Molecular Characterization and Functional Analysis of Two Steroidogenic Genes TSPO and SMAD4 in Yellow Catfish
Abstract
1. Introduction
2. Results
2.1. Molecular Characterization
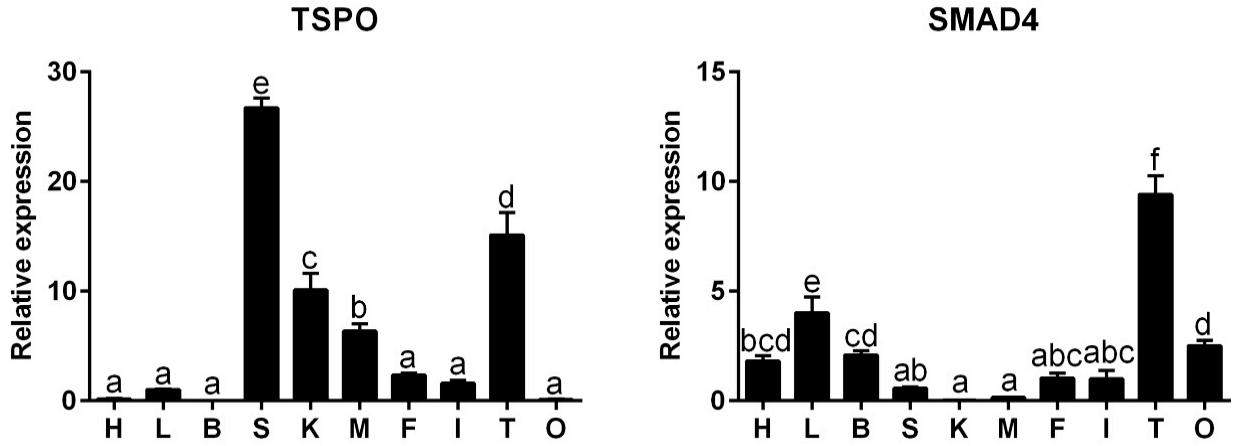
2.2. Tissue Distribution of Gene Expression
2.3. Sequence Analysis of the Promoter Regions of TSPO and SMAD4
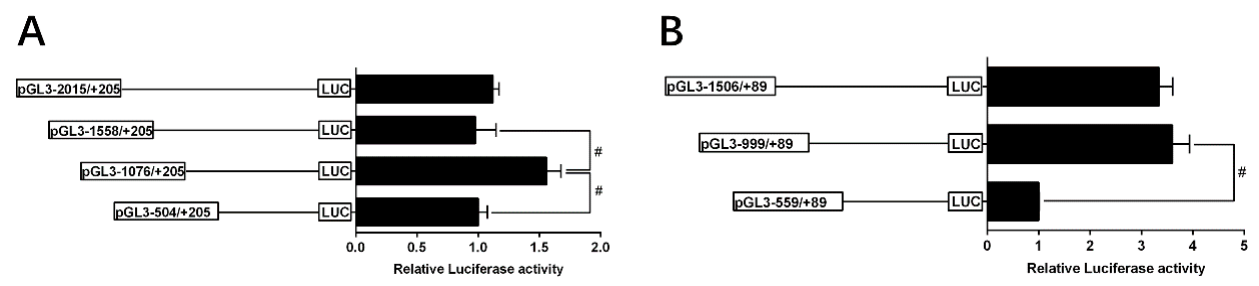
2.4. 5’-Deletion Assay of the Regions of TSPO and SAMD4 Promoters
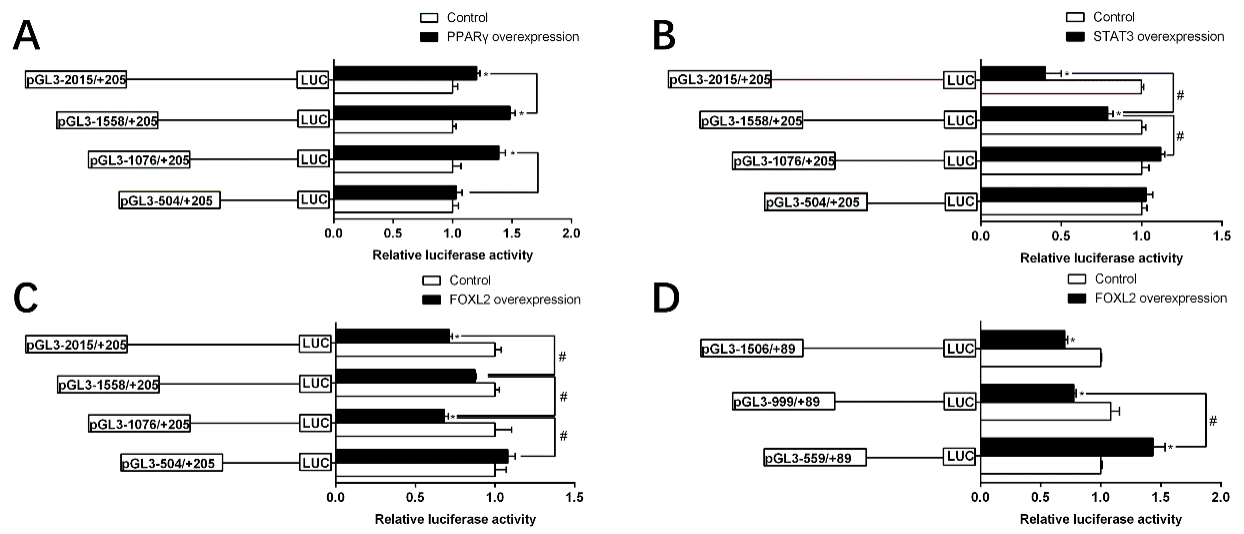
2.5. Site-Mutation Analysis of Binding Sites on the Regions of TSPO and SMAD4 Promoters
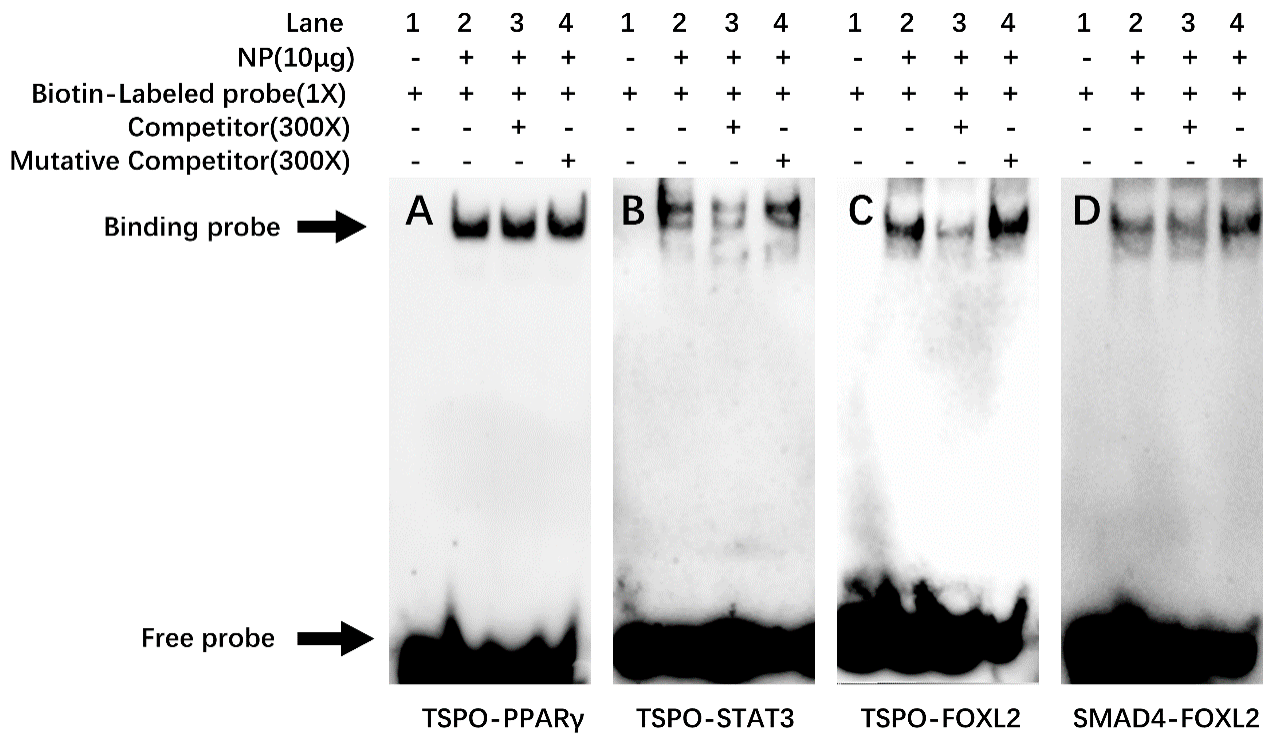
2.6. EMSA for the Confirmation of the Functional Binding of PPARγ, STAT3 and FOXL2 on the TSPO and SMAD4 Promoters
3. Discussion
4. Materials and Methods
4.1. Ethical Statement and Experimental Procedures
4.2. Experimental Animals and Reagents
4.3. Expt. 1: Cloning and Tissue Distribution of Gene Expression
4.3.1. RNA Isolation, cDNA Cloning and Sequence Editing
4.3.2. Real-Time Quantitative PCR (qPCR)
4.4. Expt. 2: Structure and Functional Analysis of Promoter
4.4.1. Cloning of Promoter and Plasmid Construction
4.4.2. Sequence Analysis
4.4.3. Transfections and Luciferase Assays
4.4.4. Site-Mutation Analysis of Binding Sites on the TSPO and SMAD4 Promoters
4.4.5. Electrophoretic Mobility-Shift Assay (EMSA)
4.5. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Lacapère, J.-J.; Papadopoulos, V. Peripheral-Type Benzodiazepine Receptor: Structure and Function of a Cholesterol-Binding Protein in Steroid and Bile Acid Biosynthesis. Steroids 2003, 68, 569–585. [Google Scholar] [CrossRef]
- Li, J.; Daly, E.; Campioli, E.; Wabitsch, M.; Papadopoulos, V. De Novo Synthesis of Steroids and Oxysterols in Adipocytes. J. Biol. Chem. 2014, 289, 747–764. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Du, X.; Pan, Z.; Zhang, L.; Li, Q. The Transcription Factor SMAD4 and miR-10b Contribute to E2 Release and Cell Apoptosis in Ovarian Granulosa Cells by Targeting CYP19A1. Mol. Cell. Endocrinol. 2018, 476, 84–95. [Google Scholar] [CrossRef] [PubMed]
- Rashid, H.; Kitano, H.; Lee, K.H.; Nii, S.; Shigematsu, T.; Kadomura, K.; Yamaguchi, A.; Matsuyama, M. Fugu (Takifugu Rubripes) Sexual Differentiation: CYP19 Regulation and Aromatase Inhibitor Induced Testicular Development. Sex. Dev. 2007, 1, 311–322. [Google Scholar] [CrossRef] [PubMed]
- Yoo, S.-Y.; Lee, J.-A.; Shin, Y.; Cho, N.-Y.; Bae, J.M.; Kang, G.H. Clinicopathological Characterization and Prognostic Implication of SMAD4 Expression in Colorectal Carcinoma. J. Pathol. Transl. Med. 2019, 53, 289–297. [Google Scholar] [CrossRef]
- Batarseh, A.; Barlow, K.D.; Martinez-Arguelles, D.B.; Papadopoulos, V. Functional Characterization of the Human Translocator Protein (18kDa) Gene Promoter in Human Breast Cancer Cell Lines. Biochim. Biophys. Acta Bioenerg. 2012, 1819, 38–56. [Google Scholar] [CrossRef]
- Nikolic, A.; Ristanovic, M.; Zivaljevic, V.; Rankov, A.D.; Radojkovic, D.; Paunovic, I. SMAD4 Gene Promoter Mutations in Patients with Thyroid Tumors. Exp. Mol. Pathol. 2015, 99, 100–103. [Google Scholar] [CrossRef]
- Chen, S.-W.; Wu, K.; Lv, W.-H.; Chen, F.; Song, C.-C.; Luo, Z. Functional Analysis of Two Zinc (Zn) Transporters (ZIP3 and ZIP8) Promoters and Their Distinct Response to MTF1 and RREB1 in the Regulation of Zn Metabolism. Int. J. Mol. Sci. 2020, 21, 6135. [Google Scholar] [CrossRef]
- Minami, R.; Kitazawa, R.; Maeda, S.; Kitazawa, S. Analysis of 5′-Flanking Region of Human Smad4 (DPC4) Gene. Biochim. Biophys. Acta Gene Struct. Expr. 1998, 1443, 182–185. [Google Scholar] [CrossRef]
- Rashid, K.; Geissl, L.; Wolf, A.; Karlstetter, M.; Langmann, T. Transcriptional Regulation of Translocator Protein (18 kDa) (TSPO) in Microglia Requires Pu.1, Ap1 and Sp Factors. Biochim. Biophys. Acta Gene Regul. Mech. 2018, 1861, 1119–1133. [Google Scholar] [CrossRef]
- Puttabyatappa, M.; Vandevoort, C.A.; Chaffin, C.L. hCG-Induced Down-Regulation of PPARγ and Liver X Receptors Promotes Periovulatory Progesterone Synthesis by Macaque Granulosa Cells. Endocrinology 2010, 151, 5865–5872. [Google Scholar] [CrossRef][Green Version]
- Wang, D.-S.; Kobayashi, T.; Zhou, L.-Y.; Paul-Prasanth, B.; Ijiri, S.; Sakai, F.; Okubo, K.; Morohashi, K.-I.; Nagahama, Y. Foxl2 Up-Regulates Aromatase Gene Transcription in a Female-Specific Manner by Binding to the Promoter as Well as Interacting with Ad4 Binding Protein/Steroidogenic Factor 1. Mol. Endocrinol. 2007, 21, 712–725. [Google Scholar] [CrossRef]
- Lv, W.-H.; Chen, G.-H.; Zhuo, M.-Q.; Xu, Y.-H.; Xu, Y.-C.; Tan, X.-Y. Functional Analysis of Steroidogenic Factor 1 (sf-1) and 17α-Hydroxylase/Lyase (cyp17α) Promoters in Yellow Catfish Pelteobagrus fulvidraco. Int. J. Mol. Sci. 2020, 22, 195. [Google Scholar] [CrossRef]
- Lejri, I.; Grimm, A.; Hallé, F.; Abarghaz, M.; Klein, C.; Maitre, M.; Schmitt, M.; Bourguignon, J.-J.; Mensah-Nyagan, A.G.; Bihel, F.; et al. TSPO Ligands Boost Mitochondrial Function and Pregnenolone Synthesis. J. Alzheimer’s Dis. 2019, 72, 1045–1058. [Google Scholar] [CrossRef]
- Zeno, S.; Veenman, L.; Katz, Y.; Bode, J.; Gavish, M.; Zaaroor, M. The 18 kDa Mitochondrial Translocator Protein (TSPO) Prevents Accumulation of Protoporphyrin IX. Involvement of Reactive Oxygen Species (ROS). Curr. Mol. Med. 2012, 12, 494–501. [Google Scholar]
- McCarthy, A.J.; Chetty, R. SMAD4/DPC4. J. Clin. Pathol. 2018, 71, 661–664. [Google Scholar] [CrossRef]
- Pangas, S.A.; Li, X.; Robertson, E.J.; Matzuk, M.M. Premature Luteinization and Cumulus Cell Defects in Ovarian-SpecificSmad4Knockout Mice. Mol. Endocrinol. 2006, 20, 1406–1422. [Google Scholar] [CrossRef]
- Iatmanen-Harbi, S.; Senicourt, L.; Papadopoulos, V.; Lequin, O.; Lacapere, J.-J. Characterization of the High-Affinity Drug Ligand Binding Site of Mouse Recombinant TSPO. Int. J. Mol. Sci. 2019, 20, 1444. [Google Scholar] [CrossRef]
- Selvaraj, V.; Stocco, D.M. The Changing Landscape in Translocator Protein (TSPO) Function. Trends Endocrinol. Metab. 2015, 26, 341–348. [Google Scholar] [CrossRef]
- Jaremko, Ł.; Jaremko, M.; Giller, K.; Becker, S.; Zweckstetter, M. Conformational Flexibility in the Transmembrane Protein TSPO. Chemistry 2015, 21, 16555–16563. [Google Scholar] [CrossRef]
- Barnes, E.; Askham, J.M.; Jones, P.F. cDNA Cloning and Molecular Characterization of Mink SMAD4. DNA Seq. 2002, 13, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Osman, A.; Niles, E.G.; LoVerde, P.T. Expression of Functional Schistosoma Mansoni SMAD4: Role in ERK-Mediated Transforming Growth Factor Beta (TGF-Beta) Down-Regulation. J. Biol. Chem. 2004, 279, 6474–6486. [Google Scholar] [CrossRef] [PubMed]
- Inman, G.J.; Nicolás, F.J.; Hill, C.S. Nucleocytoplasmic Shuttling of SMADs 2, 3, and 4 Permits Sensing of TGF-Beta Receptor Activity. Mol. Cell 2002, 10, 283–294. [Google Scholar] [CrossRef]
- Reguly, T.; Wrana, J.L. In or Out? The Dynamics of Smad Nucleocytoplasmic Shuttling. Trends Cell Biol. 2003, 13, 216–220. [Google Scholar] [CrossRef]
- Rasheeda, M.K.; Kagawa, H.; Kirubagaran, R.; Dutta-Gupta, A.; Senthilkumaran, B. Cloning, Expression and Enzyme Activity Analysis of Testicular 11beta-Hydroxysteroid Dehydrogenase during Seasonal Cycle and after hCG Induction in Air-Breathing Catfish Clarias Gariepinus. J. Steroid Biochem. Mol. Biol. 2010, 120, 1–10. [Google Scholar] [CrossRef]
- Swart, A.C.; Schloms, L.; Storbeck, K.-H.; Bloem, L.M.; Du Toit, T.; Quanson, J.L.; Rainey, W.E.; Swart, P. 11β-Hydroxyandrostenedione, the Product of Androstenedione Metabolism in the Adrenal, is Metabolized in LNCaP Cells by 5α-Reductase Yielding 11β-Hydroxy-5α-Androstanedione. J. Steroid Biochem. Mol. Biol. 2013, 138, 132–142. [Google Scholar] [CrossRef]
- Rampon, C.; Rouzaffour, M.; Ostuni, M.; Dufourcq, P.; Girard, C.; Freyssinet, J.M.; Lacapere, J.-J.; Schweizer-Groyer, G.; Vriz, S. Translocator Protein (18 kDa) is Involved in Primitive Erythropoiesis in Zebrafish. FASEB J. 2009, 23, 4181–4192. [Google Scholar] [CrossRef]
- Huang, Z.; Yuan, X.; Wang, M.; Wu, N.; Song, Y.; Chen, Y.; Zhang, Y.; Xu, Q.; Chen, G.; Zhao, W. Molecular Cloning of the SMAD4 Gene and its mRNA Expression Analysis in Ovarian Follicles of the Yangzhou Goose (Anser Cygnoides). Br. Poult. Sci. 2016, 57, 515–521. [Google Scholar] [CrossRef]
- Fortin, J.; Boehm, U.; Deng, C.; Treier, M.; Bernard, D.J. Follicle-Stimulating Hormone Synthesis and Fertility Depend on SMAD4 and FOXL2. FASEB J. 2014, 28, 3396–3410. [Google Scholar] [CrossRef]
- Goodrich, J.A.; Tjian, R. Unexpected Roles for Core Promoter Recognition Factors in Cell-Type-Specific Transcription and Gene Regulation. Nat. Rev. Genet. 2010, 11, 549–558. [Google Scholar] [CrossRef]
- Shimoyama, S.; Furukawa, T.; Ogata, Y.; Nikaido, Y.; Koga, K.; Sakamoto, Y.; Ueno, S.; Nakamura, K. Lipopolysaccharide induces Mouse Translocator Protein (18 kDa) Expression via the AP-1 Complex in the Microglial Cell Line, BV-2. PLoS ONE 2019, 14, e0222861. [Google Scholar] [CrossRef]
- Wierstra, I. Sp1: Emerging Roles—Beyond Constitutive Activation of TATA-Less Housekeeping Genes. Biochem. Biophys. Res. Commun. 2008, 372, 1–13. [Google Scholar] [CrossRef]
- Calva, D.; Dahdaleh, F.S.; Woodfield, G.; Weigel, R.J.; Carr, J.C.; Chinnathambi, S.; Howe, J.R. Discovery of SMAD4 Promoters, Transcription Factor Binding Sites and Deletions in Juvenile Polyposis Patients. Nucleic Acids Res. 2011, 39, 5369–5378. [Google Scholar] [CrossRef]
- Smale, S.T.; Kadonaga, J.T. The RNA Polymerase II Core Promoter. Annu. Rev. Biochem. 2003, 72, 449–479. [Google Scholar] [CrossRef]
- Lenhard, B.; Sandelin, A.; Carninci, P. Metazoan Promoters: Emerging Characteristics and Insights into Transcriptional Regulation. Nat. Rev. Genet. 2012, 13, 233–245. [Google Scholar] [CrossRef]
- Chan, T.-M.; Leung, K.-S.; Lee, K.-H. TFBS Identification based on Genetic Algorithm with Combined Representations and Adaptive Post-Processing. Bioinformatics 2007, 24, 341–349. [Google Scholar] [CrossRef]
- Giatzakis, C.; Papadopoulos, V. Differential Utilization of the Promoter of Peripheral-Type Benzodiazepine Receptor by Steroidogenic Versus Nonsteroidogenic Cell Lines and the Role of Sp1 and Sp3 in the Regulation of Basal Activity. Endocrinology 2004, 145, 1113–1123. [Google Scholar] [CrossRef]
- Sridevi, P.; Chaitanya, R.; Dutta-Gupta, A.; Senthilkumaran, B. FTZ-F1 and FOXL2 Up-Regulate Catfish Brain Aromatase Gene Transcription by Specific Binding to the Promoter Motifs. Biochim. Biophys. Acta Bioenerg. 2012, 1819, 57–66. [Google Scholar] [CrossRef]
- Batarseh, A.; Li, J.; Papadopoulos, V. Protein Kinase C Epsilon Regulation of Translocator Protein (18 kDa) TSPO Gene Expression is Mediated through a MAPK Pathway Targeting STAT3 and c-Jun Transcription Factors. Biochemistry 2010, 49, 4766–4778. [Google Scholar] [CrossRef]
- Wei, C.-C.; Luo, Z.; Song, Y.-F.; Pan, Y.-X.; Wu, K.; You, W.-J. Identification of Autophagy related Genes LC3 and ATG4 from Yellow Catfish Pelteobagrus Fulvidraco and their Transcriptional Responses to Waterborne and Dietborne Zinc Exposure. Chemosphere 2017, 175, 228–238. [Google Scholar] [CrossRef]
- Chen, Q.-L.; Luo, Z.; Wu, K.; Huang, C.; Zhuo, M.-Q.; Song, Y.-F.; Hu, W. Differential Effects of Dietary Copper Deficiency and Excess on Lipid Metabolism in Yellow Catfish Pelteobagrus Fulvidraco. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2015, 184, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Gong, G.; Dan, C.; Chen, N.; Gui, J.-F.; Mei, J.; Xiao, S.; Guo, W.; Huang, P.; Xiong, Y.; Wu, J.; et al. Chromosomal-Level Assembly of Yellow Catfish Genome using Third-Generation DNA Sequencing and Hi-C Analysis. GigaScience 2018, 7. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.-H.; Luo, Z.; Wu, K.; Fan, Y.-F.; You, W.-J.; Zhang, L.-H. Structure and Functional Analysis of Promoters from Two Liver Isoforms of CPT I in Grass Carp Ctenopharyngodon idella. Int. J. Mol. Sci. 2017, 18, 2405. [Google Scholar] [CrossRef] [PubMed]



| Gene | Accession No. | 5′ UTR (bp) | ORF (bp) | 3′ UTR (bp) | Full Length (bp) | Protein (aa) |
|---|---|---|---|---|---|---|
| TSPO | MN188059 | 116 | 486 | 350 | 952 | 161 |
| SMAD4 | MN188058 | 202 | 1725 | 294 | 2221 | 574 |
| Gene | Ictalurus punctatus | Danio rerio | Xenopus tropicalis | Homo sapiens | Rattus norvegicus |
|---|---|---|---|---|---|
| TSPO | 69.12 | 61.29 | 50.69 | 41.94 | 39.63 |
| SMAD4 | 94.19 | 63.93 | 65.64 | 66.32 | 66.32 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, F.; Zhong, C.-C.; Song, C.-C.; Chen, S.-W.; He, Y.; Tan, X.-Y. Molecular Characterization and Functional Analysis of Two Steroidogenic Genes TSPO and SMAD4 in Yellow Catfish. Int. J. Mol. Sci. 2021, 22, 4505. https://doi.org/10.3390/ijms22094505
Chen F, Zhong C-C, Song C-C, Chen S-W, He Y, Tan X-Y. Molecular Characterization and Functional Analysis of Two Steroidogenic Genes TSPO and SMAD4 in Yellow Catfish. International Journal of Molecular Sciences. 2021; 22(9):4505. https://doi.org/10.3390/ijms22094505
Chicago/Turabian StyleChen, Fang, Chong-Chao Zhong, Chang-Chun Song, Shu-Wei Chen, Yang He, and Xiao-Ying Tan. 2021. "Molecular Characterization and Functional Analysis of Two Steroidogenic Genes TSPO and SMAD4 in Yellow Catfish" International Journal of Molecular Sciences 22, no. 9: 4505. https://doi.org/10.3390/ijms22094505
APA StyleChen, F., Zhong, C.-C., Song, C.-C., Chen, S.-W., He, Y., & Tan, X.-Y. (2021). Molecular Characterization and Functional Analysis of Two Steroidogenic Genes TSPO and SMAD4 in Yellow Catfish. International Journal of Molecular Sciences, 22(9), 4505. https://doi.org/10.3390/ijms22094505






