HERV-Derived Ervpb1 Is Conserved in Simiiformes, Exhibiting Expression in Hematopoietic Cell Lineages Including Macrophages
Abstract
1. Introduction
2. Results
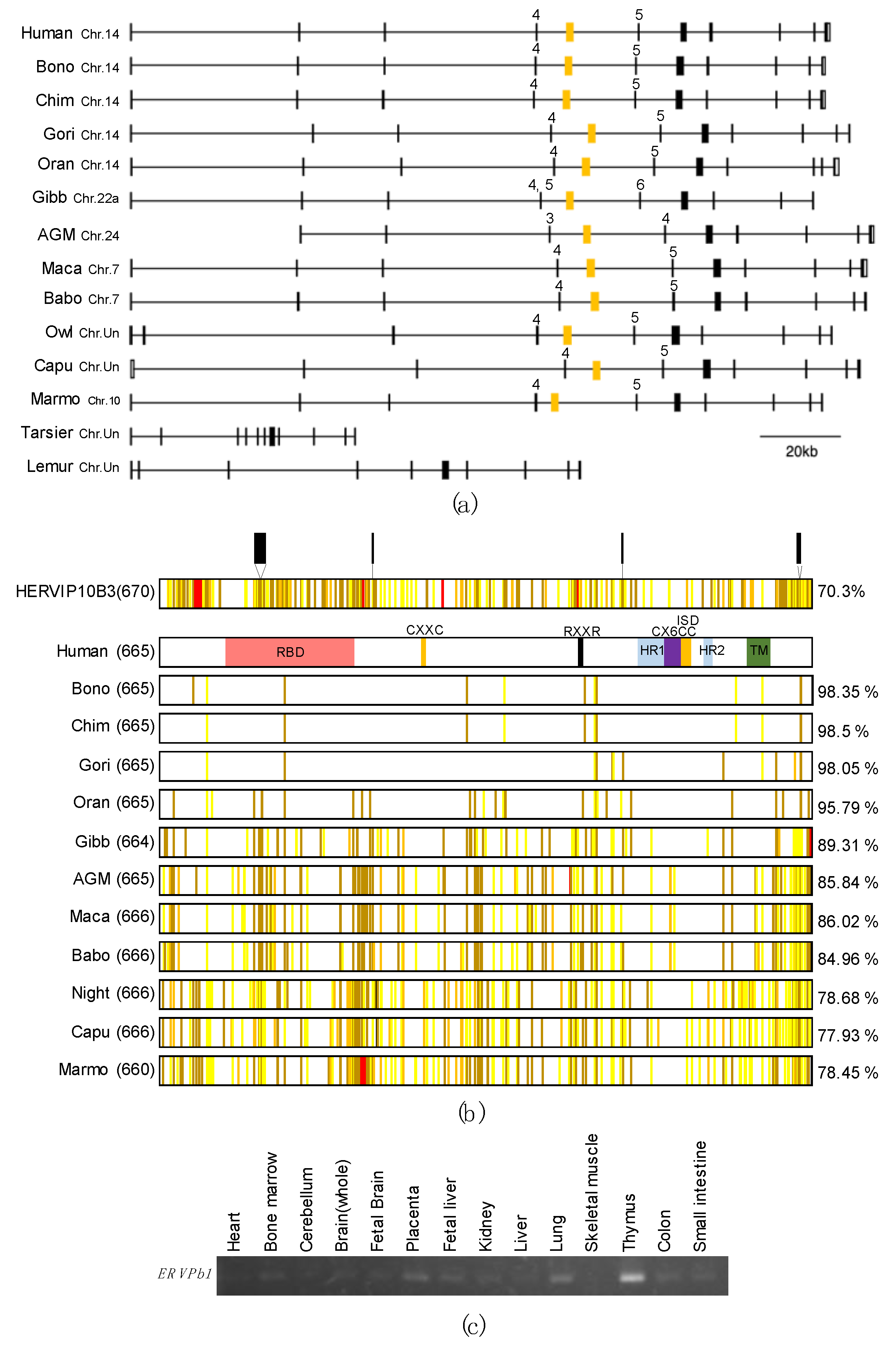
2.1. ERVPb1 Is Unique to Simiiformes and Exhibits a Quite Low Level of Expression in Limited Tissues and Organs in Humans
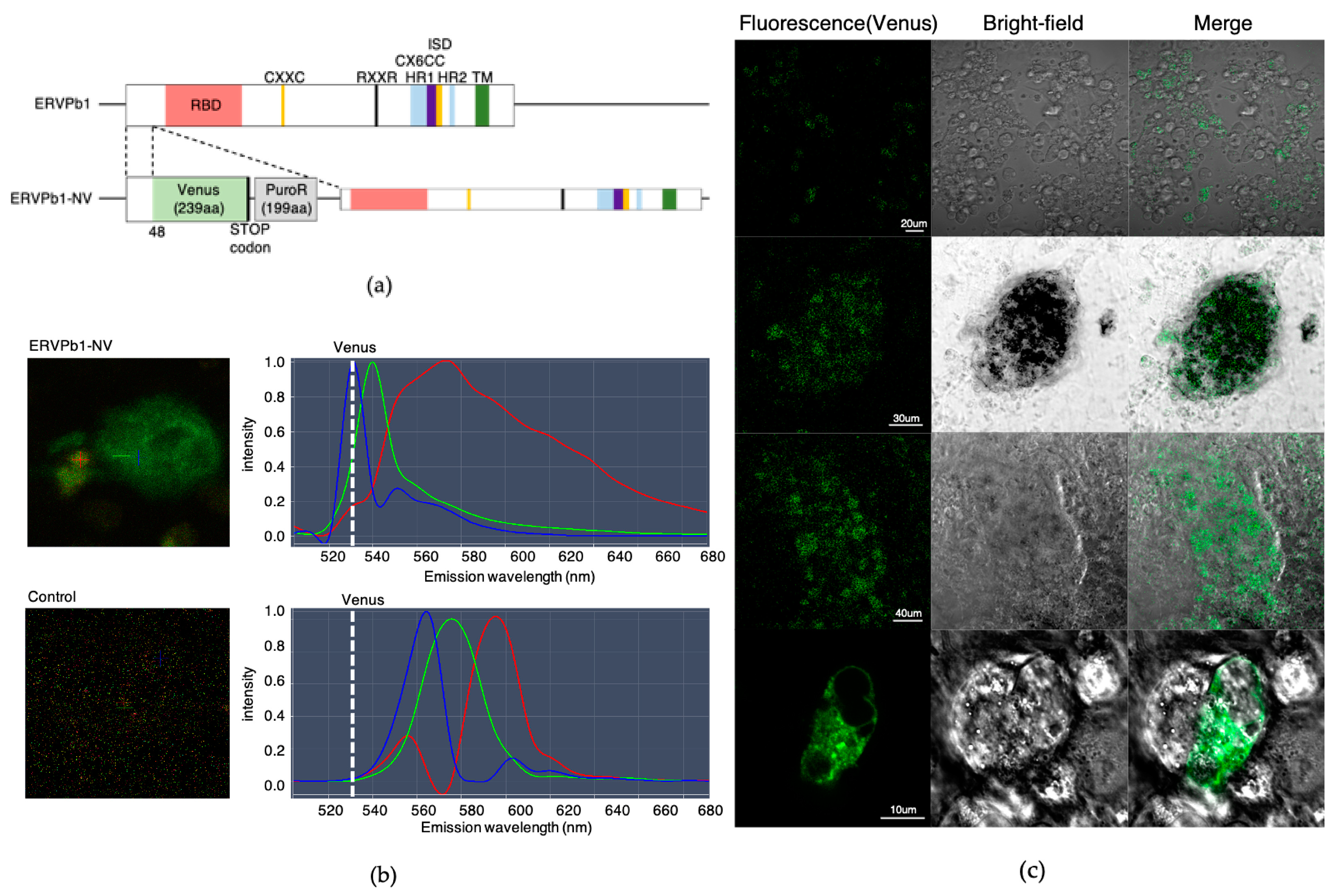
2.2. Generation of Human iPSCs with an ERVPb1–Venus Fusion Gene in Its Endogenous Locus
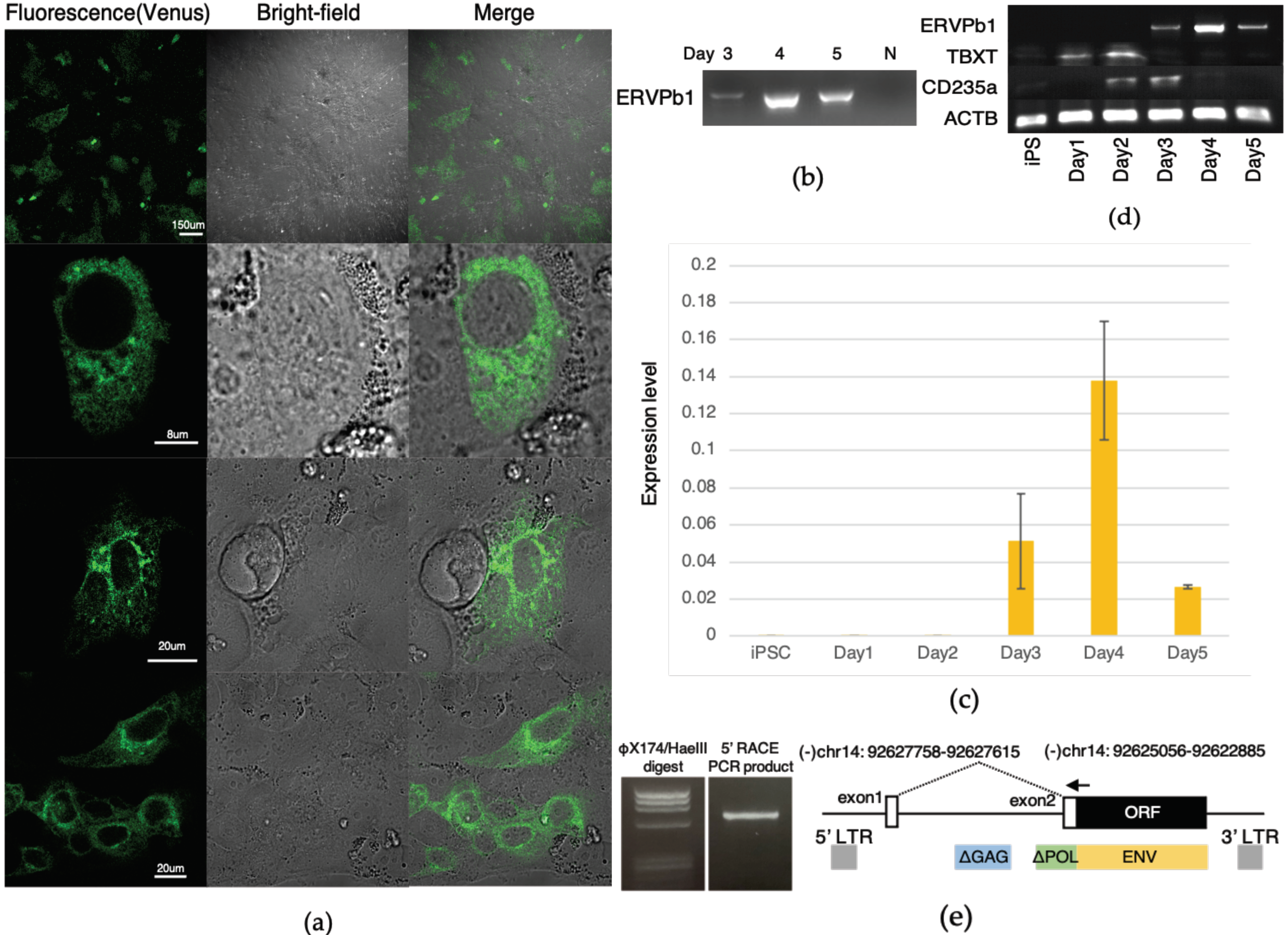
2.3. Detection of the ERVPb1–Venus Fusion Protein in a Hematopoietic Cell Lineage
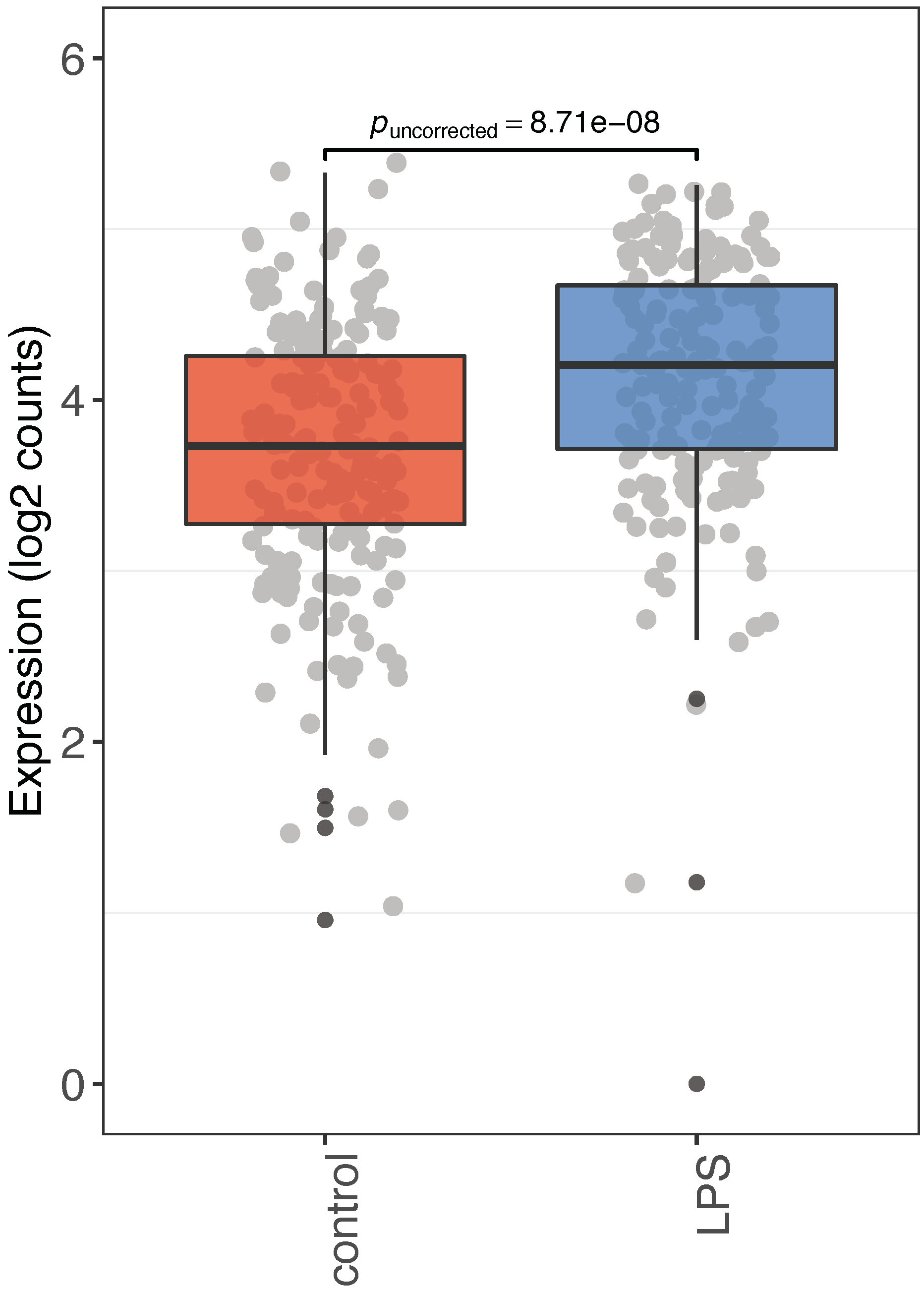
3. Discussion
4. Materials and Methods
4.1. Comparison of ERVPb1 ORF in Simiiformes
4.2. Analysis of Short Read RNA-Seq Data
4.3. Generation of Human iPSCs with an ERVPb1-NV Construct
4.4. Culture and Differentiation of Human iPSCs
4.5. Gene Expression Analysis by Real-Time PCR
4.6. Rapid Amplification of cDNA Ends (RACE)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bock, M.; Stoye, J.P. Endogenous retroviruses and the human germline. Curr. Opin. Genet. Dev. 2000, 10, 651–655. [Google Scholar] [CrossRef]
- Belshaw, R.; Pereira, V.; Katzourakis, A.; Talbot, G.; Paces, J.; Burt, A.; Tristem, M. Long-term reinfection of the human genome by endogenous retroviruses. Proc. Natl. Acad. Sci. USA 2004, 101, 4894–4899. [Google Scholar] [CrossRef]
- Villesen, P.; Aagaard, L.; Wiuf, C.; Pedersen, F.S. Identification of endogenous retroviral reading frames in the human genome. Retrovirology 2004, 1, 32. [Google Scholar] [CrossRef]
- Nakagawa, S.; Takahashi, M.U. gEVE: A genome-based endogenous viral element database provides comprehensive viral protein-coding sequences in mammalian genomes. Database 2016, 2016, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Ono, R.; Kobayashi, S.; Wagatsuma, H.; Aisaka, K.; Kohda, T.; Kaneko-Ishino, T.; Ishino, F. A retrotransposon-derived gene, PEG10, is a novel imprinted gene located on human chromosome 7q21. Genomics 2001, 73, 232–237. [Google Scholar] [CrossRef]
- Ono, R.; Shiura, H.; Aburatani, H.; Kohda, T.; Kaneko-Ishino, T.; Ishino, F. Identification of a large novel imprinted gene cluster on mouse proximal chromosome 6. Genome Res. 2003, 13, 1696–1705. [Google Scholar] [CrossRef]
- Ono, R.; Nakamura, K.; Inoue, K.; Naruse, M.; Usami, T.; Wakisaka-Saito, N.; Hino, T.; Suzuki-Migishima, R.; Ogonuki, N.; Miki, H.; et al. Deletion of Peg10, an imprinted gene acquired from a retrotransposon, causes early embryonic lethality. Nat. Genet. 2006, 38, 101–106. [Google Scholar] [CrossRef]
- Mi, S.; Lee, X.; Li, X.; Veldman, G.M.; Finnerty, H.; Racie, L.; LaVallie, E.; Tang, X.Y.; Edouard, P.; Howes, S.; et al. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 2000, 403, 785–789. [Google Scholar] [CrossRef]
- Blaise, S.; de Parseval, N.; Bénit, L.; Heidmann, T. Genomewide screening for fusogenic human endogenous retrovirus envelopes identifies syncytin 2, a gene conserved on primate evolution. Proc. Natl. Acad. Sci. USA 2003, 100, 13013–13018. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, J.; Sugimoto, M.; Bernstein, H.; Jinno, Y.; Schust, D. A novel human endogenous retroviral protein inhibits cell-cell fusion. Sci. Rep. 2013, 3, 1462. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.S.; Takenaka, O.; Crow, T.J. Isolation and phylogeny of endogenous retrovirus sequences belonging to the HERV-W family in primates. J. Gen. Virol. 1999, 80 Pt 10, 2613–2619. [Google Scholar] [CrossRef]
- Kaneko-Ishino, T.; Ishino, F. Mammalian-specific genomic functions: Newly acquired traits generated by genomic imprinting and LTR retrotransposon-derived genes in mammals. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2015, 91, 511–538. [Google Scholar] [CrossRef]
- Aagaard, L.; Villesen, P.; Kjeldbjerg, A.L.; Pedersen, F.S. The approximately 30-million-year-old ERVPb1 envelope gene is evolutionarily conserved among hominoids and Old World monkeys. Genomics 2005, 86, 685–691. [Google Scholar] [CrossRef][Green Version]
- Blaise, S.; de Parseval, N.; Heidmann, T. Functional characterization of two newly identified Human Endogenous Retrovirus coding envelope genes. Retrovirology 2005, 2, 19. [Google Scholar] [CrossRef]
- Lee, J.; Sutani, A.; Kaneko, R.; Takeuchi, J.; Sasano, T.; Kohda, T.; Ihara, K.; Takahashi, K.; Yamazoe, M.; Morio, T.; et al. In vitro generation of functional murine heart organoids via FGF4 and extracellular matrix. Nat. Commun. 2020, 11, 4283. [Google Scholar] [CrossRef]
- Huch, M.; Koo, B.K. Modeling mouse and human development using organoid cultures. Development 2015, 142, 3113–3125. [Google Scholar] [CrossRef]
- Douvaras, P.; Sun, B.; Wang, M.; Kruglikov, I.; Lallos, G.; Zimmer, M.; Terrenoire, C.; Zhang, B.; Gandy, S.; Schadt, E.; et al. Directed Differentiation of Human Pluripotent Stem Cells to Microglia. Stem. Cell Rep. 2017, 8, 1516–1524. [Google Scholar] [CrossRef] [PubMed]
- Takata, K.; Kozaki, T.; Lee, C.Z.W.; Thion, M.S.; Otsuka, M.; Lim, S.; Utami, K.H.; Fidan, K.; Park, D.S.; Malleret, B.; et al. Induced-Pluripotent-Stem-Cell-Derived Primitive Macrophages Provide a Platform for Modeling Tissue-Resident Macrophage Differentiation and Function. Immunity 2017, 47, 183–198.e186. [Google Scholar] [CrossRef]
- Steiper, M.E.; Young, N.M. The Timetree of Life; Oxford University Press: Oxford, UK, 2009; pp. 482–486. [Google Scholar]
- Quach, H.; Rotival, M.; Pothlichet, J.; Loh, Y.E.; Dannemann, M.; Zidane, N.; Laval, G.; Patin, E.; Harmant, C.; Lopez, M.; et al. Genetic Adaptation and Neandertal Admixture Shaped the Immune System of Human Populations. Cell 2016, 167, 643–656.e617. [Google Scholar] [CrossRef]
- Machinaga, A.; Ishihara, S.; Shirai, A.; Takase-Yoden, S. Splicing of Friend Murine Leukemia Virus env-mRNA Enhances Its Ability to Form Polysomes. Front. Microbiol. 2016, 7, 160. [Google Scholar] [CrossRef] [PubMed]
- Benešová, M.; Trejbalová, K.; Kovářová, D.; Vernerová, Z.; Hron, T.; Kučerová, D.; Hejnar, J. DNA hypomethylation and aberrant expression of the human endogenous retrovirus ERVWE1/syncytin-1 in seminomas. Retrovirology 2017, 14, 20. [Google Scholar] [CrossRef]
- Vargas, A.; Thiery, M.; Lafond, J.; Barbeau, B. Transcriptional and functional studies of Human Endogenous Retrovirus envelope EnvP(b) and EnvV genes in human trophoblasts. Virology 2012, 425, 1–10. [Google Scholar] [CrossRef]
- Sekita, Y.; Wagatsuma, H.; Nakamura, K.; Ono, R.; Kagami, M.; Wakisaka, N.; Hino, T.; Suzuki-Migishima, R.; Kohda, T.; Ogura, A.; et al. Role of retrotransposon-derived imprinted gene, Rtl1, in the feto-maternal interface of mouse placenta. Nat. Genet. 2008, 40, 243–248. [Google Scholar] [CrossRef]
- Naruse, M.; Ono, R.; Irie, M.; Nakamura, K.; Furuse, T.; Hino, T.; Oda, K.; Kashimura, M.; Yamada, I.; Wakana, S.; et al. Sirh7/Ldoc1 knockout mice exhibit placental P4 overproduction and delayed parturition. Development 2014, 141, 4763–4771. [Google Scholar] [CrossRef]
- Irie, M.; Yoshikawa, M.; Ono, R.; Iwafune, H.; Furuse, T.; Yamada, I.; Wakana, S.; Yamashita, Y.; Abe, T.; Ishino, F.; et al. Cognitive Function Related to the Sirh11/Zcchc16 Gene Acquired from an LTR Retrotransposon in Eutherians. PLoS Genet. 2015, 11, e1005521. [Google Scholar] [CrossRef]
- Irie, M.; Koga, A.; Kaneko-Ishino, T.; Ishino, F. An LTR Retrotransposon-Derived Gene Displays Lineage-Specific Structural and Putative Species-Specific Functional Variations in Eutherians. Front. Chem. 2016, 4, 26. [Google Scholar] [CrossRef] [PubMed]
- Kitazawa, M.; Tamura, M.; Kaneko-Ishino, T.; Ishino, F. Severe damage to the placental fetal capillary network causes mid- to late fetal lethality and reduction in placental size in Peg11/Rtl1 KO mice. Genes Cells 2017, 22, 174–188. [Google Scholar] [CrossRef] [PubMed]
- Kitazawa, M.; Hayashi, S.; Imamura, M.; Takeda, S.; Oishi, Y.; Kaneko-Ishino, T.; Ishino, F. Deficiency and overexpression of Rtl1 in the mouse cause distinct muscle abnormalities related to Temple and Kagami-Ogata syndromes. Development 2020, 147. [Google Scholar] [CrossRef] [PubMed]
- Kitazawa, M.; Sutani, A.; Kaneko-Ishino, T.; Ishino, F. The role of eutherian-specific RTL1 in the nervous system and its implications for the Kagami-Ogata and Temple syndromes. Genes Cells 2021, 26, 165–179. [Google Scholar] [CrossRef]
- Kaneko-Ishino, T.; Ishino, F. The role of genes domesticated from LTR retrotransposons and retroviruses in mammals. Front. Microbiol. 2012, 3, 262. [Google Scholar] [CrossRef]
- Kaneko-Ishino, T.; Ishino, F. Cooperation and Competition in Mammalian Evolution -Gene Domestication from LTR Retrotransposons. In Evolution, Origin of Life, Concepts and Methods; Pontarotti, P., Ed.; Springer Nature: Berlin/Heidelberg, Germany, 2019; Chapter 15; pp. 317–333. [Google Scholar]
- Ueda, M.T.; Kryukov, K.; Mitsuhashi, S.; Mitsuhashi, H.; Imanishi, T.; Nakagawa, S. Comprehensive genomic analysis reveals dynamic evolution of endogenous retroviruses that code for retroviral-like protein domains. Mob. DNA 2020, 11, 29. [Google Scholar] [CrossRef] [PubMed]
- Bao, W.; Kojima, K.K.; Kohany, O. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob. DNA 2015, 6, 11. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Shao, M.; Kingsford, C. Accurate assembly of transcripts through phase-preserving graph decomposition. Nat. Biotechnol. 2017, 35, 1167–1169. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Statisticat LLC. LaplacesDemon: Complete Environment for Bayesian Inference. Bayesian-Inference.com. R Package Version 16.1.1. 2018. Available online: https://cran.r-project.org/web/packages/LaplacesDemon/citation.html (accessed on 22 March 2019).
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Matsuzawa, A.; Lee, J.; Nakagawa, S.; Itoh, J.; Takahashi Ueda, M.; Mitsuhashi, S.; Kochi, Y.; Kaneko-Ishino, T.; Ishino, F. HERV-Derived Ervpb1 Is Conserved in Simiiformes, Exhibiting Expression in Hematopoietic Cell Lineages Including Macrophages. Int. J. Mol. Sci. 2021, 22, 4504. https://doi.org/10.3390/ijms22094504
Matsuzawa A, Lee J, Nakagawa S, Itoh J, Takahashi Ueda M, Mitsuhashi S, Kochi Y, Kaneko-Ishino T, Ishino F. HERV-Derived Ervpb1 Is Conserved in Simiiformes, Exhibiting Expression in Hematopoietic Cell Lineages Including Macrophages. International Journal of Molecular Sciences. 2021; 22(9):4504. https://doi.org/10.3390/ijms22094504
Chicago/Turabian StyleMatsuzawa, Ayumi, Jiyoung Lee, So Nakagawa, Johbu Itoh, Mahoko Takahashi Ueda, Satomi Mitsuhashi, Yuta Kochi, Tomoko Kaneko-Ishino, and Fumitoshi Ishino. 2021. "HERV-Derived Ervpb1 Is Conserved in Simiiformes, Exhibiting Expression in Hematopoietic Cell Lineages Including Macrophages" International Journal of Molecular Sciences 22, no. 9: 4504. https://doi.org/10.3390/ijms22094504
APA StyleMatsuzawa, A., Lee, J., Nakagawa, S., Itoh, J., Takahashi Ueda, M., Mitsuhashi, S., Kochi, Y., Kaneko-Ishino, T., & Ishino, F. (2021). HERV-Derived Ervpb1 Is Conserved in Simiiformes, Exhibiting Expression in Hematopoietic Cell Lineages Including Macrophages. International Journal of Molecular Sciences, 22(9), 4504. https://doi.org/10.3390/ijms22094504






