitropism. Biochem. Biophys. Res. Commun. 2018, 507, 433–436. [Google Scholar] [CrossRef] [PubMed]
- Seo, K.I.; Lee, J.H.; Nezames, C.D.; Zhong, S.; Song, E.; Byun, M.O.; Deng, X.W. ABD1 is an Arabidopsis DCAF substrate receptor for CUL4-DDB1–based E3 ligases that acts as a negative regulator of abscisic acid signaling. Plant Cell 2014, 26, 695–711. [Google Scholar] [CrossRef] [PubMed]
- Arnon, D.I. Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol. 1949, 24, 1–15. [Google Scholar] [CrossRef]
- Yang, L.; Wang, S.; Sun, L.; Ruan, M.; Li, S.; He, R.; Zhang, W.; Liang, C.; Wang, X.; Bi, Y. Involvement of G6PD5 in ABA response during seed germination and root growth in Arabidopsis. BMC Plant Biol. 2019, 19, 44. [Google Scholar] [CrossRef]
- Wang, M.; Li, P.; Ma, Y.; Nie, X.; Grebe, M.; Men, S. Membrane sterol composition in Arabidopsis thaliana affects root elongation via auxin biosynthesis. Int. J. Mol. Sci. 2021, 22, 437. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Li, Z.; Yang, Z.; Leshem, Y.; Shen, Y.; Men, S. Mitochondrial heat shock cognate protein 70 contributes to auxinmediated embryo development. Plant. Physiol. 2021. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).