Cross-TCR Antagonism Revealed by Optogenetically Tuning the Half-Life of the TCR Ligand Binding
Abstract
1. Introduction
2. Results
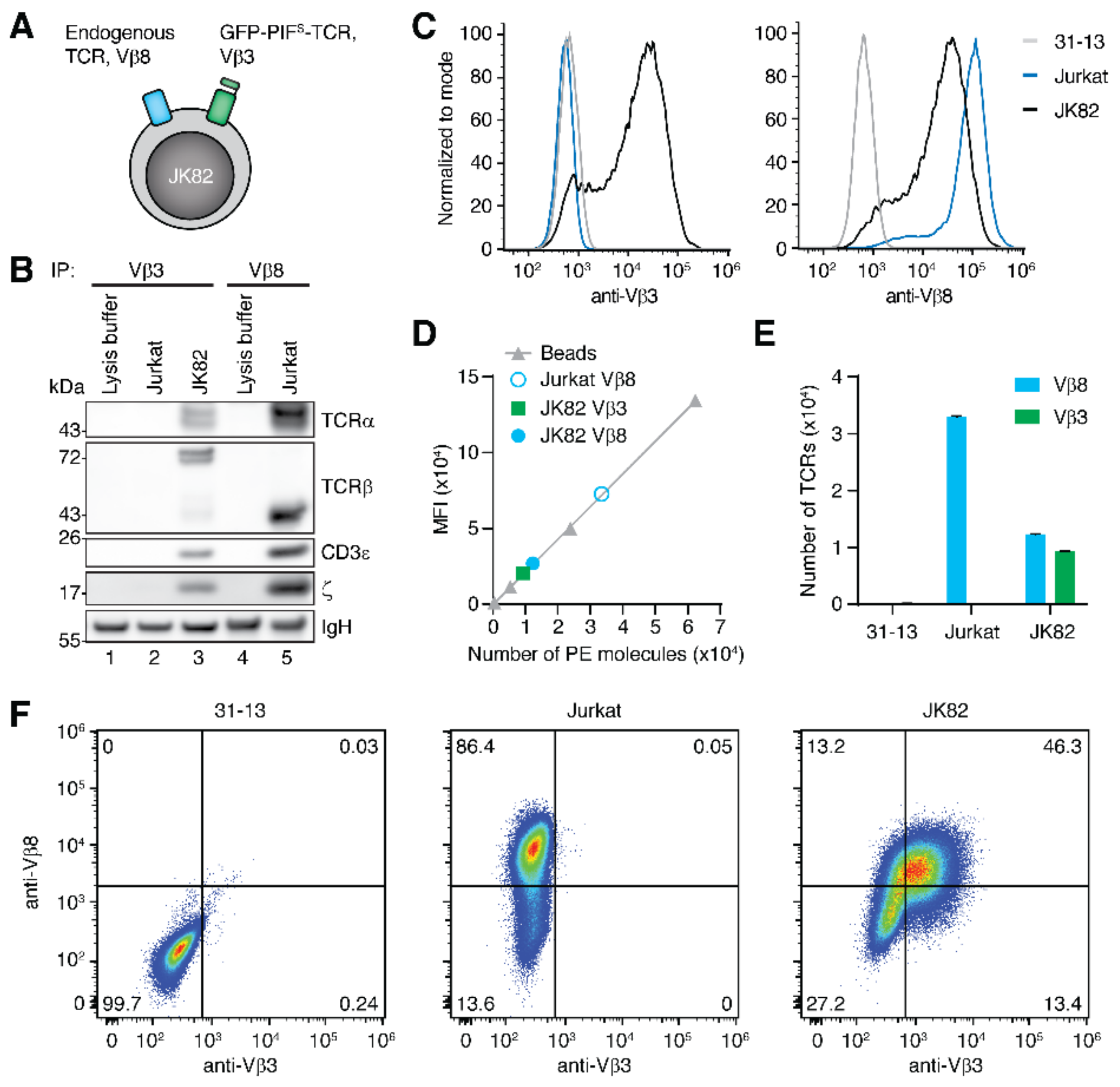
2.1. Characterization of Dual TCR Expressing Cells
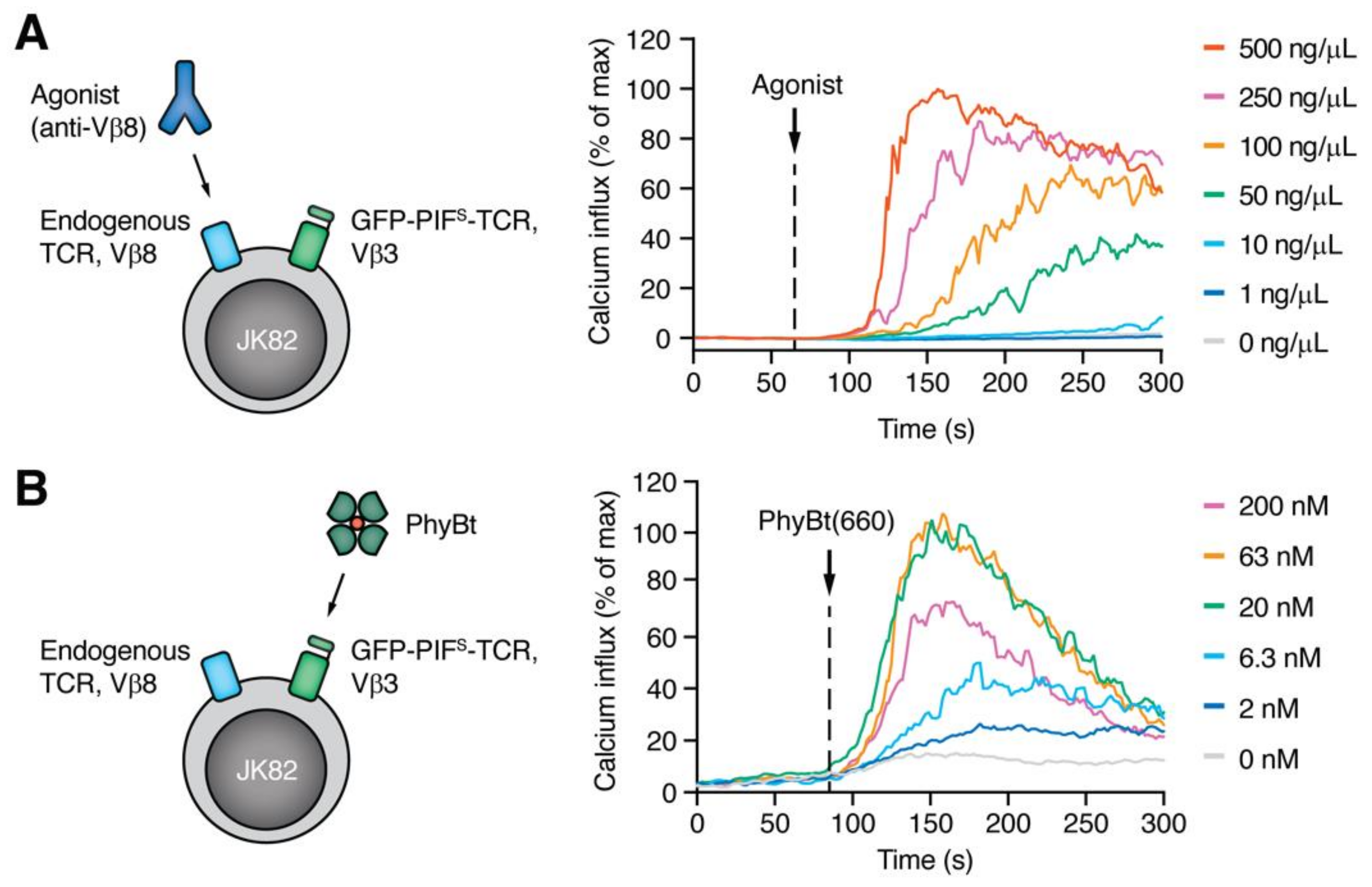
2.2. Stimulation of JK82 Cells with Agonistic Anti-Vβ8 Antibodies and PhyBt
2.3. Modulation of Agonistic Stimulation by Different Ligand Binding Half-Lives to the GFP-PIFS-TCR
2.4. Mathematical Model of Cross-Antagonism
2.5. Predictions of the Model
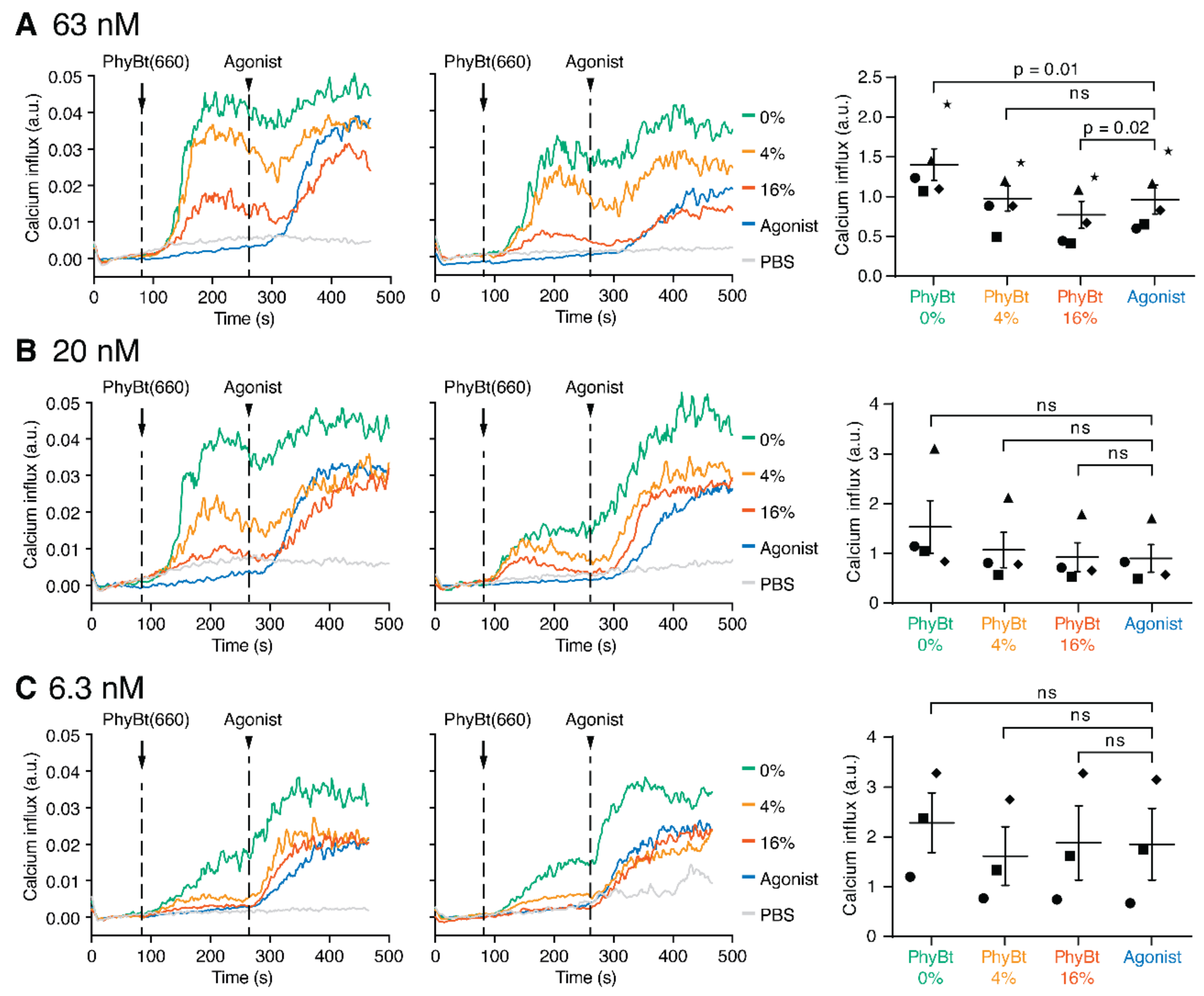
2.6. Cross-Antagonism Depends on the Concentration of the Antagonistic Ligand
3. Discussion
4. Materials and Methods
4.1. Molecular Cloning
4.2. Protein Production and Purification
4.3. Cell Line Generation and Cultivation
4.4. Calcium Influx Measurement
4.5. Cell Lysis and Immunoprecipitation
4.6. SDS-PAGE and Immunoblotting
4.7. Illumination Devices
4.8. Quantification of TCR Surface Expression
4.9. Repetition of Experiments and Data Presentation
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schamel, W.W.; Alarcon, B.; Minguet, S. The TCR is an allosterically regulated macromolecular machinery changing its conformation while working. Immunol. Rev. 2019, 291, 8–25. [Google Scholar] [CrossRef]
- Courtney, A.H.; Lo, W.L.; Weiss, A. TCR Signaling: Mechanisms of Initiation and Propagation. Trends Biochem. Sci. 2018, 43, 108–123. [Google Scholar] [CrossRef] [PubMed]
- Sloan-Lancaster, J.; Allen, P.M. Altered peptide ligand-induced partial T cell activation: Molecular mechanisms and role in T cell biology. Annu. Rev. Immunol. 1996, 14, 1–27. [Google Scholar] [CrossRef] [PubMed]
- De Magistris, M.T.; Alexander, J.; Coggeshall, M.; Altman, A.; Gaeta, F.C.; Grey, H.M.; Sette, A. Antigen analog-major histocompatibility complexes act as antagonists of the T cell receptor. Cell 1992, 68, 625–634. [Google Scholar] [CrossRef]
- Jameson, S.C.; Bevan, M.J. T cell receptor antagonists and partial agonists. Immunity 1995, 2, 1–11. [Google Scholar] [CrossRef]
- Daniels, M.A.; Schober, S.L.; Hogquist, K.A.; Jameson, S.C. Cutting edge: A test of the dominant negative signal model for TCR antagonism. J. Immunol. 1999, 162, 3761–3764. [Google Scholar]
- Stotz, S.H.; Bolliger, L.; Carbone, F.R.; Palmer, E. T cell receptor (TCR) antagonism without a negative signal: Evidence from T cell hybridomas expressing two independent TCRs. J. Exp. Med. 1999, 189, 253–264. [Google Scholar] [CrossRef]
- Dittel, B.N.; Stefanova, I.; Germain, R.N.; Janeway, C.A. Cross-antagonism of a T cell clone expressing two distinct T cell receptors. Immunity 1999, 11, 289–298. [Google Scholar] [CrossRef]
- Sykulev, Y.; Vugmeyster, Y.; Brunmark, A.; Ploegh, H.L.; Eisen, H.N. Peptide antagonism and T cell receptor interactions with peptide-MHC complexes. Immunity 1998, 9, 475–483. [Google Scholar] [CrossRef]
- Stefanova, I.; Hemmer, B.; Vergelli, M.; Martin, R.; Biddison, W.E.; Germain, R.N. TCR ligand discrimination is enforced by competing ERK positive and SHP-1 negative feedback pathways. Nat. Immunol. 2003, 4, 248–254. [Google Scholar] [CrossRef]
- Kilgore, N.E.; Carter, J.D.; Lorenz, U.; Evavold, B.D. Cutting Edge: Dependence of TCR Antagonism on Src Homology 2 Domain-Containing Protein Tyrosine Phosphatase Activity. J. Immunol. 2003, 170, 4891–4895. [Google Scholar] [CrossRef]
- Robertson, J.M.; Evavold, B.D. Cutting edge: Dueling TCRs: Peptide antagonism of CD4+ T cells with dual antigen specificities. J. Immunol. 1999, 163, 1750–1754. [Google Scholar]
- Yang, W.; Grey, H.M. Study of the Mechanism of TCR Antagonism Using Dual-TCR-Expressing T Cells. J. Immunol. 2003, 170, 4532–4538. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.S.; Reichardt, P.; Ford, M.L.; Edwards, L.J.; Evavold, B.D. TCR Antagonism by Peptide Requires High TCR Expression. J. Immunol. 2008, 181, 1760–1766. [Google Scholar] [CrossRef] [PubMed]
- Sykulev, Y.; Brunmark, A.; Jackson, M.; Cohen, R.J.; Peterson, P.A.; Eisen, H.N. Kinetics and affinity of reactions between an antigen-specific T cell receptor and peptide-MHC complexes. Immunity 1994, 1, 15–22. [Google Scholar] [CrossRef]
- Davis, M.M.; Boniface, J.J.; Reich, Z.; Lyons, D.; Hampl, J.; Arden, B.; Chien, Y. Ligand recognition by αβ T cell receptors. Annu. Rev. Immunol. 1998, 16, 523–544. [Google Scholar] [CrossRef]
- Matsui, K.; Boniface, J.J.; Steffner, P.; Reay, P.A.; Davis, M.M. Kinetics of T-cell receptor binding to peptide/I-Ek complexes: Correlation of the dissociation rate with T-cell responsiveness. Proc. Natl. Acad. Sci. USA 1994, 91, 12862–12866. [Google Scholar] [CrossRef] [PubMed]
- Alam, S.M.; Travers, P.J.; Wung, J.L.; Nasholds, W.; Redpath, S.J.S.C.; Gascoigne, N.R. T-cell-receptor affinity and thymocyte positive selection. Nature 1996, 381, 616–620. [Google Scholar] [CrossRef]
- Yoon, S.T.; Dianzani, U.; Bottomly, K.; Janeway, C.A.J. Both high and low avidity antibodies to the T cell receptor can have agonist or antagonist activity. Immunity 1994, 1, 563–569. [Google Scholar] [CrossRef]
- McKeithan, T.W. Kinetic proofreading in T-cell receptor signal transduction. Proc. Natl. Acad. Sci. USA 1995, 92, 5042–5046. [Google Scholar] [CrossRef] [PubMed]
- Aleksic, M.; Dushek, O.; Zhang, H.; Shenderov, E.; Chen, J.L.; Cerundolo, V.; Coombs, D.; van der Merwe, P.A. Dependence of T cell antigen recognition on T cell receptor-peptide MHC confinement time. Immunity 2010, 32, 163–174. [Google Scholar] [CrossRef]
- Govern, C.C.; Paczosa, M.K.; Chakraborty, A.K.; Huseby, E.S. Fast on-rates allow short dwell time ligands to activate T cells. Proc. Natl. Acad. Sci. USA 2010, 107, 8724–8729. [Google Scholar] [CrossRef]
- Lin, J.J.Y.; Low-Nam, S.T.; Alfieri, K.N.; McAffee, D.B.; Fay, N.C.; Groves, J.T. Mapping the stochastic sequence of individual ligand-receptor binding events to cellular activation: T cells act on the rare events. Sci. Signal. 2019, 12. [Google Scholar] [CrossRef] [PubMed]
- Adams, J.J.; Narayanan, S.; Liu, B.; Birnbaum, M.E.; Kruse, A.C.; Bowerman, N.A.; Chen, W.; Levin, A.M.; Connolly, J.M.; Zhu, C.; et al. T Cell Receptor Signaling Is Limited by Docking Geometry to Peptide-Major Histocompatibility Complex. Immunity 2011, 35, 681–693. [Google Scholar] [CrossRef] [PubMed]
- Gil, D.; Schamel, W.W.; Montoya, M.; Sanchez-Madrid, F.; Alarcon, B. Recruitment of Nck by CD3 epsilon reveals a ligand-induced conformational change essential for T cell receptor signaling and synapse formation. Cell 2002, 109, 901–912. [Google Scholar] [CrossRef]
- Risueno, R.M.; van Santen, H.M.; Alarcon, B. A conformational change senses the strength of T cell receptor-ligand interaction during thymic selection. Proc. Natl. Acad. Sci. USA 2006, 103, 9625–9630. [Google Scholar] [CrossRef]
- Dopfer, E.P.; Hartl, F.A.; Oberg, H.H.; Siegers, G.M.; Yousefi, O.S.; Kock, S.; Fiala, G.J.; Garcillan, B.; Sandstrom, A.; Alarcon, B.; et al. The CD3 Conformational Change in the gammadelta T Cell Receptor Is Not Triggered by Antigens but Can Be Enforced to Enhance Tumor Killing. Cell Rep. 2014, 7, 1704–1715. [Google Scholar] [CrossRef] [PubMed]
- Swamy, M.; Beck-Garcia, K.; Beck-Garcia, E.; Hartl, F.A.; Morath, A.; Yousefi, O.S.; Dopfer, E.P.; Molnar, E.; Schulze, A.K.; Blanco, R.; et al. A Cholesterol-Based Allostery Model of T Cell Receptor Phosphorylation. Immunity 2016, 44, 1091–1101. [Google Scholar] [CrossRef]
- Kim, S.T.; Takeuchi, K.; Sun, Z.Y.; Touma, M.; Castro, C.E.; Fahmy, A.; Lang, M.J.; Wagner, G.; Reinherz, E.L. The alphabeta T cell receptor is an anisotropic mechanosensor. J. Biol. Chem. 2009, 284, 31028–31037. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Chen, W.; Evavold, B.D.; Zhu, C. Accumulation of dynamic catch bonds between TCR and agonist peptide-MHC triggers T cell signaling. Cell 2014, 157, 357–368. [Google Scholar] [CrossRef]
- Yousefi, O.S.; Gunther, M.; Horner, M.; Chalupsky, J.; Wess, M.; Brandl, S.M.; Smith, R.W.; Fleck, C.; Kunkel, T.; Zurbriggen, M.D.; et al. Optogenetic control shows that kinetic proofreading regulates the activity of the T cell receptor. eLife 2019, 8, e42475. [Google Scholar] [CrossRef]
- Levskaya, A.; Weiner, O.D.; Lim, W.A.; Voigt, C.A. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature 2009, 461, 997–1001. [Google Scholar] [CrossRef] [PubMed]
- Toettcher, J.E.; Weiner, O.D.; Lim, W.A. Using optogenetics to interrogate the dynamic control of signal transmission by the Ras/Erk module. Cell 2013, 155, 1422–1434. [Google Scholar] [CrossRef] [PubMed]
- Bae, G.; Choi, G. Decoding of light signals by plant phytochromes and their interacting proteins. Annu. Rev. Plant Biol. 2008, 59, 281–311. [Google Scholar] [CrossRef] [PubMed]
- Kolar, K.; Knobloch, C.; Stork, H.; Znidaric, M.; Weber, W. OptoBase: A Web Platform for Molecular Optogenetics. ACS Synth. Biol. 2018, 7, 1825–1828. [Google Scholar] [CrossRef]
- Smith, R.W.; Helwig, B.; Westphal, A.H.; Pel, E.; Hörner, M.; Beyer, H.M.; Samodelov, S.L.; Weber, W.; Zurbriggen, M.D.; Borst, J.W.; et al. Unearthing the transition rates between photoreceptor conformers. BMC Syst. Biol. 2016, 10, 110. [Google Scholar] [CrossRef]
- Mancinelli, A.L. The physiology of phytochrome action. In Photomorphogenesis in Plants; Kendrick, R.E., Kronenberg, G.M.H., Eds.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 1994; pp. 211–269. [Google Scholar]
- Tischer, D.K.; Weiner, O.D. Light-based tuning of ligand half-life supports kinetic proofreading model of T cell signaling. eLife 2019, 8, e42498. [Google Scholar] [CrossRef]
- Baeuerle, P.A.; Ding, J.; Patel, E.; Thorausch, N.; Horton, H.; Gierut, J.; Scarfo, I.; Choudhary, R.; Kiner, O.; Krishnamurthy, J.; et al. Synthetic TRuC receptors engaging the complete T cell receptor for potent anti-tumor response. Nat. Commun. 2019, 10, 2087. [Google Scholar] [CrossRef]
- Hardy, I.R.; Schamel, W.W.; Baeuerle, P.A.; Getts, D.R.; Hofmeister, R. Implications of T cell receptor biology on the development of new T cell therapies for cancer. Immunotherapy 2020, 12, 89–103. [Google Scholar] [CrossRef] [PubMed]
- Minami, Y.; Samelson, L.E.; Klausner, R.D. Internalization and cycling of the T cell antigen receptor. Role of protein kinase C. J. Biol. Chem. 1987, 262, 13342–13347. [Google Scholar] [CrossRef]
- De Waal Malefyt, R.; Alarcon, B.; Yssel, H.; Sancho, J.; Miyajima, A.; Terhorst, C.P.; Spits, H.; De Vries, J.E. Introduction of T cell receptor (TCR)-alpha cDNA has differential effects on TCR-gamma delta/CD3 expression by PEER and Lyon-1 cells. J. Immunol. 1989, 142, 3634–3642. [Google Scholar]
- Hörner, M.; Yousefi, O.S.; Schamel, W.W.; Weber, W. Production, Purification and Characterization of Recombinant Biotinylated Phytochrome B for Extracellular Optogenetics. Bio-Protocol 2020, 10, e3541. [Google Scholar] [CrossRef]
- Yousefi, O.S.; Hörner, M.; Wess, M.; Idstein, V.; Weber, W.; Schamel, W.W. Optogenetic Tuning of Ligand Binding to The Human T cell Receptor Using The opto-ligand-TCR System. Bio-Protocol 2020, 10, e3540. [Google Scholar] [CrossRef]
- Chan, A.C.; Iwashima, M.; Turck, C.W.; Weiss, A. ZAP-70: A 70 kd protein-tyrosine kinase that associates with the TCR zeta chain. Cell 1992, 71, 649–662. [Google Scholar] [CrossRef]
- Chakraborty, A.K.; Weiss, A. Insights into the initiation of TCR signaling. Nat. Immunol. 2014, 15, 798–807. [Google Scholar] [CrossRef] [PubMed]
- Rabinowitz, J.D.; Beeson, C.; Lyons, D.S.; Davis, M.M.; McConnell, H.M. Kinetic discrimination in T-cell activation. Proc. Natl. Acad. Sci. USA 1996, 93, 1401–1405. [Google Scholar] [CrossRef] [PubMed]
- Pettmann, J.; Abu-Shah, E.; Kutuzov, M.; Wilson, D.B.; Dustin, M.L.; Davis, S.J.; van der Merwe, P.A.; Dushek, O. T cells exhibit unexpectedly low discriminatory power and can respond to ultra-low affinity peptide-MHC ligands. bioRxiv 2021. [Google Scholar] [CrossRef]
- Lever, M.; Maini, P.K.; van der Merwe, P.A.; Dushek, O. Phenotypic models of T cell activation. Nat. Rev. Immunol. 2014, 14, 619–629. [Google Scholar] [CrossRef]
- Francois, P.; Voisinne, G.; Siggia, E.D.; Altan-Bonnet, G.; Vergassola, M. Phenotypic model for early T-cell activation displaying sensitivity, specificity, and antagonism. Proc. Natl. Acad. Sci. USA 2013, 110, E888–E897. [Google Scholar] [CrossRef]
- Schamel, W.W.; Arechaga, I.; Risueno, R.M.; van Santen, H.M.; Cabezas, P.; Risco, C.; Valpuesta, J.M.; Alarcon, B. Coexistence of multivalent and monovalent TCRs explains high sensitivity and wide range of response. J. Exp. Med. 2005, 202, 493–503. [Google Scholar] [CrossRef] [PubMed]
- Lillemeier, B.F.; Mortelmaier, M.A.; Forstner, M.B.; Huppa, J.B.; Groves, J.T.; Davis, M.M. TCR and Lat are expressed on separate protein islands on T cell membranes and concatenate during activation. Nat. Immunol. 2010, 11, 90–96. [Google Scholar] [CrossRef]
- Schamel, W.W.; Alarcon, B. Organization of the resting TCR in nanoscale oligomers. Immunol. Rev. 2013, 251, 13–20. [Google Scholar] [CrossRef]
- Wylie, D.C.; Das, J.; Chakraborty, A.K. Sensitivity of T cells to antigen and antagonism emerges from differential regulation of the same molecular signaling module. Proc. Natl. Acad. Sci. USA 2007, 104, 5533–5538. [Google Scholar] [CrossRef]
- Altan-Bonnet, G.; Germain, R.N. Modeling T cell antigen discrimination based on feedback control of digital ERK responses. PLoS Biol. 2005, 3, e356. [Google Scholar] [CrossRef] [PubMed]
- Carreno, L.J.; Riquelme, E.M.; Gonzalez, P.A.; Espagnolle, N.; Riedel, C.A.; Valitutti, S.; Kalergis, A.M. T-cell antagonism by short half-life pMHC ligands can be mediated by an efficient trapping of T-cell polarization toward the APC. Proc. Natl. Acad. Sci. USA 2009, 107, 210–215. [Google Scholar] [CrossRef] [PubMed]
- Lyons, D.; Lieberman, S.; Hampl, J.; Boniface, J.; Chien, Y.; Berg, L.; Davis, M. A TCR binds to antagonist ligands with lower affinities and faster dissociation rates than to agonists. Immunity 1996, 5, 53–61. [Google Scholar] [CrossRef]
- Stone, J.D.; Aggen, D.H.; Chervin, A.S.; Narayanan, S.; Schmitt, T.M.; Greenberg, P.D.; Kranz, D.M. Opposite Effects of Endogenous Peptide–MHC Class I on T Cell Activity in the Presence and Absence of CD8. J. Immunol. 2011, 186, 5193–5200. [Google Scholar] [CrossRef] [PubMed]
- Purbhoo, M.A.; Irvine, D.J.; Huppa, J.B.; Davis, M.M. T cell killing does not require the formation of a stable mature immunological synapse. Nat. Immunol. 2004, 5, 524–530. [Google Scholar] [CrossRef]
- Minguet, S.; Swamy, M.; Alarcon, B.; Luescher, I.F.; Schamel, W.W. Full activation of the T cell receptor requires both clustering and conformational changes at CD3. Immunity 2007, 26, 43–54. [Google Scholar] [CrossRef]
- Lever, M.; Lim, H.S.; Kruger, P.; Nguyen, J.; Trendel, N.; Abu-Shah, E.; Maini, P.K.; van der Merwe, P.A.; Dushek, O. Architecture of a minimal signaling pathway explains the T-cell response to a 1 million-fold variation in antigen affinity and dose. Proc. Natl. Acad. Sci. USA 2016, 113, E6630–E6638. [Google Scholar] [CrossRef]
- Letourneur, F.; Gabert, J.; Cosson, P.; Blanc, D.; Davoust, J.; Malissen, B. A signaling role for the cytoplasmic segment of the CD8 alpha chain detected under limiting stimulatory conditions. Proc. Natl. Acad. Sci. USA 1990, 87, 2339–2343. [Google Scholar] [CrossRef] [PubMed]
- Zamoyska, R.; Derham, P.; Gorman, S.D.; von Hoegen, P.; Bolen, J.B.; Veillette, A.; Parnes, J.R. Inability of CD8 alpha’ polypeptides to associate with p56lck correlates with impaired function in vitro and lack of expression in vivo. Nature 1989, 342, 278–281. [Google Scholar] [CrossRef]
- Wang, R.; Natarajan, K.; Margulies, D.H. Structural basis of the CD8αβ/MHCI interaction: Focused recognition orients CD8β to a T cell proximal position1. J. Immunol. 2009, 183, 2554–2564. [Google Scholar] [CrossRef]
- Hartl, F.A.; Beck-Garcia, E.; Woessner, N.M.; Flachsmann, L.J.; Cardenas, R.M.V.; Brandl, S.M.; Taromi, S.; Fiala, G.J.; Morath, A.; Yousefi, O.S.; et al. Noncanonical binding of Lck to CD3ε promotes TCR signaling and CAR function. Nat. Immunol. 2020, 21, 902–913. [Google Scholar] [CrossRef] [PubMed]
- Katz, Z.B.; Novotna, L.; Blount, A.; Lillemeier, B.F. A cycle of Zap70 kinase activation and release from the TCR amplifies and disperses antigenic stimuli. Nat. Immunol. 2017, 18, 86–95. [Google Scholar] [CrossRef]
- Weiss, A.; Stobo, J.D. Requirement for the coexpression of T3 and the T cell antigen receptor on a malignant human T cell line. J. Exp. Med. 1984, 160, 1284–1299. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yousefi, O.S.; Ruggieri, M.; Idstein, V.; von Prillwitz, K.U.; Herr, L.A.; Chalupsky, J.; Köhn, M.; Weber, W.; Timmer, J.; Schamel, W.W.A. Cross-TCR Antagonism Revealed by Optogenetically Tuning the Half-Life of the TCR Ligand Binding. Int. J. Mol. Sci. 2021, 22, 4920. https://doi.org/10.3390/ijms22094920
Yousefi OS, Ruggieri M, Idstein V, von Prillwitz KU, Herr LA, Chalupsky J, Köhn M, Weber W, Timmer J, Schamel WWA. Cross-TCR Antagonism Revealed by Optogenetically Tuning the Half-Life of the TCR Ligand Binding. International Journal of Molecular Sciences. 2021; 22(9):4920. https://doi.org/10.3390/ijms22094920
Chicago/Turabian StyleYousefi, Omid Sascha, Matias Ruggieri, Vincent Idstein, Kai Uwe von Prillwitz, Laurenz A. Herr, Julia Chalupsky, Maja Köhn, Wilfried Weber, Jens Timmer, and Wolfgang W. A. Schamel. 2021. "Cross-TCR Antagonism Revealed by Optogenetically Tuning the Half-Life of the TCR Ligand Binding" International Journal of Molecular Sciences 22, no. 9: 4920. https://doi.org/10.3390/ijms22094920
APA StyleYousefi, O. S., Ruggieri, M., Idstein, V., von Prillwitz, K. U., Herr, L. A., Chalupsky, J., Köhn, M., Weber, W., Timmer, J., & Schamel, W. W. A. (2021). Cross-TCR Antagonism Revealed by Optogenetically Tuning the Half-Life of the TCR Ligand Binding. International Journal of Molecular Sciences, 22(9), 4920. https://doi.org/10.3390/ijms22094920





