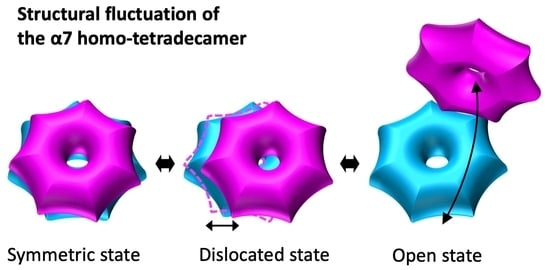
Structural Fluctuations of the Human Proteasome α7 Homo-Tetradecamer Double Ring Imply the Proteasomal α-Ring Assembly Mechanism
Abstract
1. Introduction
2. Results
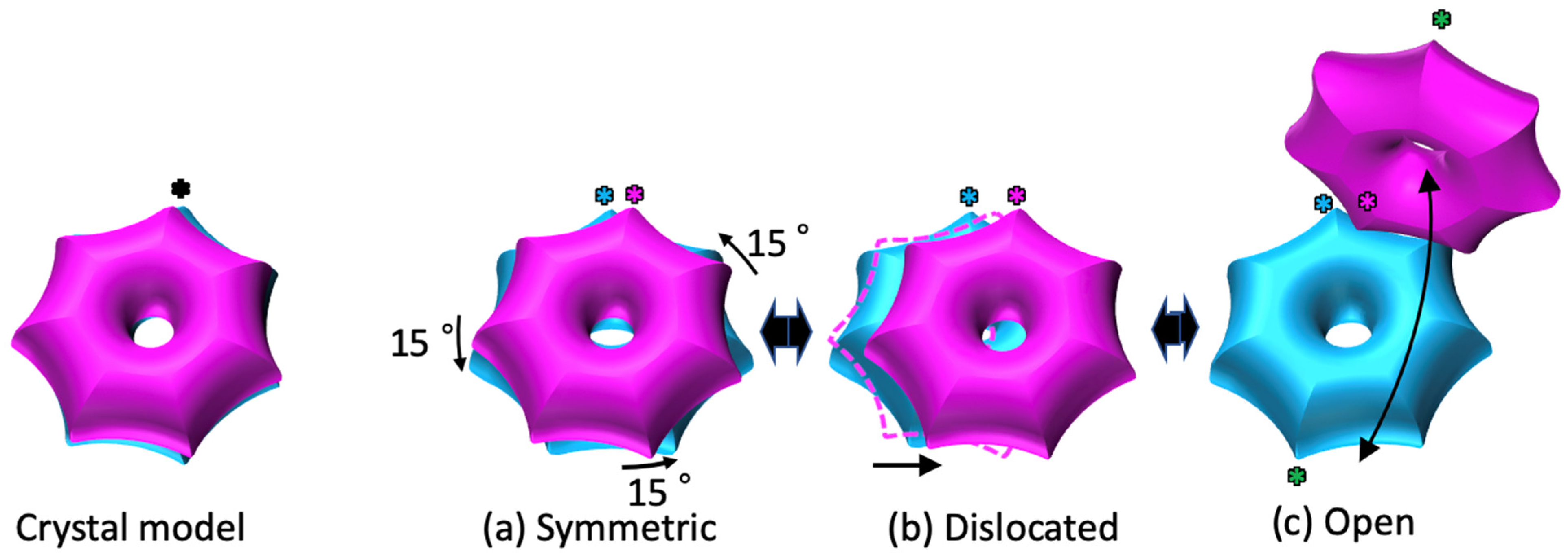
2.1. The α7 Tetradecamer Double Ring Shows Three Different Structures in Solution
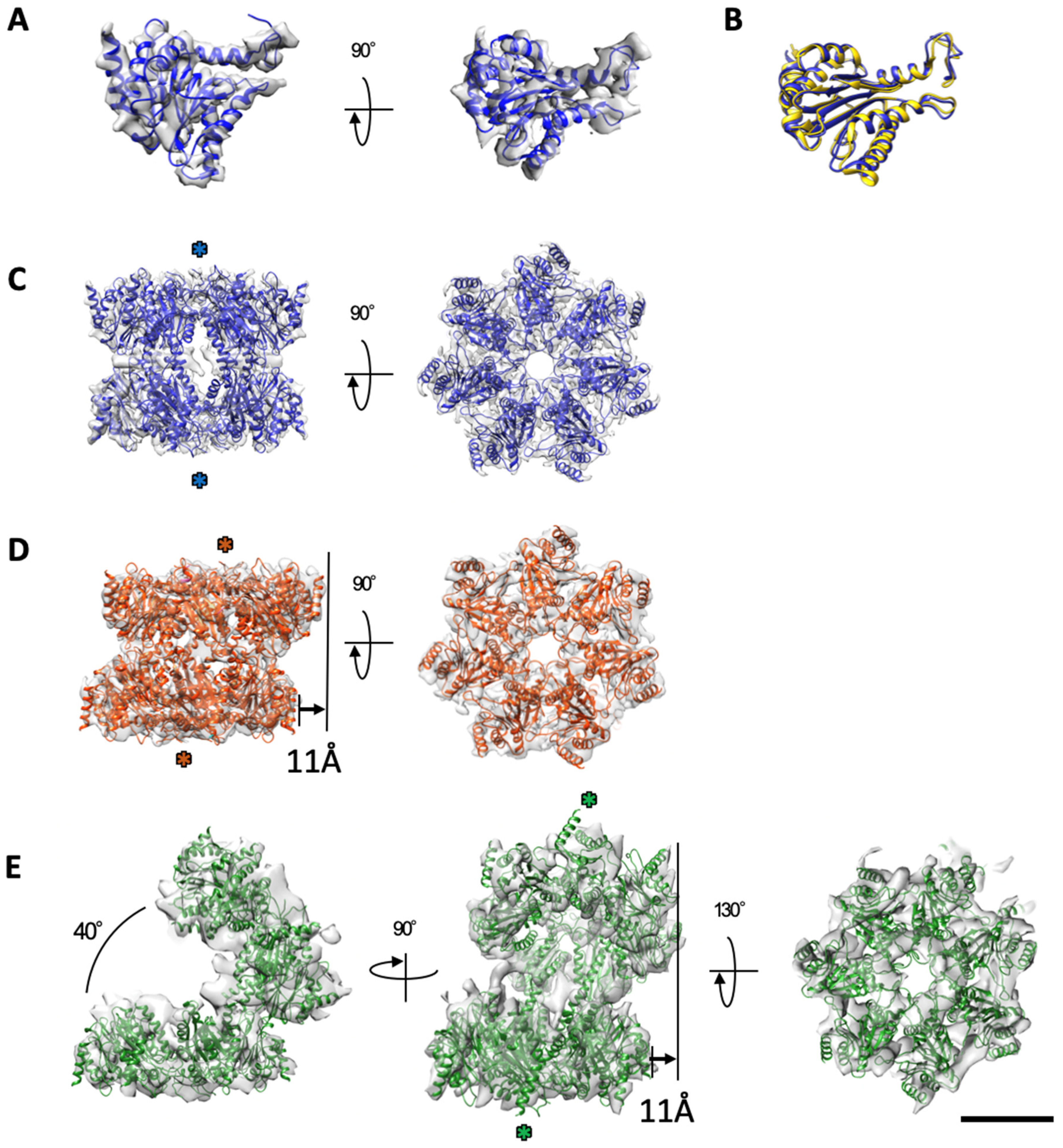
2.2. The α7 Subunit Models Were Fitted to the Densities in Each Cryo-EM Map
2.3. Cryo-EM Structures of the α7 Double Ring in Solution
3. Discussion
4. Materials and Methods
4.1. Sample Preparation for Cryo-EM
4.2. Image Processing for Cryo-EM
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Collins, G.A.; Goldberg, A.L. The Logic of the 26S Proteasome. Cell 2017, 169, 792–806. [Google Scholar] [CrossRef]
- Finley, D.; Chen, X.; Walters, K.J. Gates, Channels, and Switches: Elements of the Proteasome Machine. Trends Biochem. Sci. 2016, 41, 77–93. [Google Scholar] [CrossRef]
- Tanaka, K. The proteasome: Overview of structure and functions. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2009, 85, 12–36. [Google Scholar] [CrossRef] [PubMed]
- Groll, M.; Ditzel, L.; Löwe, J.; Stock, D.; Bochtler, M.; Bartunik, H.D.; Huber, R. Structure of 20S proteasome from yeast at 2.4 A resolution. Nature 2017, 386, 463–471. [Google Scholar] [CrossRef]
- Unno, M.; Mizushima, T.; Morimoto, Y.; Tomisugi, Y.; Tanaka, K.; Yasuoka, N.; Tsukihara, T. The structure of the mammalian 20S proteasome at 2.75 A resolution. Structure 2002, 10, 609–618. [Google Scholar] [CrossRef]
- Lowe, J.; Stock, D.; Jap, B.; Zwickl, P.; Baumeister, W.; Huber, R. Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 A resolution. Science 1995, 268, 533–539. [Google Scholar] [CrossRef]
- Rabl, J.; Smith, D.M.; Yu, Y.; Chang, S.C.; Goldberg, A.L.; Cheng, Y. Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases. Mol. Cell 2008, 30, 360–368. [Google Scholar] [CrossRef] [PubMed]
- Kumoi, K.; Satoh, T.; Murata, K.; Hiromoto, T.; Mizushima, T.; Kamiya, Y.; Noda, M.; Uchiyama, S.; Yagi, H.; Kato, K. An archaeal homolog of proteasome assembly factor functions as a proteasome activator. PLoS ONE 2013, 8, e60294. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Wu, J.; Lu, Y.; Ma, Y.B.; Lee, B.H.; Yu, Z.; Ouyang, Q.; Finley, D.J.; Kirschner, M.W.; Mao, Y. Structural basis for dynamic regulation of the human 26S proteasome. Proc. Natl. Acad. Sci. USA 2016, 113, 12991–12996. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Wang, W.L.; Yu, D.; Ouyang, Q.; Lu, Y.; Mao, Y. Structural mechanism for nucleotide-driven remodeling of the AAA-ATPase unfoldase in the activated human 26S proteasome. Nat. Commun. 2018, 9, 1360. [Google Scholar] [CrossRef]
- Dong, Y.; Zhang, S.; Wu, Z.; Li, X.; Wang, W.L.; Zhu, Y.; Stoilova-McPhie, S.; Lu, Y.; Finley, D.; Mao, Y. Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome. Nature 2019, 565, 49–55. [Google Scholar] [CrossRef]
- Yu, Z.; Yu, Y.; Wang, F.; Myasnikov, A.G.; Coffino, P.; Cheng, Y. Allosteric coupling between α-rings of the 20S proteasome. Nat. Commun. 2020, 11, 4580. [Google Scholar] [CrossRef]
- Dorn, I.T.; Eschrich, R.; Seemöller, E.; Guckenberger, R.; Tampé, R. High-resolution AFM-imaging and mechanistic analysis of the 20 S proteasome. J. Mol. Biol. 1999, 288, 1027–1036. [Google Scholar] [CrossRef]
- Osmulski, P.A.; Hochstrasser, M.; Gaczynska, M. A tetrahedral transition state at the active sites of the 20S proteasome is coupled to opening of the alpha-ring channel. Structure 2009, 17, 1137–1147. [Google Scholar] [CrossRef]
- Yagi-Utsumi, M.; Sikdar, A.; Song, C.; Park, J.; Inoue, R.; Watanabe, H.; Burton-Smith, R.N.; Kozai, T.; Suzuki, T.; Kodama, A.; et al. Supramolecular tholos-like architecture constituted by archaeal proteins without functional annotation. Sci. Rep. 2020, 10, 1540. [Google Scholar] [CrossRef] [PubMed]
- Religa, T.L.; Sprangers, R.; Kay, L.E. Dynamic regulation of archaeal proteasome gate opening as studied by TROSY NMR. Science 2010, 328, 98–102. [Google Scholar] [CrossRef]
- Rennella, E.; Huang, R.; Yu, Z.; Kay, L.E. Exploring long-range cooperativity in the 20S proteasome core particle from Thermoplasma acidophilum using methyl-TROSY-based NMR. Proc. Natl. Acad. Sci. USA 2020, 117, 5298–5309. [Google Scholar] [CrossRef]
- Sprangers, R.; Kay, L.E. Quantitative dynamics and binding studies of the 20S proteasome by NMR. Nature 2007, 445, 618–622. [Google Scholar] [CrossRef] [PubMed]
- Budenholzer, L.; Cheng, C.L.; Li, Y.; Hochstrasser, M. Proteasome Structure and Assembly. J. Mol. Biol. 2017, 429, 3500–3524. [Google Scholar] [CrossRef] [PubMed]
- Kato, K.; Satoh, T. Structural insights on the dynamics of proteasome formation. Biophys. Rev. 2018, 10, 597–604. [Google Scholar] [CrossRef]
- Gerards, W.L.; Enzlin, J.; Haner, M.; Hendriks, I.L.; Aebi, U.; Bloemendal, H.; Boelens, W. The human alpha-type proteasomal subunit HsC8 forms a double ringlike structure, but does not assemble into proteasome-like particles with the beta-type subunits HsDelta or HsBPROS26. J. Biol. Chem. 2017, 272, 10080–10086. [Google Scholar] [CrossRef] [PubMed]
- Ishii, K.; Noda, M.; Yagi, H.; Thammaporn, R.; Seetaha, S.; Satoh, T.; Kato, K.; Uchiyama, S. Disassembly of the self-assembled, double-ring structure of proteasome alpha7 homo-tetradecamer by alpha6. Sci. Rep. 2015, 5, 18167. [Google Scholar] [CrossRef] [PubMed]
- Sugiyama, M.; Hamada, K.; Kato, K.; Kurimoto, E.; Okamoto, K.; Morimoto, Y.; Ikeda, S.; Naito, S.; Furusaka, M.; Itoh, K.; et al. SANS simulation of aggregated protein in aqueous solution. Nucl. Instrum. Methods. Phys. Res. A 2009, 600, 272–274. [Google Scholar] [CrossRef]
- Sugiyama, M.; Kurimoto, E.; Yagi, H.; Mori, K.; Fukunaga, T.; Hirai, M.; Zaccai, G.; Kato, K. Kinetic asymmetry of subunit exchange of homooligomeric protein as revealed by deuteration-assisted small-angle neutron scattering. Biophys. J. 2011, 101, 2037–2042. [Google Scholar] [CrossRef]
- Sekiguchi, T.; Satoh, T.; Kurimoto, E.; Song, C.; Kozai, T.; Watanabe, H.; Ishii, K.; Yagi, H.; Yanaka, S.; Uchiyama, S.; et al. Mutational and Combinatorial Control of Self-Assembling and Disassembling of Human Proteasome alpha Subunits. Int. J. Mol. Sci. 2019, 20, 2308. [Google Scholar] [CrossRef]
- Kozai, T.; Sekiguchi, T.; Satoh, T.; Yagi, H.; Kato, K.; Uchihashi, T. Two-step process for disassembly mechanism of proteasome alpha7 homo-tetradecamer by alpha6 revealed by high-speed atomic force microscopy. Sci. Rep. 2017, 7, 15373. [Google Scholar] [CrossRef] [PubMed]
- Kimanius, D.; Forsberg, B.O.; Scheres, S.; Lindahl, E. Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. eLife 2016, 5, e18722. [Google Scholar] [CrossRef]
- Rohou, A.; Grigorieff, N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 2015, 192, 216–221. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comp. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef]
- Adams, P.D.; Afonine, P.V.; Bunkoczi, G.; Chen, V.B. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 213–221. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, C.; Satoh, T.; Sekiguchi, T.; Kato, K.; Murata, K. Structural Fluctuations of the Human Proteasome α7 Homo-Tetradecamer Double Ring Imply the Proteasomal α-Ring Assembly Mechanism. Int. J. Mol. Sci. 2021, 22, 4519. https://doi.org/10.3390/ijms22094519
Song C, Satoh T, Sekiguchi T, Kato K, Murata K. Structural Fluctuations of the Human Proteasome α7 Homo-Tetradecamer Double Ring Imply the Proteasomal α-Ring Assembly Mechanism. International Journal of Molecular Sciences. 2021; 22(9):4519. https://doi.org/10.3390/ijms22094519
Chicago/Turabian StyleSong, Chihong, Tadashi Satoh, Taichiro Sekiguchi, Koichi Kato, and Kazuyoshi Murata. 2021. "Structural Fluctuations of the Human Proteasome α7 Homo-Tetradecamer Double Ring Imply the Proteasomal α-Ring Assembly Mechanism" International Journal of Molecular Sciences 22, no. 9: 4519. https://doi.org/10.3390/ijms22094519
APA StyleSong, C., Satoh, T., Sekiguchi, T., Kato, K., & Murata, K. (2021). Structural Fluctuations of the Human Proteasome α7 Homo-Tetradecamer Double Ring Imply the Proteasomal α-Ring Assembly Mechanism. International Journal of Molecular Sciences, 22(9), 4519. https://doi.org/10.3390/ijms22094519