Trypsin Induced Degradation of Amyloid Fibrils
Abstract
1. Introduction
2. Results
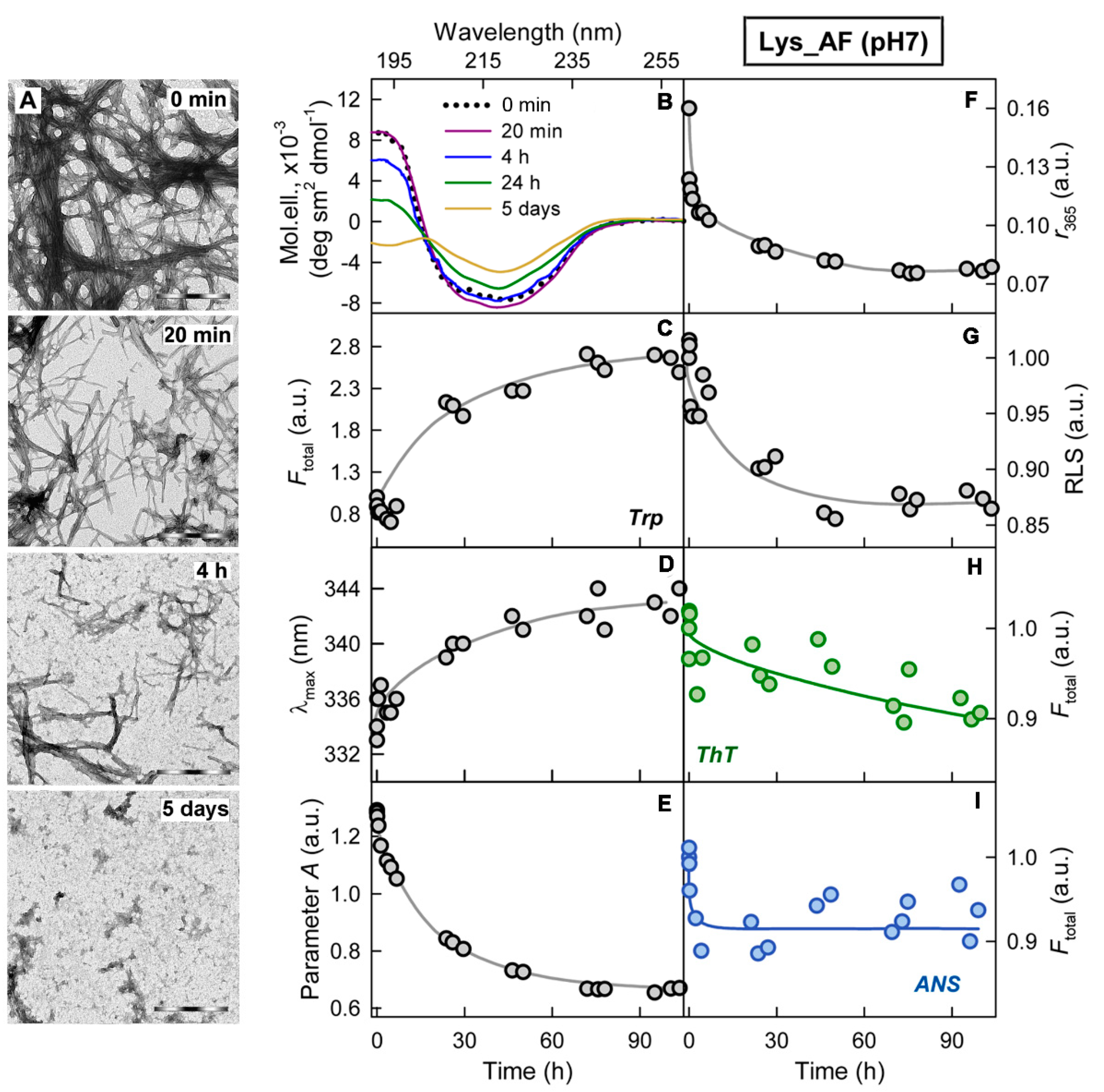
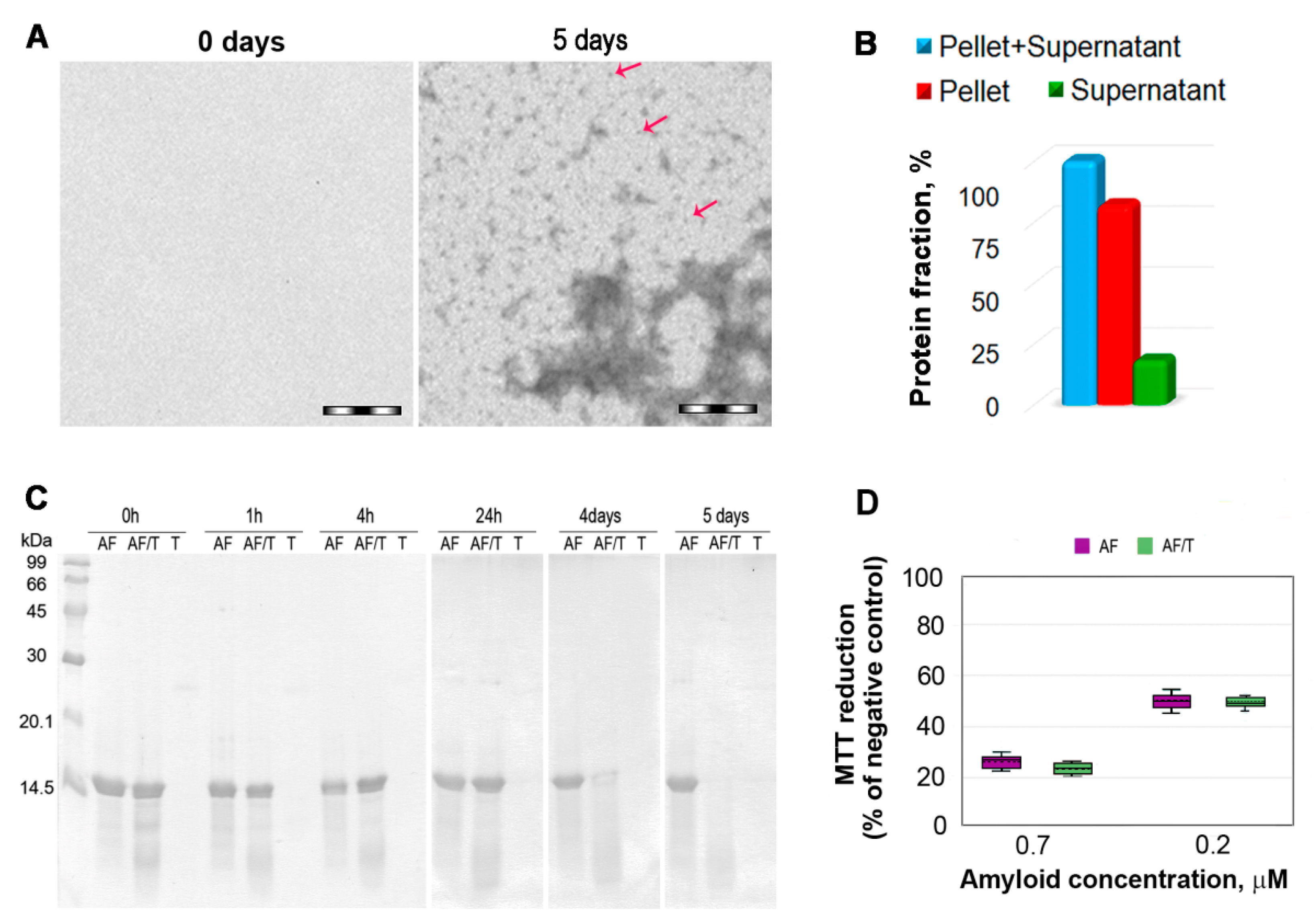
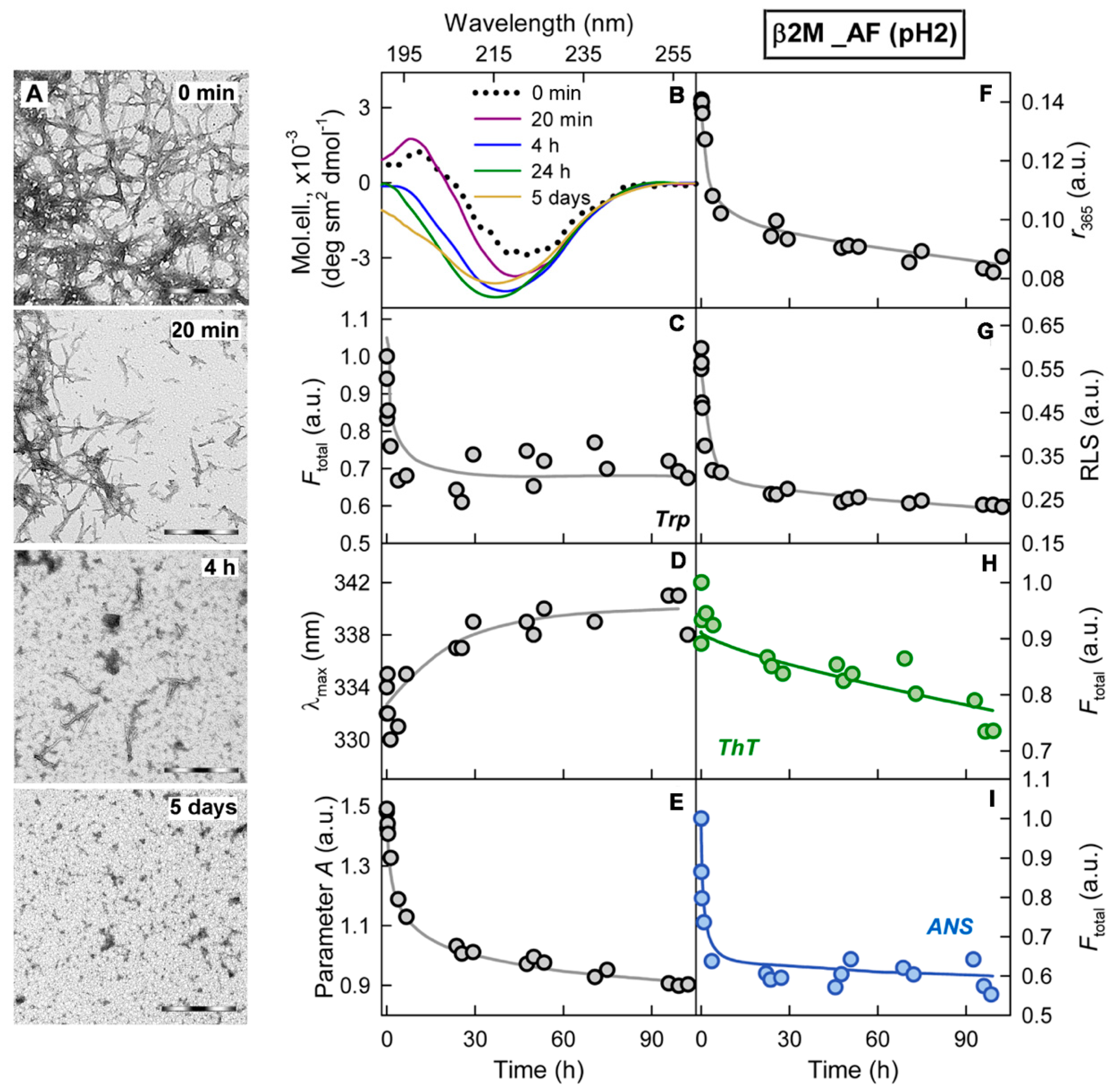
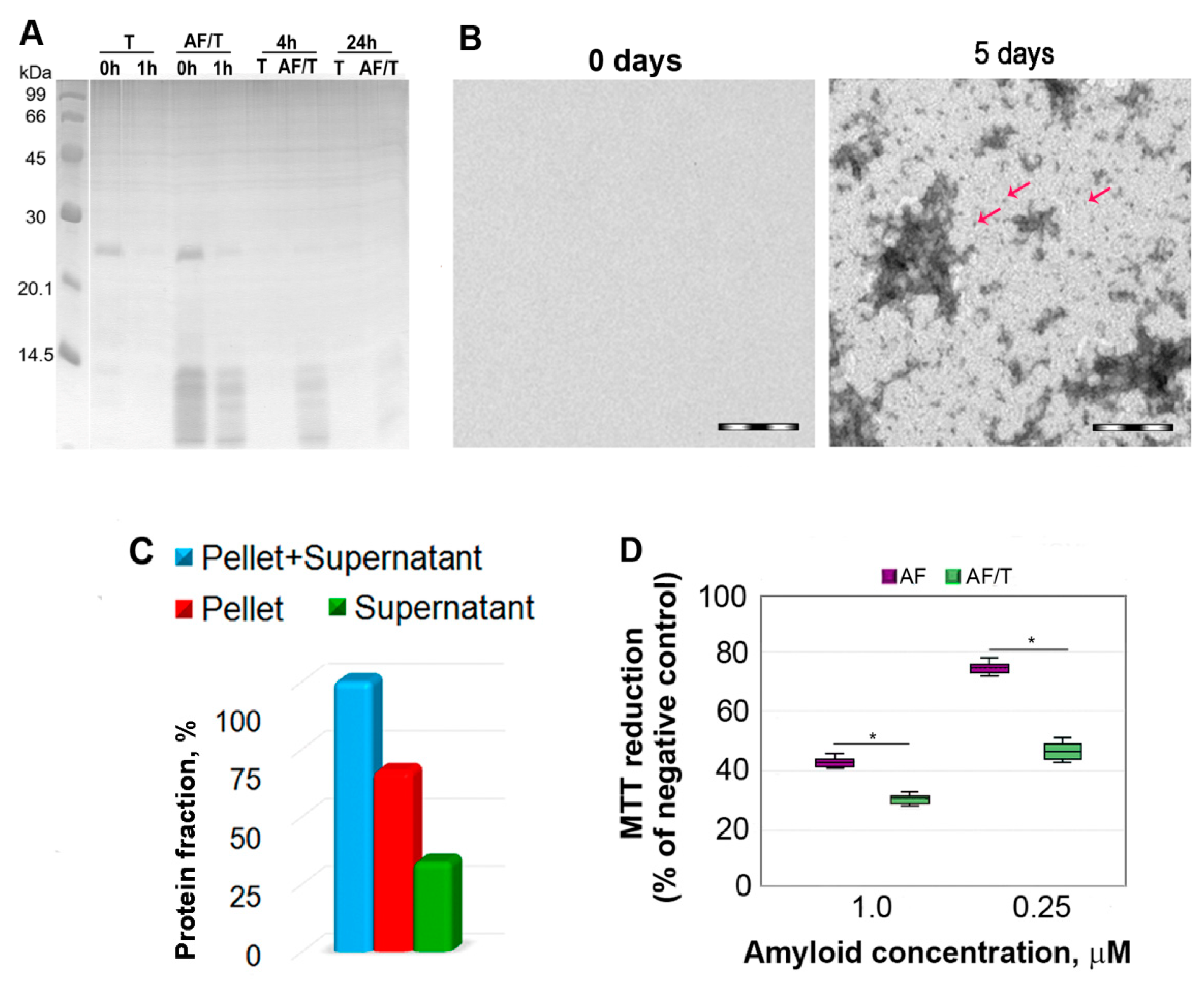
2.1. Mechanism of Amyloid Fibril Degradation Induced by Trypsin: Fragmentation, Disordering, and Monomerization
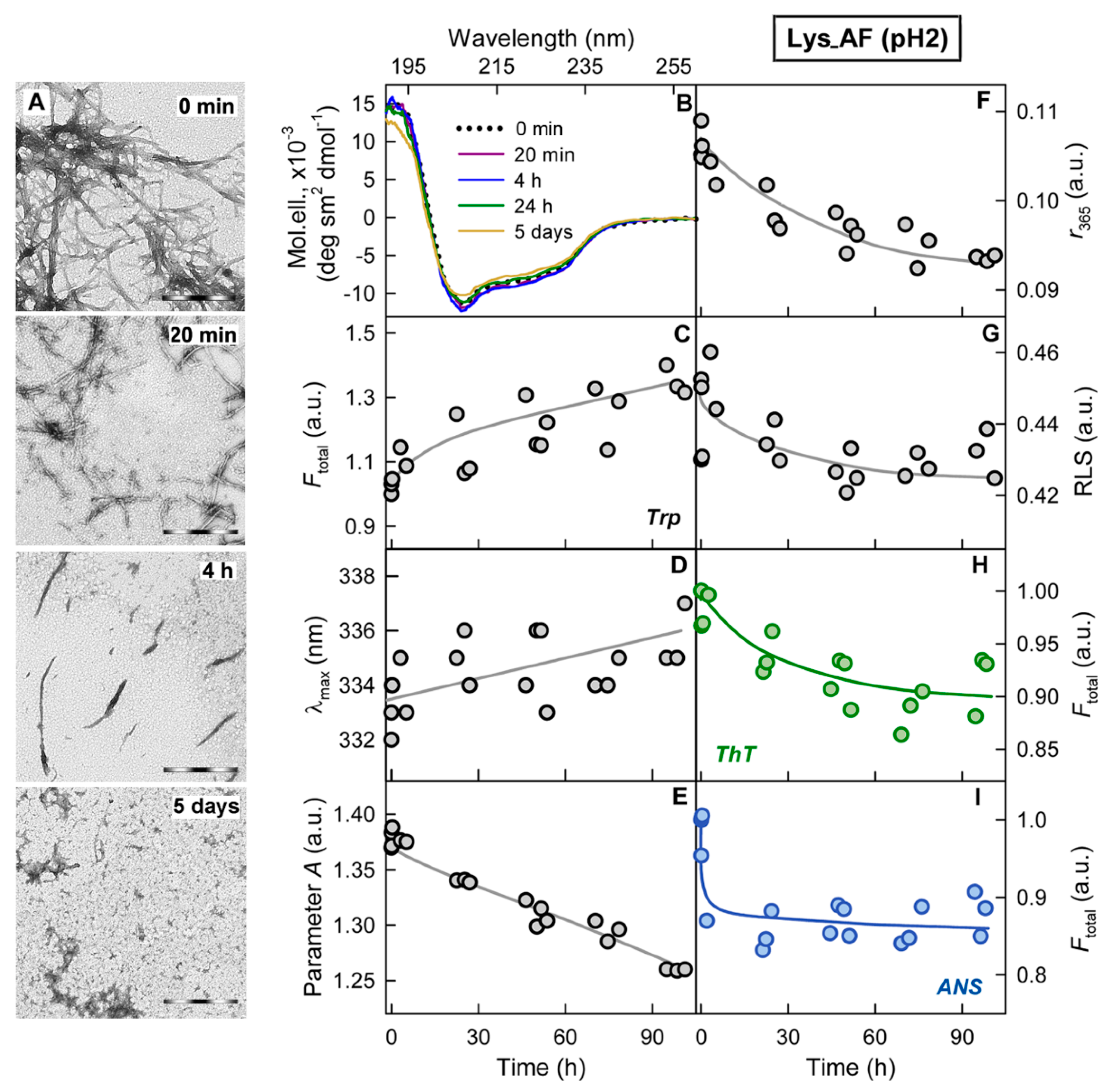
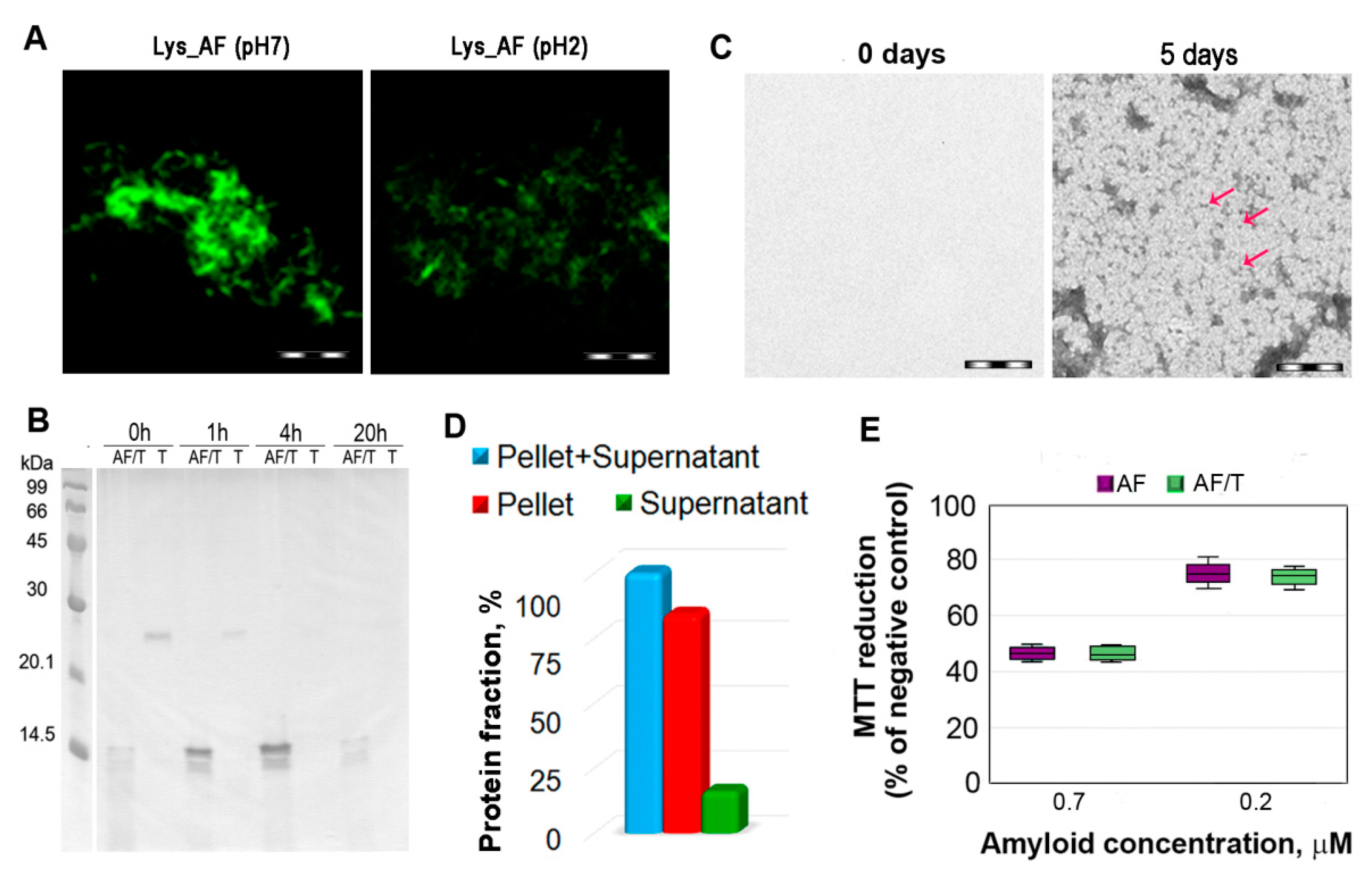
2.2. Relationship of Amyloid Fibrils Degradation Induced by Trypsin and Their Polymorphism
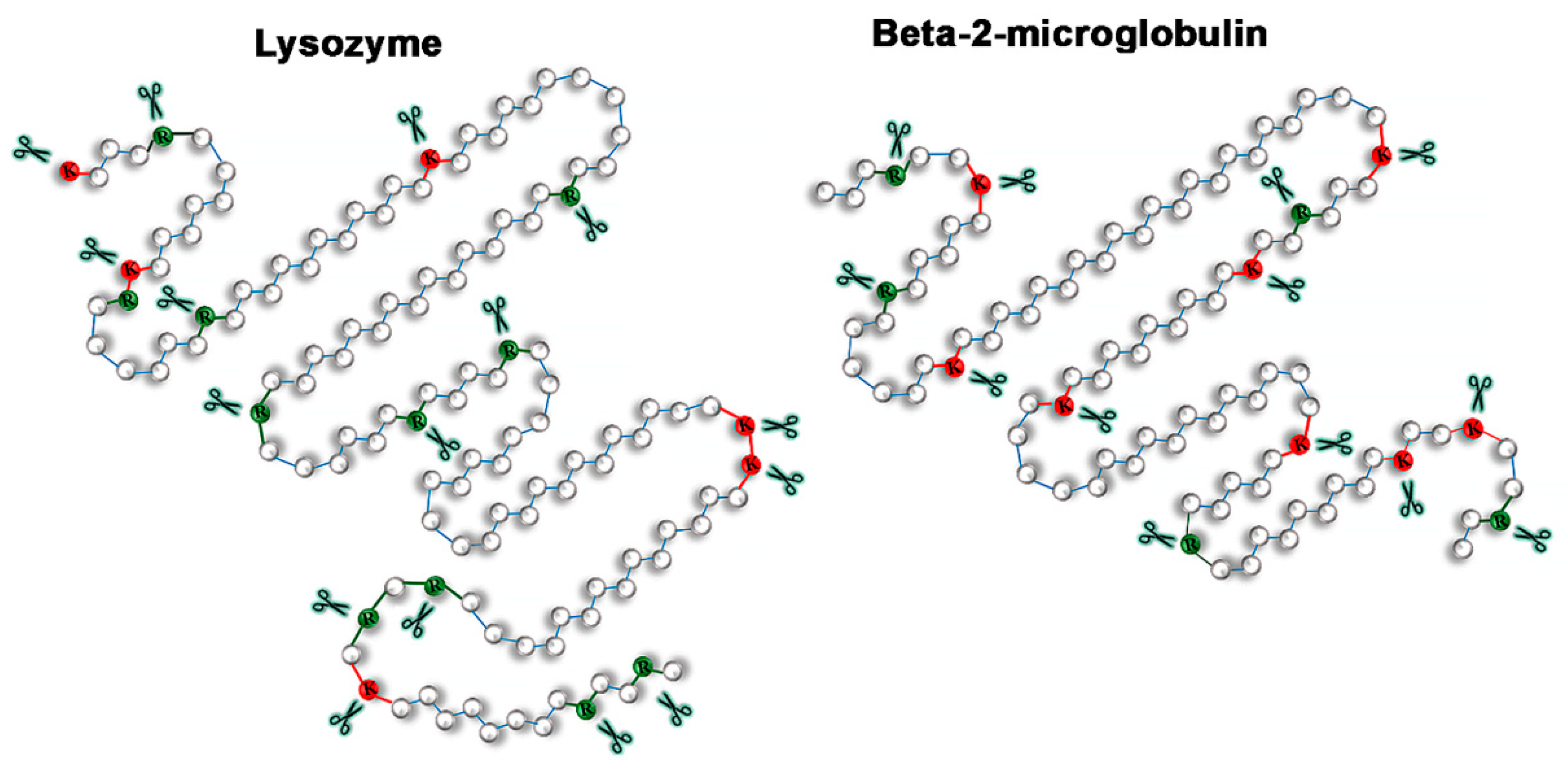
2.3. Relationship of Amyloid Fibrils Degradation Induced by Trypsin and Primary Structure of Amyloid-Forming Protein
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Amyloid Fibrils Preparation
4.3. Amyloid Fibrils Degradation by Trypsin
4.4. Transmission Electron Microscopy
4.5. Confocal Microscopy
4.6. Spectral Measurements
4.7. MTT Assay
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Merlini, G.; Bellotti, V. Molecular mechanisms of amyloidosis. N. Engl. J. Med. 2003, 349, 583–596. [Google Scholar] [CrossRef] [PubMed]
- Recchia, A.; Debetto, P.; Negro, A.; Guidolin, D.; Skaper, S.D.; Giusti, P. Alpha-synuclein and Parkinson’s disease. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2004, 18, 617–626. [Google Scholar] [CrossRef]
- Lu, J.X.; Qiang, W.; Yau, W.M.; Schwieters, C.D.; Meredith, S.C.; Tycko, R. Molecular structure of beta-amyloid fibrils in Alzheimer’s disease brain tissue. Cell 2013, 154, 1257–1268. [Google Scholar] [CrossRef] [PubMed]
- Naiki, H.; Okoshi, T.; Ozawa, D.; Yamaguchi, I.; Hasegawa, K. Molecular pathogenesis of human amyloidosis: Lessons from beta2 -microglobulin-related amyloidosis. Pathol. Int. 2016, 66, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Sipe, J.D.; Benson, M.D.; Buxbaum, J.N.; Ikeda, S.I.; Merlini, G.; Saraiva, M.J.; Westermark, P. Amyloid fibril proteins and amyloidosis: Chemical identification and clinical classification International Society of Amyloidosis 2016 Nomenclature Guidelines. Amyloid Int. J. Exp. Clin. Investig. Off. J. Int. Soc. Amyloidosis 2016, 23, 209–213. [Google Scholar] [CrossRef] [PubMed]
- Esposito, G.; Michelutti, R.; Verdone, G.; Viglino, P.; Hernandez, H.; Robinson, C.V.; Amoresano, A.; Dal Piaz, F.; Monti, M.; Pucci, P.; et al. Removal of the N-terminal hexapeptide from human beta2-microglobulin facilitates protein aggregation and fibril formation. Protein Sci. A Publ. Protein Soc. 2000, 9, 831–845. [Google Scholar] [CrossRef] [PubMed]
- Sattianayagam, P.T.; Gibbs, S.D.; Rowczenio, D.; Pinney, J.H.; Wechalekar, A.D.; Gilbertson, J.A.; Hawkins, P.N.; Lachmann, H.J.; Gillmore, J.D. Hereditary lysozyme amyloidosis—phenotypic heterogeneity and the role of solid organ transplantation. J. Intern. Med. 2012, 272, 36–44. [Google Scholar] [CrossRef]
- Wilquet, V.; De Strooper, B. Amyloid-beta precursor protein processing in neurodegeneration. Curr. Opin. Neurobiol. 2004, 14, 582–588. [Google Scholar] [CrossRef] [PubMed]
- Gebbink, M.F.; Voest, E.E.; Reijerkerk, A. Do antiangiogenic protein fragments have amyloid properties? Blood 2004, 104, 1601–1605. [Google Scholar] [CrossRef]
- Zhang, H.; Ma, Q.; Zhang, Y.W.; Xu, H. Proteolytic processing of Alzheimer’s beta-amyloid precursor protein. J. Neurochem. 2012, 120, 9–21. [Google Scholar] [CrossRef]
- Hogl, S.; Kuhn, P.H.; Colombo, A.; Lichtenthaler, S.F. Determination of the proteolytic cleavage sites of the amyloid precursor-like protein 2 by the proteases ADAM10, BACE1 and gamma-secretase. PLoS ONE 2011, 6, e21337. [Google Scholar] [CrossRef] [PubMed]
- Mirgorodskaia, O.A.; Kazanina, G.A.; Mirgorodskaia, E.P.; Shevchenko, A.A.; Mal’tsev, K.V.; Miroshnikov, A.I.; Roipstorff, P. Proteolysis of human proinsulin catalysed by native, modified, and immobilized trypsin. Bioorg. Khimiia 1997, 23, 91–97. [Google Scholar]
- Gao, Y.Z.; Xu, H.H.; Ju, T.T.; Zhao, X.H. The effect of limited proteolysis by different proteases on the formation of whey protein fibrils. J. Dairy Sci. 2013, 96, 7383–7392. [Google Scholar] [CrossRef]
- Mangione, P.P.; Porcari, R.; Gillmore, J.D.; Pucci, P.; Monti, M.; Porcari, M.; Giorgetti, S.; Marchese, L.; Raimondi, S.; Serpell, L.C.; et al. Proteolytic cleavage of Ser52Pro variant transthyretin triggers its amyloid fibrillogenesis. Proc. Natl. Acad. Sci. USA 2014, 111, 1539–1544. [Google Scholar] [CrossRef] [PubMed]
- Marcoux, J.; Mangione, P.P.; Porcari, R.; Degiacomi, M.T.; Verona, G.; Taylor, G.W.; Giorgetti, S.; Raimondi, S.; Sanglier-Cianferani, S.; Benesch, J.L.; et al. A novel mechano-enzymatic cleavage mechanism underlies transthyretin amyloidogenesis. EMBO Mol. Med. 2015, 7, 1337–1349. [Google Scholar] [CrossRef] [PubMed]
- Shin, T.M.; Isas, J.M.; Hsieh, C.L.; Kayed, R.; Glabe, C.G.; Langen, R.; Chen, J. Formation of soluble amyloid oligomers and amyloid fibrils by the multifunctional protein vitronectin. Mol. Neurodegener. 2008, 3, 16. [Google Scholar] [CrossRef]
- Poepsel, S.; Sprengel, A.; Sacca, B.; Kaschani, F.; Kaiser, M.; Gatsogiannis, C.; Raunser, S.; Clausen, T.; Ehrmann, M. Determinants of amyloid fibril degradation by the PDZ protease HTRA1. Nat. Chem. Biol. 2015, 11, 862–869. [Google Scholar] [CrossRef] [PubMed]
- Soderberg, L.; Dahlqvist, C.; Kakuyama, H.; Thyberg, J.; Ito, A.; Winblad, B.; Naslund, J.; Tjernberg, L.O. Collagenous Alzheimer amyloid plaque component assembles amyloid fibrils into protease resistant aggregates. FEBS J. 2005, 272, 2231–2236. [Google Scholar] [CrossRef]
- Chander, H.; Chauhan, A.; Wegiel, J.; Malik, M.; Sheikh, A.; Chauhan, V. Binding of trypsin to fibrillar amyloid beta-protein. Brain Res. 2006, 1082, 173–181. [Google Scholar] [CrossRef]
- Azevedo, E.P.; Guimaraes-Costa, A.B.; Torezani, G.S.; Braga, C.A.; Palhano, F.L.; Kelly, J.W.; Saraiva, E.M.; Foguel, D. Amyloid fibrils trigger the release of neutrophil extracellular traps (NETs), causing fibril fragmentation by NET-associated elastase. J. Biol. Chem. 2012, 287, 37206–37218. [Google Scholar] [CrossRef]
- Emeson, E.E.; Kikkawa, Y.; Gueft, B. New features of amyloid found after digestion with trypsin. J. Cell Biol. 1966, 28, 570–577. [Google Scholar] [CrossRef]
- Bezawork-Geleta, A.; Brodie, E.J.; Dougan, D.A.; Truscott, K.N. LON is the master protease that protects against protein aggregation in human mitochondria through direct degradation of misfolded proteins. Sci. Rep. 2015, 5, 17397. [Google Scholar] [CrossRef] [PubMed]
- Wagner, I.; Arlt, H.; van Dyck, L.; Langer, T.; Neupert, W. Molecular chaperones cooperate with PIM1 protease in the degradation of misfolded proteins in mitochondria. EMBO J. 1994, 13, 5135–5145. [Google Scholar] [CrossRef] [PubMed]
- Ninagawa, S.; Mori, K. Trypsin Sensitivity Assay to Study the Folding Status of Proteins. BIO-PROTOCOL 2016, 6. [Google Scholar] [CrossRef]
- Kay, J. A possible role for neutral proteolysis in the degradation of intracellular proteins. Ciba Found. Symp. 1979, 219–225. [Google Scholar] [CrossRef]
- Sanchez-Ruiz, J.M. Protein kinetic stability. Biophys. Chem. 2010, 148, 1–15. [Google Scholar] [CrossRef]
- Merulla, J.; Fasana, E.; Solda, T.; Molinari, M. Specificity and regulation of the endoplasmic reticulum-associated degradation machinery. Traffic 2013, 14, 767–777. [Google Scholar] [CrossRef]
- Pleyer, C.; Flesche, J.; Saeed, F. Lysozyme amyloidosis-a case report and review of the literature. Clin. Nephrol. Case Stud. 2015, 3, 42–45. [Google Scholar] [CrossRef]
- Scarpioni, R.; Ricardi, M.; Albertazzi, V.; De Amicis, S.; Rastelli, F.; Zerbini, L. Dialysis-related amyloidosis: Challenges and solutions. Int. J. Nephrol. Renov. Dis. 2016, 9, 319–328. [Google Scholar] [CrossRef]
- Sulatskaya, A.I.; Rodina, N.P.; Kuznetsova, I.M.; Turoverov, K.K. Different conditions of fibrillogenesis cause polymorphysm of lysozyme amyloid fibrils. J. Mol. Struct. 2017, 1140, 52–58. [Google Scholar] [CrossRef]
- Sipos, T.; Merkel, J.R. An effect of calcium ions on the activity, heat stability, and structure of trypsin. Biochemistry 1970, 9, 2766–2775. [Google Scholar] [CrossRef]
- Kolodzeiskaia, M.V. Serine proteinases of lower vertebrates. Ukr. Biokhimicheskii Zhurnal 1986, 58, 90–104. [Google Scholar]
- Buck, F.F.; Vithayathil, A.J.; Bier, M.; Nord, F.F. On the mechanism of enzyme action. 73. Studies on trypsins from beef, sheep and pig pancreas. Arch. Biochem. Biophys. 1962, 97, 417–424. [Google Scholar] [CrossRef]
- Stepanenko, O.V.; Sulatsky, M.I.; Mikhailova, E.V.; Stepanenko, O.V.; Povarova, O.I.; Kuznetsova, I.M.; Turoverov, K.K.; Sulatskaya, A.I. Alpha-B-Crystallin Effect on Mature Amyloid Fibrils: Different Degradation Mechanisms and Changes in Cytotoxicity. Int. J. Mol. Sci. 2020, 21, 7659. [Google Scholar] [CrossRef] [PubMed]
- Myers, S.L.; Thomson, N.H.; Radford, S.E.; Ashcroft, A.E. Investigating the structural properties of amyloid-like fibrils formed in vitro from beta2-microglobulin using limited proteolysis and electrospray ionisation mass spectrometry. Rapid Commun. Mass Spectrom. RCM 2006, 20, 1628–1636. [Google Scholar] [CrossRef]
- Monti, M.; Amoresano, A.; Giorgetti, S.; Bellotti, V.; Pucci, P. Limited proteolysis in the investigation of beta2-microglobulin amyloidogenic and fibrillar states. Biochim. Biophys. Acta 2005, 1753, 44–50. [Google Scholar] [CrossRef]
- Relini, A.; De Stefano, S.; Torrassa, S.; Cavalleri, O.; Rolandi, R.; Gliozzi, A.; Giorgetti, S.; Raimondi, S.; Marchese, L.; Verga, L.; et al. Heparin strongly enhances the formation of beta2-microglobulin amyloid fibrils in the presence of type I collagen. J. Biol. Chem. 2008, 283, 4912–4920. [Google Scholar] [CrossRef]
- Turoverov, K.K.; Verkhusha, V.V.; Shavlovsky, M.M.; Biktashev, A.G.; Povarova, O.I.; Kuznetsova, I.M. Kinetics of actin unfolding induced by guanidine hydrochloride. Biochemistry 2002, 41, 1014–1019. [Google Scholar] [CrossRef]
- Kuznetsova, I.M.; Stepanenko, O.V.; Stepanenko, O.V.; Povarova, O.I.; Biktashev, A.G.; Verkhusha, V.V.; Shavlovsky, M.M.; Turoverov, K.K. The place of inactivated actin and its kinetic predecessor in actin folding-unfolding. Biochemistry 2002, 41, 13127–13132. [Google Scholar] [CrossRef]
- Mine, Y.; Ma, F.; Lauriau, S. Antimicrobial peptides released by enzymatic hydrolysis of hen egg white lysozyme. J. Agric. Food Chem. 2004, 52, 1088–1094. [Google Scholar] [CrossRef]
- Proctor, V.A.; Cunningham, F.E. The chemistry of lysozyme and its use as a food preservative and a pharmaceutical. Crit. Rev. Food Sci. Nutr. 1988, 26, 359–395. [Google Scholar] [CrossRef] [PubMed]
- Naiki, H.; Higuchi, K.; Hosokawa, M.; Takeda, T. Fluorometric determination of amyloid fibrils in vitro using the fluorescent dye, thioflavin T1. Anal. Biochem. 1989, 177, 244–249. [Google Scholar] [CrossRef]
- LeVine, H., 3rd. Quantification of beta-sheet amyloid fibril structures with thioflavin T. Methods Enzym. 1999, 309, 274–284. [Google Scholar]
- LeVine, H., 3rd. Thioflavine T interaction with synthetic Alzheimer’s disease beta-amyloid peptides: Detection of amyloid aggregation in solution. Protein Sci. 1993, 2, 404–410. [Google Scholar] [CrossRef]
- Hawe, A.; Sutter, M.; Jiskoot, W. Extrinsic fluorescent dyes as tools for protein characterization. Pharm. Res. 2008, 25, 1487–1499. [Google Scholar] [CrossRef]
- Lindgren, M.; Sorgjerd, K.; Hammarstrom, P. Detection and characterization of aggregates, prefibrillar amyloidogenic oligomers, and protofibrils using fluorescence spectroscopy. Biophys. J. 2005, 88, 4200–4212. [Google Scholar] [CrossRef]
- Kuznetsova, I.M.; Sulatskaya, A.I.; Povarova, O.I.; Turoverov, K.K. Reevaluation of ANS binding to human and bovine serum albumins: Key role of equilibrium microdialysis in ligand-receptor binding characterization. PLoS ONE 2012, 7, e40845. [Google Scholar] [CrossRef]
- Sulatsky, M.I.; Sulatskaya, A.I.; Povarova, O.I.; Antifeeva, I.A.; Kuznetsova, I.M.; Turoverov, K.K. Effect of the fluorescent probes ThT and ANS on the mature amyloid fibrils. Prion 2020, 14, 67–75. [Google Scholar] [CrossRef]
- Sulatsky, M.I.; Sulatskaya, A.I.; Stepanenko, O.V.; Povarova, O.I.; Kuznetsova, I.M.; Turoverov, K.K. Denaturant effect on amyloid fibrils: Declasterization, depolymerization, denaturation and reassembly. Int. J. Biol. Macromol. 2020, 150, 681–694. [Google Scholar] [CrossRef]
- Lindberg, D.J.; Wranne, M.S.; Gilbert Gatty, M.; Westerlund, F.; Esbjorner, E.K. Steady-state and time-resolved Thioflavin-T fluorescence can report on morphological differences in amyloid fibrils formed by Abeta(1-40) and Abeta(1-42). Biochem. Biophys. Res. Commun. 2015, 458, 418–423. [Google Scholar] [CrossRef]
- Sidhu, A.; Vaneyck, J.; Blum, C.; Segers-Nolten, I.; Subramaniam, V. Polymorph-specific distribution of binding sites determines thioflavin-T fluorescence intensity in alpha-synuclein fibrils. Amyloid Int. J. Exp. Clin. Investig. Off. J. Int. Soc. Amyloidosis 2018, 25, 189–196. [Google Scholar] [CrossRef]
- Stepanenko, O.V.; Povarova, O.I.; Sulatskaya, A.I.; Ferreira, L.A.; Zaslavsky, B.Y.; Kuznetsova, I.M.; Turoverov, K.K.; Uversky, V.N. Protein unfolding in crowded milieu: What crowding can do to a protein undergoing unfolding? J. Biomol. Struct. Dyn. 2016, 34, 2155–2170. [Google Scholar] [CrossRef] [PubMed]
- Kuznetsova, I.M.; Stepanenko, O.V.; Turoverov, K.K.; Zhu, L.; Zhou, J.M.; Fink, A.L.; Uversky, V.N. Unraveling multistate unfolding of rabbit muscle creatine kinase. Biochim. Biophys. Acta 2002, 1596, 138–155. [Google Scholar] [CrossRef]
- Stepanenko, O.V.; Kuznetsova, I.M.; Turoverov, K.K.; Huang, C.; Wang, C.C. Conformational change of the dimeric DsbC molecule induced by GdnHCl. A study by intrinsic fluorescence. Biochemistry 2004, 43, 5296–5303. [Google Scholar] [CrossRef]
- Staiano, M.; D’Auria, S.; Varriale, A.; Rossi, M.; Marabotti, A.; Fini, C.; Stepanenko, O.V.; Kuznetsova, I.M.; Turoverov, K.K. Stability and dynamics of the porcine odorant-binding protein. Biochemistry 2007, 46, 11120–11127. [Google Scholar] [CrossRef]
- Staiano, M.; Scognamiglio, V.; Rossi, M.; D’Auria, S.; Stepanenko, O.V.; Kuznetsova, I.M.; Turoverov, K.K. Unfolding and refolding of the glutamine-binding protein from Escherichia coli and its complex with glutamine induced by guanidine hydrochloride. Biochemistry 2005, 44, 5625–5633. [Google Scholar] [CrossRef]
- Stepanenko, O.V.; Stepanenko, O.V.; Kuznetsova, I.M.; Shcherbakova, D.M.; Verkhusha, V.V.; Turoverov, K.K. Distinct effects of guanidine thiocyanate on the structure of superfolder GFP. PLoS ONE 2012, 7, e48809. [Google Scholar] [CrossRef]
- Sreerama, N.; Woody, R.W. Estimation of protein secondary structure from circular dichroism spectra: Comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 2000, 287, 252–260. [Google Scholar] [CrossRef]
- Krebs, M.R.; Bromley, E.H.; Donald, A.M. The binding of thioflavin-T to amyloid fibrils: Localisation and implications. J. Struct. Biol. 2005, 149, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Bolognesi, B.; Kumita, J.R.; Barros, T.P.; Esbjorner, E.K.; Luheshi, L.M.; Crowther, D.C.; Wilson, M.R.; Dobson, C.M.; Favrin, G.; Yerbury, J.J. ANS binding reveals common features of cytotoxic amyloid species. ACS Chem. Biol. 2010, 5, 735–740. [Google Scholar] [CrossRef] [PubMed]
- Yanushevich, Y.G.; Staroverov, D.B.; Savitsky, A.P.; Fradkov, A.F.; Gurskaya, N.G.; Bulina, M.E.; Lukyanov, K.A.; Lukyanov, S.A. A strategy for the generation of non-aggregating mutants of Anthozoa fluorescent proteins. FEBS Lett. 2002, 511, 11–14. [Google Scholar] [CrossRef]
- Leissring, M.A.; Farris, W.; Chang, A.Y.; Walsh, D.M.; Wu, X.; Sun, X.; Frosch, M.P.; Selkoe, D.J. Enhanced proteolysis of beta-amyloid in APP transgenic mice prevents plaque formation, secondary pathology, and premature death. Neuron 2003, 40, 1087–1093. [Google Scholar] [CrossRef]
- Marr, R.A.; Rockenstein, E.; Mukherjee, A.; Kindy, M.S.; Hersh, L.B.; Gage, F.H.; Verma, I.M.; Masliah, E. Neprilysin gene transfer reduces human amyloid pathology in transgenic mice. J. Neurosci. Off. J. Soc. Neurosci. 2003, 23, 1992–1996. [Google Scholar] [CrossRef]
- Sulatskaya, A.I.; Rodina, N.P.; Polyakov, D.S.; Sulatsky, M.I.; Artamonova, T.O.; Khodorkovskii, M.A.; Shavlovsky, M.M.; Kuznetsova, I.M.; Turoverov, K.K. Structural Features of Amyloid Fibrils Formed from the Full-Length and Truncated Forms of Beta-2-Microglobulin Probed by Fluorescent Dye Thioflavin T. Int. J. Mol. Sci. 2018, 19, 2762. [Google Scholar] [CrossRef]
- Walsh, K.A. Trypsinogens and trypsins of various species. Methods Enzymol. 1970, 19, 41–63. [Google Scholar] [CrossRef]
- Vladimirov, Y.A.; Litvin, F.F. Photobiology and spectroscopic methods. In Handbook of General Biophisics; High School: Moscow, Russia, 1964. [Google Scholar]
- Turoverov, K.K.; Biktashev, A.G.; Dorofeiuk, A.V.; Kuznetsova, I.M. A complex of apparatus and programs for the measurement of spectral, polarization and kinetic characteristics of fluorescence in solution. Tsitologiia 1998, 40, 806–817. [Google Scholar]
- Fonin, A.V.; Sulatskaya, A.I.; Kuznetsova, I.M.; Turoverov, K.K. Fluorescence of dyes in solutions with high absorbance. Inner filter effect correction. PLoS ONE 2014, 9, e103878. [Google Scholar] [CrossRef]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Nishimura, S.; Murasugi, T.; Kubo, T.; Kaneko, I.; Meguro, M.; Marumoto, S.; Kogen, H.; Koyama, K.; Oda, T.; Nakagami, Y. RS-4252 inhibits amyloid beta-induced cytotoxicity in HeLa cells. Pharmacol. Toxicol. 2003, 93, 29–32. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stepanenko, O.V.; Sulatsky, M.I.; Mikhailova, E.V.; Stepanenko, O.V.; Kuznetsova, I.M.; Turoverov, K.K.; Sulatskaya, A.I. Trypsin Induced Degradation of Amyloid Fibrils. Int. J. Mol. Sci. 2021, 22, 4828. https://doi.org/10.3390/ijms22094828
Stepanenko OV, Sulatsky MI, Mikhailova EV, Stepanenko OV, Kuznetsova IM, Turoverov KK, Sulatskaya AI. Trypsin Induced Degradation of Amyloid Fibrils. International Journal of Molecular Sciences. 2021; 22(9):4828. https://doi.org/10.3390/ijms22094828
Chicago/Turabian StyleStepanenko, Olga V., Maksim I. Sulatsky, Ekaterina V. Mikhailova, Olesya V. Stepanenko, Irina M. Kuznetsova, Konstantin K. Turoverov, and Anna I. Sulatskaya. 2021. "Trypsin Induced Degradation of Amyloid Fibrils" International Journal of Molecular Sciences 22, no. 9: 4828. https://doi.org/10.3390/ijms22094828
APA StyleStepanenko, O. V., Sulatsky, M. I., Mikhailova, E. V., Stepanenko, O. V., Kuznetsova, I. M., Turoverov, K. K., & Sulatskaya, A. I. (2021). Trypsin Induced Degradation of Amyloid Fibrils. International Journal of Molecular Sciences, 22(9), 4828. https://doi.org/10.3390/ijms22094828









