Hydroxyurea and Caffeine Impact pRb-like Protein-Dependent Chromatin Architecture Profiles in Interphase Cells of Vicia faba
Abstract
1. Introduction
2. Results
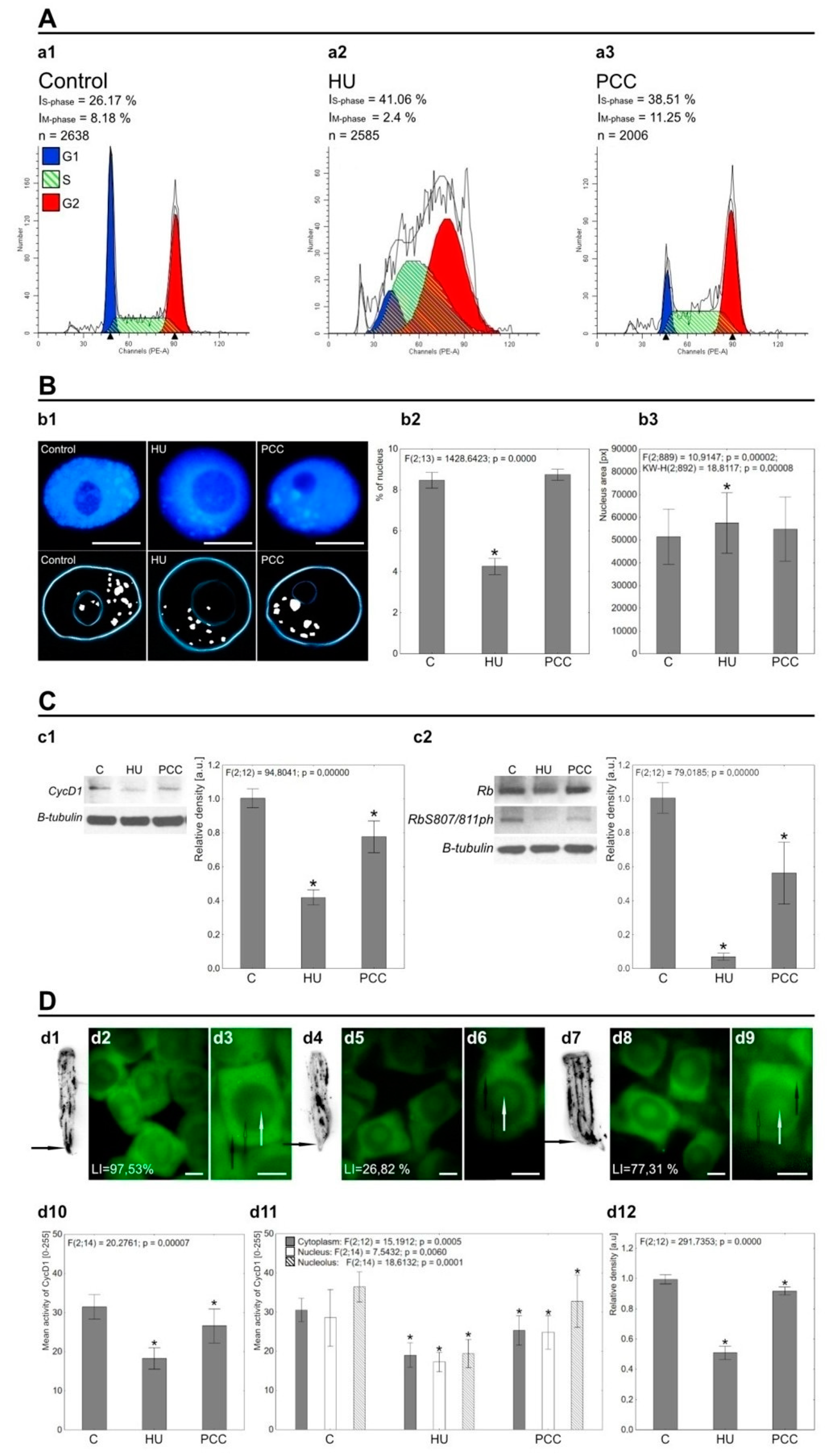
2.1. The Effects of Prolonged HU and HU/CF Incubation on Heterochromatin Areas, Replication, and the Activity of CycD1 and RbS807/811
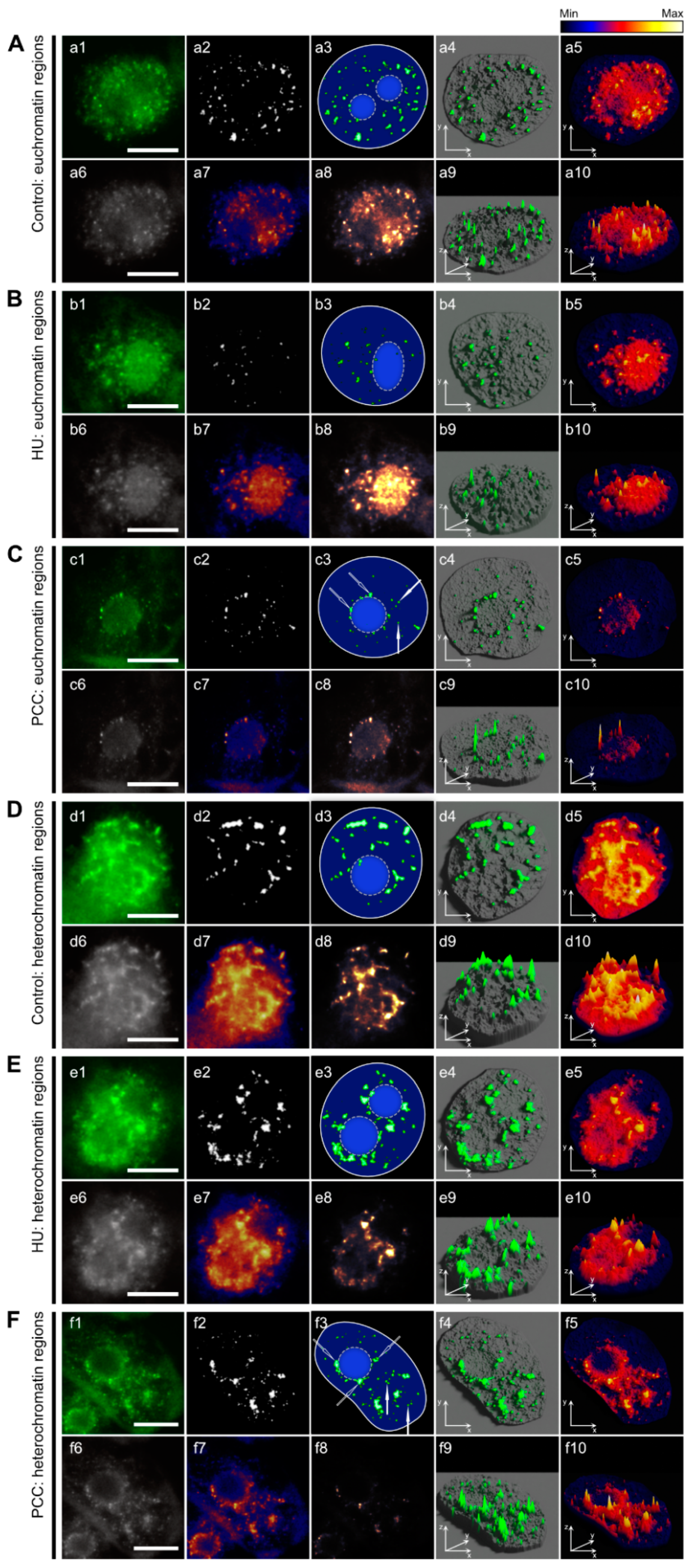
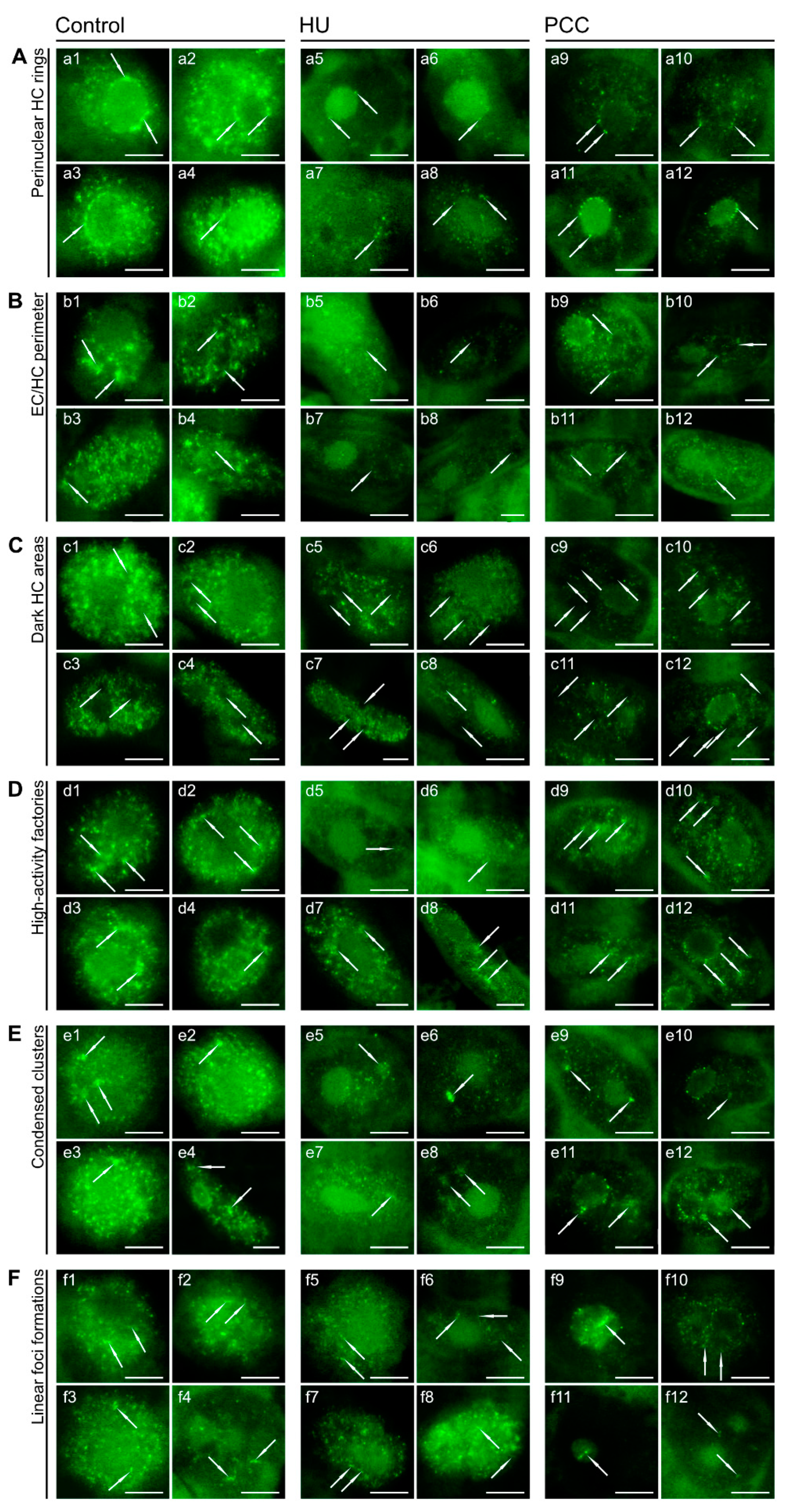
2.2. Nucleus and Foci Analysis Leading to the Identification of Marking Types
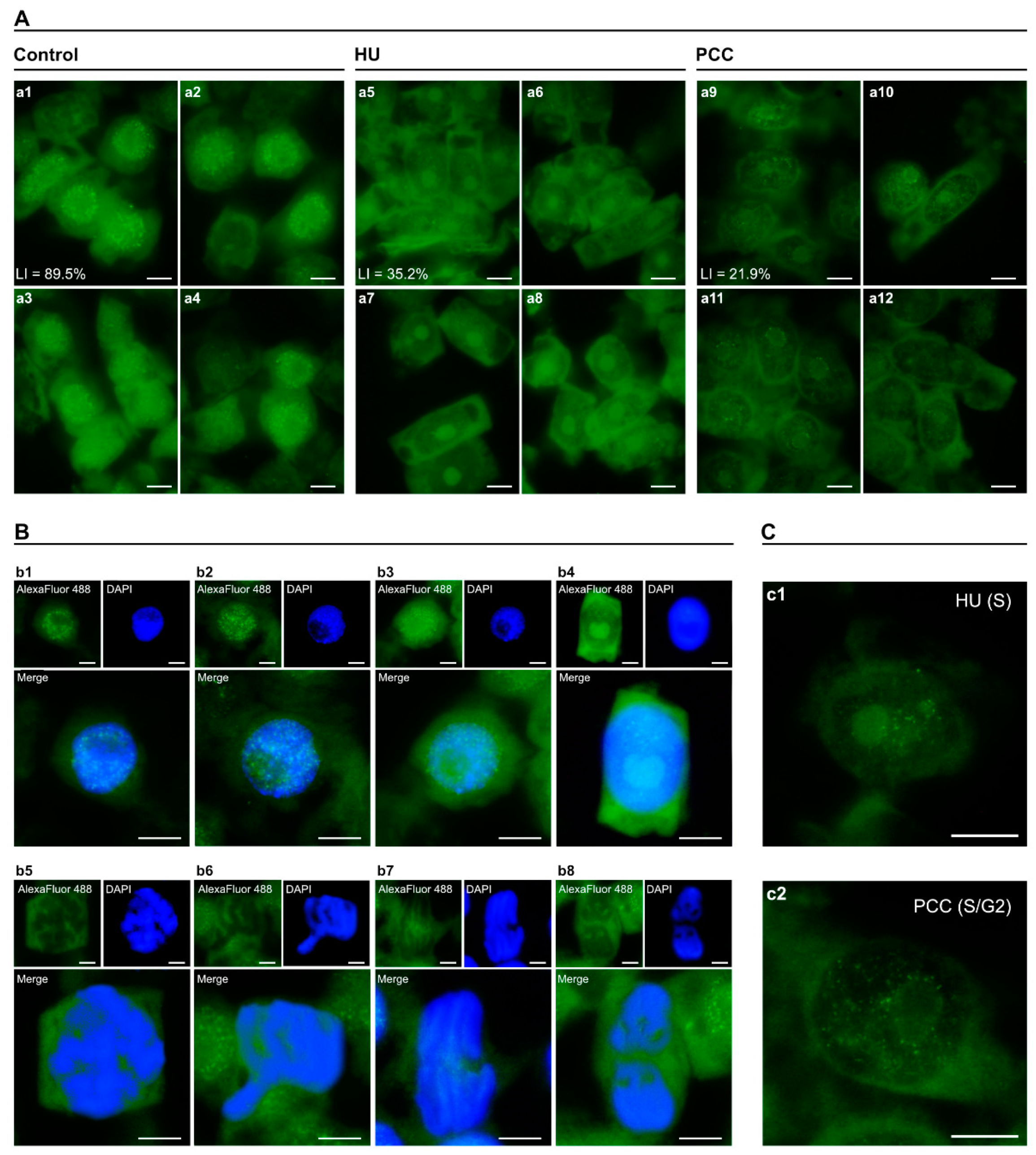
2.3. Retinoblastoma ProteinHas Five Labeling Profiles in Interphase Cells
2.4. Retinoblastoma Protein Patterns Alterations Caused by HU and PCC
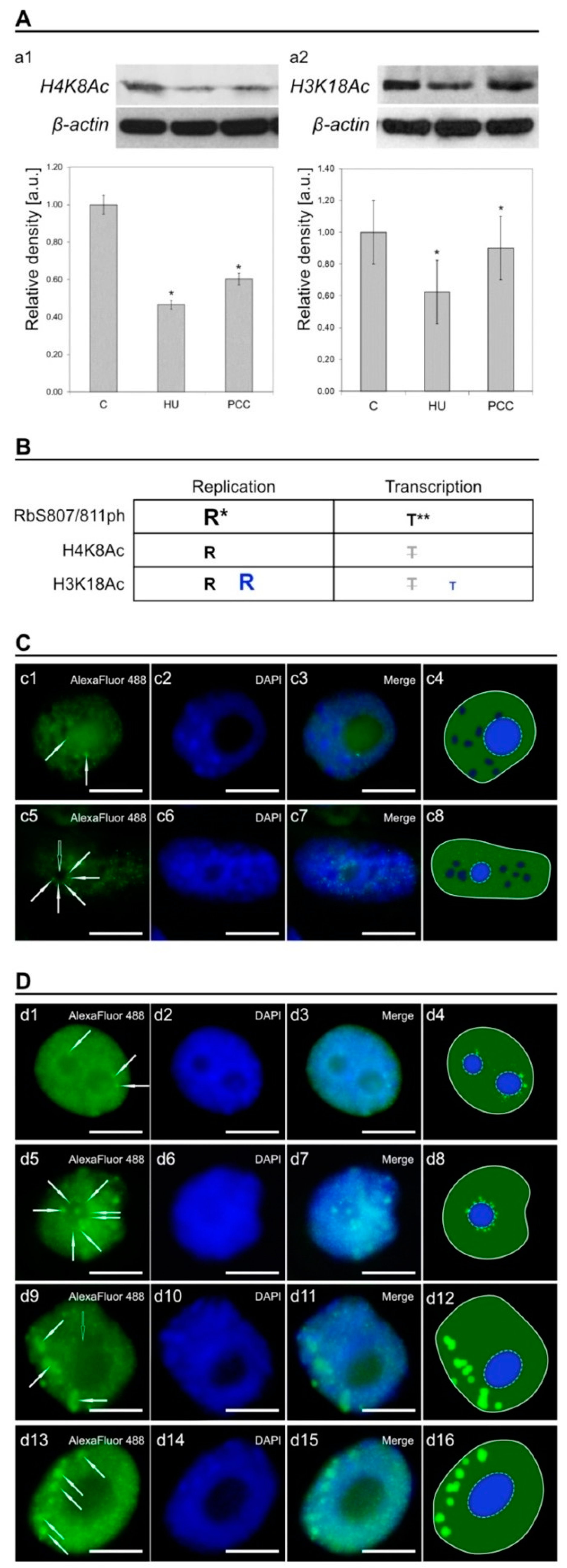
2.5. Histones
3. Discussion
3.1. Prolonged Hydroxyurea Incubation Causes Heterochromatin Under-replication, Suppresses Mitotic Divisions, and Diminishes CycD1 and RbS807/811 Activity
3.2. Retinoblastoma ProteinActivity Profiles Do Not Change Despite Induction of Replication Stress and PCC
3.3. Heterochromatin
3.4. Histones
4. Material and methods
4.1. Plant Material, Growth Conditions, Hydroxyurea, and Caffeine Treatment
4.2. DAPI Staining and Quantitative Heterochromatin Measurements
4.3. Cell Cycle Analysis
4.4. Western Blotting
4.5. Tissue Printing
4.6. Immunocytochemistry
4.7. Image Analysis and 3D Modeling
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ATR | ataxia telangiectasia and Rad3-related kinase |
| CF | caffeine |
| dNTPs | deoxyribonucleoside triphosphates |
| EC | euchromatin |
| HC | heterochromatin |
| HU | hydroxyurea |
| PCC | premature chromosome condensation |
| pRb | retinoblastoma protein |
| RNR | ribonucleotide reductase |
References
- Hornykiewicz, O. A brief history of levodopa. J. Neurol. 2010, 257, 249–252. [Google Scholar] [CrossRef]
- Rybaczek, D.; Kowalewicz-Kulbat, M. Premature chromosome condensation induced by caffeine, 2-aminopurine, staurosporine and sodium metavanadate in S-phase arrested HeLa cells is associated with a decrease in Chk1 phosphorylation, formation of phospho-H2AX and minor cytoskeletal rearrangement. Histochem. Cell Biol. 2011, 135, 263–280. [Google Scholar] [CrossRef]
- Rybaczek, D. Ultrastructural changes associated with the induction of premature chromosome condensation in Vicia faba root meristem cells. Plant Cell Rep. 2014, 33, 1547–1564. [Google Scholar] [CrossRef] [PubMed]
- Rybaczek, D.; Musialek, M.W.; Balcerczyk, A. Caffeine-induced premature chromosome condensation results in the apoptosis-like programmed cell death in root meristems of Vicia faba. PLoS ONE 2015, 10, e0142307. [Google Scholar] [CrossRef]
- Hübner, B.; Strickfaden, H.; Müller, S.; Cremer, M.; Cremer, T. Chromosome shattering: A mitotic catastrophe due to chromosome condensation failure. Eur. Biophys. J. 2009, 38, 729–747. [Google Scholar] [CrossRef] [PubMed]
- Forment, J.V.; Kaidi, A.; Jackson, S.P. Chromothripsis and cancer: Causes and consequences of chromosome shattering. Nat. Rev. Cancer 2012, 12, 663–670. [Google Scholar] [CrossRef]
- Huang, M.E.; Facca, C.; Fatmi, Z.; Baïlle, D.; Bénakli, S.; Vernis, L. DNA replication inhibitor hydroxyurea alters Fe-S centers by producing reactive oxygen species in vivo. Sci. Rep. 2016, 6, 29361–29372. [Google Scholar] [CrossRef] [PubMed]
- Rybaczek, D.; Musiałek, M.W.; Vrána, J.; Petrovská, B.; Pikus, E.G.; Doležel, J. Kinetics of DNA repair in Vicia faba meristem regeneration following replication stress. Cells 2021, 10, 88. [Google Scholar]
- Budirahardja, Y.; Gönczy, P. Coupling the cell cycle to development. Development 2009, 136, 2861–2872. [Google Scholar] [CrossRef]
- Harashima, H.; Dissmeyer, N.; Schnittger, A. Cell cycle control across the eukaryotic kingdom. Trends Cell Biol. 2013, 23, 345–356. [Google Scholar] [CrossRef]
- Barnum, K.J.; O’Connell, M.J. Cell cycle regulation by checkpoints. Methods Mol. Biol. 2014, 1170, 29–40. [Google Scholar]
- Bertoli, C.; Skotheim, J.M.; De Bruin, R.A.M. Control of cell cycle transcription during G1 and S phases. Nat. Rev. Mol. Cell Biol. 2013, 14, 518–528. [Google Scholar] [CrossRef]
- Giacinti, C.; Giordano, A. RB and cell cycle progression. Oncogene 2006, 25, 5220–5227. [Google Scholar] [CrossRef] [PubMed]
- Polit, J.T.; Kaźmierczak, A.; Walczak-Drzewiecka, A. Cell cycle-dependent phosphorylation of pRb-like protein in root meristem cells of Vicia faba. Protoplasma 2012, 249, 131–137. [Google Scholar] [CrossRef]
- Desvoyes, B.; de Mendoza, A.; Ruiz-Trillo, I.; Gutierrez, C. Novel roles of plant RETINOBLASTOMA-RELATED (RBR) protein in cell proliferation and asymmetric cell division. J. Exp. Bot. 2014, 65, 2657–2666. [Google Scholar] [CrossRef]
- Zluhan-Martínez, E.; Pérez-Koldenkova, V.; Ponce-Castañeda, M.V.; Sánchez, M.d.l.P.; García-Ponce, B.; Miguel-Hernández, S.; Álvarez-Buylla, E.R.; Garay-Arroyo, A. Beyond what your retina can see: Similarities of retinoblastoma function between plants and animals, from developmental processes to epigenetic regulation. Int. J. Mol. Sci. 2020, 21, 4925. [Google Scholar]
- Grant, G.D.; Cook, J.G. The temporal regulation of S phase proteins during G1. Adv. Exp. Med. Biol. 2017, 1042, 335–369. [Google Scholar] [PubMed]
- Lee, B.K.; Bhinge, A.A.; Iyer, V.R. Wide-ranging functions of E2F4 in transcriptional activation and repression revealed by genome-wide analysis. Nucleic Acids Res. 2011, 39, 3558–3573. [Google Scholar] [CrossRef]
- Kato, J.; Matsushime, H.; Hiebert, S.W.; Ewen, M.E.; Sherr, C.J. Direct binding of cyclin D to the retinoblastoma gene product (pRb) and pRb phosphorylation by the cyclin D-dependent kinase CDK4. Genes Dev. 1993, 7, 331–342. [Google Scholar]
- Meijer, M.; Murray, J.A.H. The role and regulation of D-type cyclins in the plant cell cycle. Plant Mol. Biol. 2000, 43, 621–633. [Google Scholar] [CrossRef] [PubMed]
- Simone, C.; Bagella, L.; Bellan, C.; Giordano, A. Physical interaction between pRb and cdk9/cyclinT2 complex. Oncogene 2002, 21, 4158–4165. [Google Scholar] [CrossRef]
- De Veylder, L.; Joubès, J.; Inzé, D. Plant cell cycle transitions. Curr. Opin. Plant Biol. 2003, 6, 536–543. [Google Scholar] [CrossRef] [PubMed]
- Paternot, S.; Arsenijevic, T.; Coulonval, K.; Bockstaele, L.; Dumont, J.E.; Roger, P.P. Distinct specificities of pRb phosphorylation by CDK4 activated by cyclin D1 or cyclin D3: Differential involvement in the distinct mitogenic modes of thyroid epithelial cells. Cell Cycle 2006, 5, 61–70. [Google Scholar] [CrossRef][Green Version]
- Uemukai, K.; Iwakawa, H.; Kosugi, S.; de Uemukai, S.; Kato, K.; Kondorosi, E.; Murray, J.A.; Ito, M.; Shinmyo, A.; Sekine, M. Transcriptional activation of tobacco E2F is repressed by co-transfection with the retinoblastoma-related protein: Cyclin D expression overcomes this repressor activity. Plant Mol Biol. 2005, 57, 83–100. [Google Scholar] [CrossRef]
- Ewens, K.G.; Bhatti, T.R.; Moran, K.A.; Richards-Yutz, J.; Shields, C.L.; Eagle, R.C.; Ganguly, A. Phosphorylation of pRb: Mechanism for RB pathway inactivation in MYCN-amplified retinoblastoma. Cancer Med. 2017, 6, 619–630. [Google Scholar] [CrossRef] [PubMed]
- Bouyer, D.; Heese, M.; Chen, P.; Harashima, H.; Roudier, F.; Grüttner, C.; Schnittger, A. Genome-wide identification of RETINOBLASTOMA RELATED 1 binding sites in Arabidopsis reveals novel DNA damage regulators. PLoS Genet. 2018, 14, e1007797. [Google Scholar]
- Shen, W.H. The plant cell cycle: G1/S regulation. Euphytica 2001, 118, 223–236. [Google Scholar] [CrossRef]
- Menges, M.; Samland, A.K.; Planchais, S.; Murray, J.A. The D-type cyclin CYCD3;1 is limiting for the G1-to-S-phase transition in Arabidopsis. Plant Cell 2006, 18, 893–906. [Google Scholar] [CrossRef] [PubMed]
- Markey, M.P.; Bergseid, J.; Bosco, E.E.; Stengel, K.; Xu, H.; Mayhew, C.N.; Schwemberger, S.J.; Braden, W.A.; Jiang, Y.; Babcock, G.F.; et al. Loss of the retinoblastoma tumor suppressor: Differential action on transcriptional programs related to cell cycle control and immune function. Oncogene 2007, 26, 6307–6318. [Google Scholar]
- Hinds, P.W. A little pRB can lead to big problems. Cancer Discov. 2014, 4, 764–765. [Google Scholar] [CrossRef][Green Version]
- Uchida, C. Roles of pRB in the regulation of nucleosome and chromatin structures. BioMed Res. Int. 2016, 7, 1–11. [Google Scholar] [CrossRef]
- Ishak, C.A.; Coschi, C.H.; Roes, M.V.; Dick, F.A. Disruption of CDK-resistant chromatin association by pRB causes DNA damage, mitotic errors, and reduces condensin II recruitment. Cell Cycle 2017, 16, 1430–1439. [Google Scholar] [CrossRef][Green Version]
- Jiang, Y.; Yam, J.C.; Tham, C.C.; Pang, C.P.; Chu, W.K. RB reguates DNA double strand break repair pathway choise by mediating CtIP dependent end resection. Int. J. Mol. Sci. 2020, 21, 9176. [Google Scholar] [CrossRef]
- Knudsen, E.S.; Pruitt, S.C.; Hershberger, P.A.; Witkiewicz, A.K.; Goodrich, D.W. Cell cycle and beyond: Exploiting new RB1 controlled mechanisms for cancer therapy. Trends Cancer 2019, 5, 308–324. [Google Scholar] [CrossRef]
- Tarang, S.; Pyakurel, U.; Weston, M.D.; Vijayakumar, S.; Jones, T.; Wagner, K.U.; Rocha-Sanchez, S.M. Spatiotemporally controlled overexpression of cyclin D1 triggers generation of supernumerary cells in the postnatal mouse inner ear. Hear. Res. 2020, 390, 107951. [Google Scholar] [CrossRef] [PubMed]
- Longworth, M.S.; Dyson, N.J. pRb, a local chromatin organizer with global possibilities. Bone 2008, 23, 1–7. [Google Scholar] [CrossRef]
- Munro, S.; Hookway, E.S.; Floderer, M.; Carr, S.M.; Konietzny, R.; Kessler, B.M.; Oppermann, U.; La Thangue, N.B. Linker Histone H1.2 Directs genome-wide chromatin association of the retinoblastoma tumor suppressor protein and facilitates its function. Cell Rep. 2017, 19, 2193–2201. [Google Scholar] [CrossRef][Green Version]
- Brownlee, P.M.; Meisenberg, C.; Downs, J.A. The SWI/SNF chromatin remodelling complex: Its role in maintaining genome stability and preventing tumourigenesis. DNA Repair 2015, 32, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Trouche, D.; Le Chalony, C.; Muchardt, C.; Yaniv, M.; Kouzarides, T. RB and hbrm cooperate to repress the activation functions of E2F1. Proc. Natl. Acad. Sci. USA 1997, 94, 11268–11273. [Google Scholar] [CrossRef]
- Morrison, A.J.; Sardet, C.; Herrera, R.E. Retinoblastoma protein transcriptional repression through histone deacetylation of a single nucleosome. Mol. Cell. Biol. 2002, 22, 856–865. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Talluri, S.; Dick, F.A. Regulation of transcription and chromatin structure by pRB: Here, there and everywhere. Cell Cycle 2012, 11, 189–3198. [Google Scholar] [CrossRef]
- Isaac, C.E.; Francis, S.M.; Martens, A.L.; Julian, L.M.; Seifried, L.A.; Erdmann, N.; Binné, U.K.; Harrington, L.; Sicinski, P.; Bérubé, N.G.; et al. The retinoblastoma protein regulates pericentric heterochromatin. Mol. Cell. Biol. 2006, 26, 3659–3671. [Google Scholar]
- Schotta, G.; Lachner, M.; Sarma, K.; Ebert, A.; Sengupta, R.; Reuter, G.; Reinberg, D.; Jenuwein, T. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 2004, 18, 1251–1262. [Google Scholar] [CrossRef]
- Uchida, C.; Hattori, T.; Takahashi, H.; Yamamoto, N.; Kitagawa, M.; Taya, Y. Interaction between RB protein and NuMA is required for proper alignment of spindle microtubules. Genes Cells 2014, 19, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Conroy, L.R.; Dougherty, S.; Kruer, T.; Metcalf, S.; Lorkiewicz, P.; He, L.; Yin, X.; Zhang, X.; Arumugam, S.; Young, L.E.A.; et al. Loss of Rb1 enhances glycolytic metabolism in Kras-driven lung tumors in vivo. Cancers 2020, 12, 237. [Google Scholar]
- Manning, A.L.; Longworth, M.S.; Dyson, N.J. Loss of pRB causes centromere dysfunction and chromosomal instability. Genes Dev. 2010, 24, 1364–1376. [Google Scholar] [CrossRef]
- Hall, M.A.; Shundrovsky, A.; Bai, L.; Fulbright, R.M.; Lis, J.T.; Wang, M.D. High-resolution dynamic mapping of histone-DNA interactions in a nucleosome. Nat. Struct. Mol. Biol. 2009, 16, 124–129. [Google Scholar]
- Jasencakova, Z.; Groth, A. Replication stress, a source of epigenetic aberrations in cancer? BioEssays 2010, 32, 847–855. [Google Scholar] [CrossRef]
- Jasencakova, Z.; Scharf, A.N.; Ask, K.; Corpet, A.; Imhof, A.; Almouzni, G.; Growth, A. Replication stress interferes with histone recycling and predeposition marking of new histones. Mol. Cell 2010, 37, 736–743. [Google Scholar] [CrossRef]
- Huh, M.S.; Ivanochko, D.; Hashem, L.E.; Curtin, M.; Delorme, M.; Goodall, E.; Yan, K.; Picketts, D.J. Stalled replication forks within heterochromatin require ATRX for protection. Cell Death Dis. 2016, 7, e2220. [Google Scholar]
- Fortuny, A.; Polo, S.E. The response to DNA damage in heterochromatin domains. Chromosoma 2018, 127, 291–300. [Google Scholar] [CrossRef] [PubMed]
- Jasencakova, Z.; Meister, A.; Walter, J.; Turner, B.M.; Schubert, I. Histone H4 acetylation of euchromatin and heterochromatin is cell cycle dependent and correlated with replication rather than with transcription. Plant Cell 2000, 12, 2087–2100. [Google Scholar] [CrossRef]
- Koç, A.; Wheeler, L.J.; Mathews, C.K.; Merrill, G.F. Hydroxyurea arrests DNA replication by a mechanism that preserves basal dNTP pools. J. Biol. Chem. 2004, 279, 223–230. [Google Scholar]
- Ercilla, A.; Feu, S.; Aranda, S.; Llopis, A.; Brynjólfsdóttir, S.H.; Sørensen, C.S.; Toledo, L.I.; Agell, N. Acute hydroxyurea-induced replication blockade results in replisome components disengagement from nascent DNA without causing fork collapse. Cell. Mol. Life Sci. 2020, 77, 735–749. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Yi, J.; Agbu, P.; Zhou, Z.; Kelley, R.L.; Kallgren, S.; Jia, S.; He, X. Replication stress affects the fidelity of nucleosome-mediated epigenetic inheritance. PLoS Genet. 2017, 13, e1006900. [Google Scholar] [CrossRef] [PubMed]
- Singh, G.; Klar, A.J.S. Mutations in deoxyribonucleotide biosynthesis pathway cause spreading of silencing across heterochromatic barriers at the mating-type region of the fission yeast. Yeast 2008, 25, 117–128. [Google Scholar] [CrossRef]
- Li, B.; Zhao, H.; Rybak, P.; Dobrucki, J.W.; Darzynkiewicz, Z.; Kimmel, M. Different rates of DNA replication at early versus late S-phase sections: Multiscale modeling of stochastic events related to DNA content/EdU (5-ethynyl-2’deoxyuridine) incorporation distributions. Cytometry A 2014, 85, 785–797. [Google Scholar]
- Maya-Miles, D.; Andújar, E.; Pérez-Alegre, M.; Murillo-Pineda, M.; Barrientos-Moreno, M.; Cabello-Lobato, M.J.; Gómez-Marín, E.; Morillo-Huesca, M.; Prado, F. Crosstalk between chromatin structure, cohesin activity and transcription. Epigenetics Chromatin 2019, 12, 47. [Google Scholar]
- Bester, A.C.; Roniger, M.; Oren, Y.S.; Im, M.M.; Sarni, D.; Chaoat, M.; Bensimon, A.; Zamir, G.; Shewach, D.S.; Kerem, B. Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell 2011, 145, 435–446. [Google Scholar] [CrossRef]
- Musiałek, M.W.; Rybaczek, D. Behavior of replication origins in Eukaryota—spatio-temporal dynamics of licensing and firing. Cell Cycle 2015, 14, 2251–2264. [Google Scholar] [CrossRef] [PubMed]
- Yousefi, R.; Rowicka, M. Stochasticity of replication forks’ speeds plays a key role in the dynamics of DNA replication. PLOS Comput. Biol. 2019, 15, e1007519. [Google Scholar] [CrossRef]
- Lee, T.J.; Pascuzzi, P.E.; Settlage, S.B.; Shultz, R.W.; Tanurdzic, M.; Rabinowicz, P.D.; Menges, M.; Zheng, P.; Main, D.; Murray, J.A.; et al. Arabidopsis thaliana chromosome 4 replicates in two phases that correlate with chromatin state. PLoS Genet. 2010, 6, e1000982. [Google Scholar]
- Zou, L.; Elledge, S.J. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 2003, 300, 1542–1548. [Google Scholar] [CrossRef]
- Wojtala, M.; Dąbek, A.; Rybaczek, D.; Śliwińska, A.; Świderska, E.; Słapek, K.; El-Osta, A.; Balcerczyk, A. Silencing lysine-specific histone demethylase 1 (LSD1) causes increased HP1-positive chromatin, stimulation of DNA repair processes, and dysregulation of proliferation by Chk1 phosphorylation in human endothelial cells. Cells 2019, 8, 1212. [Google Scholar] [CrossRef]
- Sarkaria, J.N.; Busby, E.C.; Tibbetts, R.S.; Roos, P.; Taya, Y.; Karnitz, L.M.; Abraham, R.T. Inhibition of ATM and ATR kinase activities by the radiosensitizing agent, caffeine. Cancer Res. 1999, 59, 4375–4382. [Google Scholar]
- McKinley, K.L.; Cheeseman, I.M. The molecular basis for centromere identity and function. Nat. Rev. Mol. Cell Biol. 2016, 17, 16–29. [Google Scholar] [CrossRef]
- Osmanagic-Myers, S.; Foisner, R. The structural and gene expression hypotheses in laminopathic diseases - not so different after all. Mol. Biol. Cell. 2019, 30, 1786–1790. [Google Scholar] [CrossRef] [PubMed]
- Penagos-Puig, A.; Furlan-Magaril, M. Heterochromatin as an important driver of genome organization. Front. Cell Dev. Biol. 2020, 8, 1–10. [Google Scholar] [CrossRef]
- Ragoczy, T.; Telling, A.; Scalzo, D.; Kooperberg, C.; Groudine, M. Functional redundancy in the nuclear compartmentalization of the late-replicating genome. Nucleus 2014, 5, 626–635. [Google Scholar] [CrossRef] [PubMed]
- Cremer, T.; Cremer, M.; Cremer, C. The 4D nucleome: Genome compartmentalization in an evolutionary context. Biochemistry 2018, 83, 313–325. [Google Scholar] [CrossRef]
- Girelli, G.; Custodio, J.; Kallas, T.; Agostini, F.; Wernersson, E.; Spanjaard, B.; Mota, A.; Kolbeinsdottir, S.; Gelali, E.; Crosetto, N.; et al. GPSeq reveals the radial organization of chromatin in the cell nucleus. Nat. Biotechnol. 2020, 38, 1184–1193. [Google Scholar]
- Nozaki, T.; Imai, R.; Tanbo, M.; Nagashima, R.; Tamura, S.; Tani, T.; Joti, Y.; Tomita, M.; Hibino, K.; Kanemaki, M.T.; et al. Dynamic Organization of Chromatin Domains Revealed by Super-Resolution Live-Cell Imaging. Mol. Cell 2017, 67, 282–293.e3. [Google Scholar] [CrossRef]
- Vertii, A.; Ou, J.; Yu, J.; Yan, A.; Pagès, H.; Liu, H.; Zhu, L.J.; Kaufman, P.D. Two contrasting classes of nucleolus-associated domains in mouse fibroblast heterochromatin. Genome Res. 2019, 29, 1235–1249. [Google Scholar] [CrossRef]
- Growth, A.; Rocha, W.; Verreault, A.; Almouzni, G. Chromatin challenges during DNA replication and repair. Cell 2007, 128, 721–733. [Google Scholar] [CrossRef]
- Jasencakova, Z.; Soppe, W.J.; Meister, A.; Gernand, D.; Turner, B.M.; Schubert, I. Histone modifications in Arabidopsis- high methylation of H3 lysine 9 is dispensable for constitutive heterochromatin. Plant J. 2003, 33, 471–480. [Google Scholar] [CrossRef] [PubMed]
- Van’t Hoff, J. DNA fiber replication in chromosomes of a higher plant (Pisum sativum). Exp. Cell Res. 1975, 93, 95–104. [Google Scholar] [CrossRef]
- Cassab, G.I. Localization of cell wall proteins using tissue-print western blot techniques. In Recombinant DNA Part I; Wu, R.B., Ed.; Academic Press: London, UK, 1993; pp. 682–688. [Google Scholar]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longaair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Musiałek, M.W.; Deckert, J.; Rybaczek, D. Hydroxyurea and Caffeine Impact pRb-like Protein-Dependent Chromatin Architecture Profiles in Interphase Cells of Vicia faba. Int. J. Mol. Sci. 2021, 22, 4572. https://doi.org/10.3390/ijms22094572
Musiałek MW, Deckert J, Rybaczek D. Hydroxyurea and Caffeine Impact pRb-like Protein-Dependent Chromatin Architecture Profiles in Interphase Cells of Vicia faba. International Journal of Molecular Sciences. 2021; 22(9):4572. https://doi.org/10.3390/ijms22094572
Chicago/Turabian StyleMusiałek, Marcelina W., Joanna Deckert, and Dorota Rybaczek. 2021. "Hydroxyurea and Caffeine Impact pRb-like Protein-Dependent Chromatin Architecture Profiles in Interphase Cells of Vicia faba" International Journal of Molecular Sciences 22, no. 9: 4572. https://doi.org/10.3390/ijms22094572
APA StyleMusiałek, M. W., Deckert, J., & Rybaczek, D. (2021). Hydroxyurea and Caffeine Impact pRb-like Protein-Dependent Chromatin Architecture Profiles in Interphase Cells of Vicia faba. International Journal of Molecular Sciences, 22(9), 4572. https://doi.org/10.3390/ijms22094572







