Barley Seeds miRNome Stability during Long-Term Storage and Aging
Abstract
1. Introduction
2. Results
2.1. Overview of Small RNA Library Sequencing
2.2. Identification of Known and Novel miRNAs
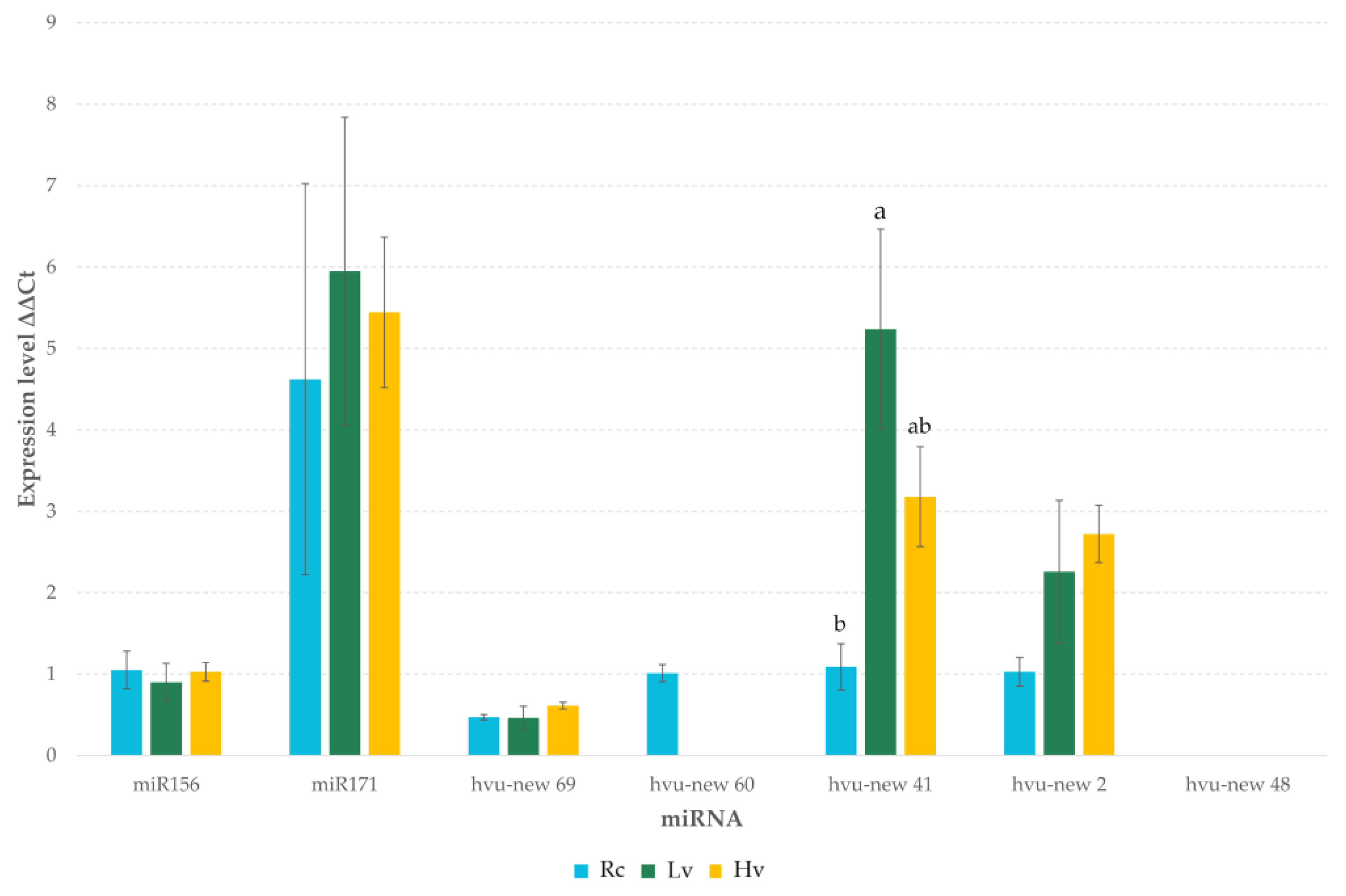
2.3. Differential Expression of miRNAs
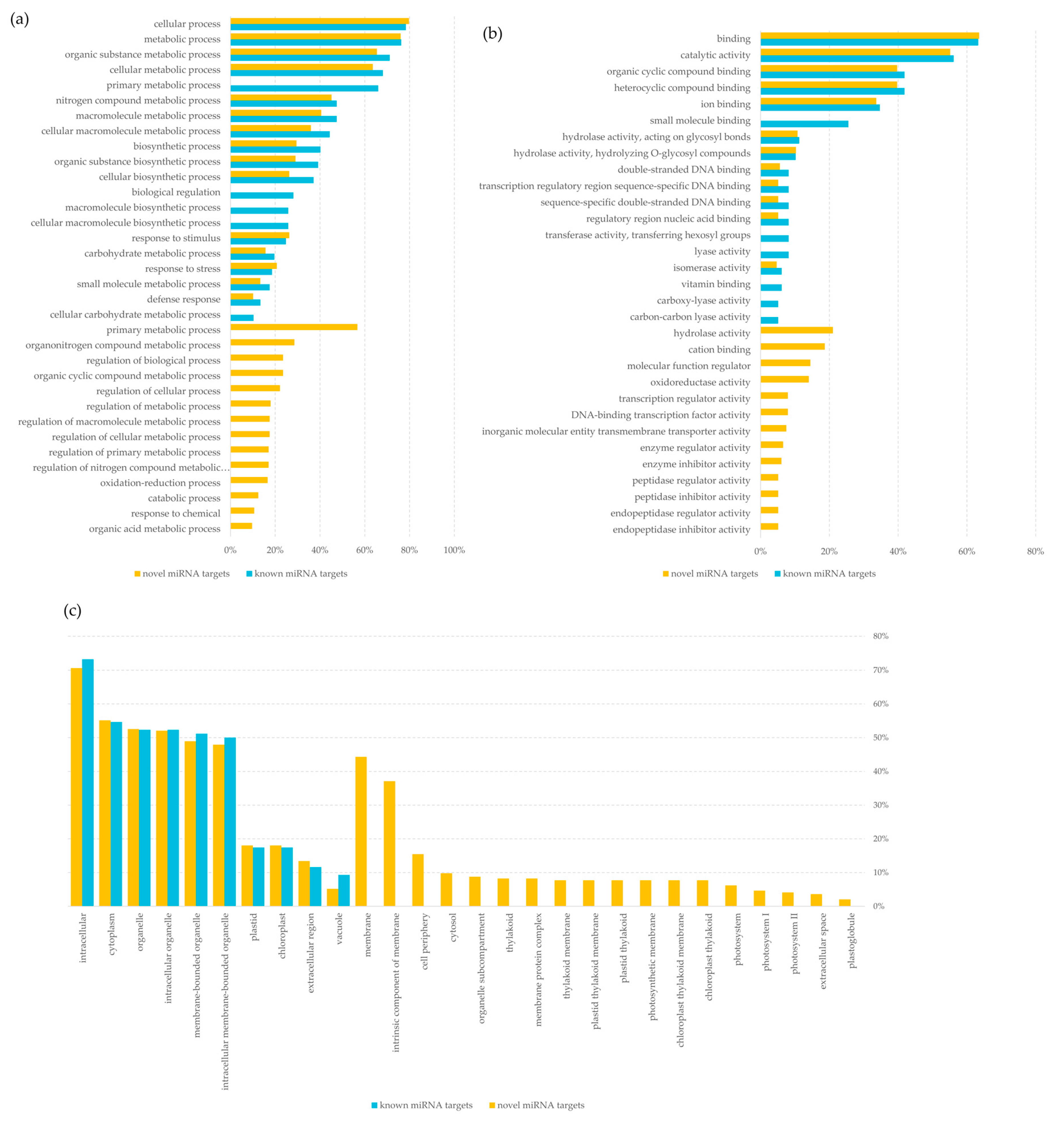
2.4. Prediction Targets of miRNA
3. Discussion
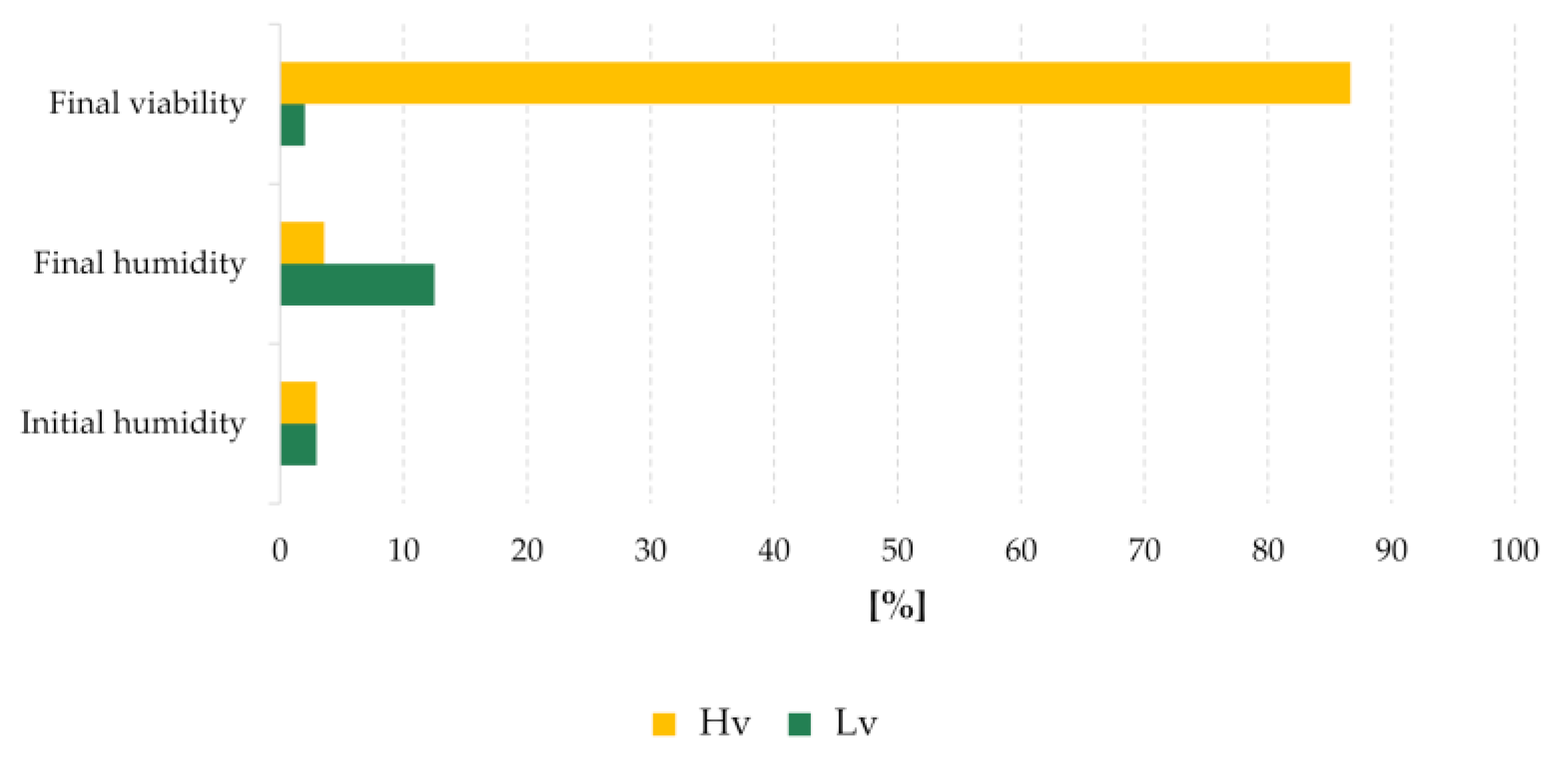
3.1. miRNA and Seed Aging
3.2. miRNome of Barley Seeds
3.3. miRNA in Regulating Gene Expression Based on in Silico Analysis
4. Methods
4.1. Plant Material
4.2. miRNA Extraction and Library Preparation
4.3. Bioinformatic and Statistical Analysis
4.4. RT-qPCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| miRNA | microRNA |
| sRNA | small RNA |
| siRNA | small interfering RNA |
| ta-siRNA | trans-acting siRNA |
| Rc | renewed seeds |
| Hv | seeds with high viability |
| Lv | seeds with low viability |
| rRNA | ribosomal RNA |
| tRNA | transfer RNA |
| NGS | next generation sequencing |
| RT-qPCR | quantitative reverse transcription-PCR |
References
- Roberts, E.H. Viability of Seeds; Springer: Dordrecht, The Netherlands, 1972. [Google Scholar]
- Currey, D.R. An ancient bristlecone pine stand in eastern Nevada. Ecology 1965, 46, 564–566. [Google Scholar] [CrossRef]
- Shen-Miller, J. Sacred lotus, the long-living fruits of China Antique. Seed Sci. Res. 2002, 12, 131. [Google Scholar] [CrossRef]
- Graeber, K.; Nakabayashi, K.; Miatton, E.; Leubner-Metzger, G.; Soppe, W.J. Molecular mechanisms of seed dormancy. Plant Cell Environ. 2012, 35, 1769–1786. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Liu, H.; Li, D.; Chen, H. Identification and characterization of maize microRNAs involved in the very early stage of seed germination. BMC Genom. 2011, 12, 1–10. [Google Scholar] [CrossRef]
- Das, S.S.; Yadav, S.; Singh, A.; Gautam, V.; Sarkar, A.K.; Nandi, A.K.; Karmakar, P.; Majee, M.; Sanan-Mishra, N. Expression dynamics of miRNAs and their targets in seed germination conditions reveals miRNA-ta-siRNA crosstalk as regulator of seed germination. Sci. Rep. 2018, 8, 1–13. [Google Scholar]
- Lv, S.; Nie, X.; Wang, L.; Du, X.; Biradar, S.S.; Jia, X.; Weining, S. Identification and characterization of microRNAs from barley (Hordeum vulgare L.) by high-throughput sequencing. Int. J. Mol. Sci. 2012, 13, 2973–2984. [Google Scholar] [CrossRef]
- Datta, R.; Paul, S. Plant microRNAs: Master regulator of gene expression mechanism. Cell Biol. Int. 2015, 39, 1185–1190. [Google Scholar] [CrossRef]
- Sun, F.; Guo, G.; Du, J.; Guo, W.; Peng, H.; Ni, Z.; Sun, Q.; Yao, Y. Whole-genome discovery of miRNAs and their targets in wheat (Triticum aestivum L.). BMC Plant Biol. 2014, 14, 1–17. [Google Scholar] [CrossRef]
- Taylor, R.S.; Tarver, J.E.; Hiscock, S.J.; Donoghue, P.C. Evolutionary history of plant microRNAs. Trends Plant Sci. 2014, 19, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef]
- Sunkar, R.; Jagadeeswaran, G. In silicoidentification of conserved microRNAs in large number of diverse plant species. BMC Plant Biol. 2008, 8, 37. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Jin, J.; Qian, Q.; Huang, K.; Ding, Y. Small RNA and degradome profiling reveals miRNA regulation in the seed germination of ancient eudicot Nelumbo nucifera. BMC Genom. 2016, 17, 684. [Google Scholar] [CrossRef]
- Deng, P.; Bian, J.; Yue, H.; Feng, K.; Wang, M.; Du, X.; Weining, S.; Nie, X. Characterization of microRNAs and their targets in wild barley (Hordeum vulgare subsp. spontaneum) using deep sequencing. Genome 2016, 59, 339–348. [Google Scholar] [CrossRef] [PubMed]
- FAO. Faostat, Statistical Databases; Food and Agriculture Organization of the United Nations: Rome, Italy, 2019. [Google Scholar]
- Haberer, G.; Mayer, K.F. Barley: From brittle to stable harvest. Cell 2015, 162, 469–471. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Lee, T.; Yang, S.; Lee, I. BarleyNet: A Network-Based Functional Omics Analysis Server for Cultivated Barley, Hordeum vulgare L. Front. Plant Sci. 2020, 11, 98. [Google Scholar] [CrossRef]
- Martí, E.; Pantano, L.; Bañez-Coronel, M.; Llorens, F.; Miñones-Moyano, E.; Porta, S.; Sumoy, L.; Ferrer, I.; Estivill, X. A myriad of miRNA variants in control and Huntington’s disease brain regions detected by massively parallel sequencing. Nucleic Acids Res. 2010, 38, 7219–7235. [Google Scholar] [CrossRef]
- Morin, R.D.; O’Connor, M.D.; Griffith, M.; Kuchenbauer, F.; Delaney, A.; Prabhu, A.-L.; Zhao, Y.; McDonald, H.; Zeng, T.; Hirst, M. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008, 18, 610–621. [Google Scholar] [CrossRef]
- Axtell, M.J.; Meyers, B.C. Revisiting criteria for plant microRNA annotation in the era of big data. Plant Cell 2018, 30, 272–284. [Google Scholar] [CrossRef]
- Dai, X.; Zhuang, Z.; Zhao, P.X. psRNATarget: A plant small RNA target analysis server (2017 release). Nucleic Acids Res. 2018, 46, W49–W54. [Google Scholar] [CrossRef]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. g: Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef]
- Li, C.; Zhang, B. MicroRNAs in control of plant development. J. Cell. Physiol. 2016, 231, 303–313. [Google Scholar] [CrossRef]
- Song, Q.-X.; Liu, Y.-F.; Hu, X.-Y.; Zhang, W.-K.; Ma, B.; Chen, S.-Y.; Zhang, J.-S. Identification of miRNAs and their target genes in developing soybean seeds by deep sequencing. BMC Plant Biol. 2011, 11, 5. [Google Scholar] [CrossRef]
- Yu, J.-Y.; Zhang, Z.-G.; Huang, S.-Y.; Han, X.; Wang, X.-Y.; Pan, W.-J.; Qin, H.-T.; Qi, H.-D.; Yin, Z.-G.; Qu, K.-X. Analysis of miRNAs Targeted Storage Regulatory Genes during Soybean Seed Development Based on Transcriptome Sequencing. Genes 2019, 10, 408. [Google Scholar] [CrossRef]
- Gong, S.; Ding, Y.; Zhu, C. Role of miRNA in plant seed development. Yi Chuan 2015, 37, 554–560. [Google Scholar]
- Das, S.S.; Karmakar, P.; Nandi, A.K.; Sanan-Mishra, N. Small RNA mediated regulation of seed germination. Front. Plant Sci. 2015, 6, 828. [Google Scholar] [CrossRef]
- Li, D.; Wang, L.; Liu, X.; Cui, D.; Chen, T.; Zhang, H.; Jiang, C.; Xu, C.; Li, P.; Li, S. Deep sequencing of maize small RNAs reveals a diverse set of microRNA in dry and imbibed seeds. PLoS ONE 2013, 8, e55107. [Google Scholar] [CrossRef]
- He, D.; Wang, Q.; Wang, K.; Yang, P. Genome-wide dissection of the microRNA expression profile in rice embryo during early stages of seed germination. PLoS ONE 2015, 10, e0145424. [Google Scholar] [CrossRef]
- Sattler, S.E.; Gilliland, L.U.; Magallanes-Lundback, M.; Pollard, M.; DellaPenna, D. Vitamin E is essential for seed longevity and for preventing lipid peroxidation during germination. Plant Cell 2004, 16, 1419–1432. [Google Scholar] [CrossRef] [PubMed]
- França, M.; Panek, A.; Eleutherio, E. Oxidative stress and its effects during dehydration. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2007, 146, 621–631. [Google Scholar] [CrossRef] [PubMed]
- Kranner, I.; Chen, H.; Pritchard, H.W.; Pearce, S.R.; Birtić, S. Inter-nucleosomal DNA fragmentation and loss of RNA integrity during seed ageing. Plant Growth Regul. 2011, 63, 63–72. [Google Scholar] [CrossRef]
- Green, E.J.; Speller, C.F. Novel substrates as sources of ancient DNA: Prospects and hurdles. Genes 2017, 8, 180. [Google Scholar] [CrossRef]
- Waterworth, W.M.; Bray, C.M.; West, C.E. The importance of safeguarding genome integrity in germination and seed longevity. J. Exp. Bot. 2015, 66, 3549–3558. [Google Scholar] [CrossRef]
- Halliwell, B.; Gutteridge, J.M. Free Radicals in Biology and Medicine; Oxford University Press: Oxford, UK, 2015. [Google Scholar]
- Oracz, K.; Bouteau, H.E.M.; Farrant, J.M.; Cooper, K.; Belghazi, M.; Job, C.; Job, D.; Corbineau, F.; Bailly, C. ROS production and protein oxidation as a novel mechanism for seed dormancy alleviation. Plant J. 2007, 50, 452–465. [Google Scholar] [CrossRef]
- Lima, J.J.P.; Buitink, J.; Lalanne, D.; Rossi, R.F.; Pelletier, S.; da Silva, E.A.A.; Leprince, O. Molecular characterization of the acquisition of longevity during seed maturation in soybean. PLoS ONE 2017, 12, e0180282. [Google Scholar] [CrossRef]
- Fernández-Marín, B.; Kranner, I.; Sebastián, M.S.; Artetxe, U.; Laza, J.M.; Vilas, J.L.; Pritchard, H.W.; Nadajaran, J.; Míguez, F.; Becerril, J.M. Evidence for the absence of enzymatic reactions in the glassy state. A case study of xanthophyll cycle pigments in the desiccation-tolerant moss Syntrichia ruralis. J. Exp. Bot. 2013, 64, 3033–3043. [Google Scholar] [CrossRef] [PubMed]
- Miyata, T.; Kurokawa, K.; De Strihou, C.V.Y. Advanced glycation and lipoxidation end products: Role of reactive carbonyl compounds generated during carbohydrate and lipid metabolism. J. Am. Soc. Nephrol. 2000, 11, 1744–1752. [Google Scholar]
- Murthy, U.; Sun, W. Cell and Molecular Biology, Biochemistry and Molecular Physiology-Protein modification by Amadori and Maillard reactions during seed storage: Roles of sugar hydrolysis and lipid peroxidation. J. Exp. Bot. 2000, 51, 1221–1228. [Google Scholar] [CrossRef]
- Held, P. An introduction to reactive oxygen species. Tech Resour. App Guides 2012, 802, 5–9. [Google Scholar]
- Wurtmann, E.J.; Wolin, S.L. A role for a bacterial ortholog of the Ro autoantigen in starvation-induced rRNA degradation. Proc. Natl. Acad. Sci. USA 2010, 107, 4022–4027. [Google Scholar] [CrossRef] [PubMed]
- Sano, N.; Ono, H.; Murata, K.; Yamada, T.; Hirasawa, T.; Kanekatsu, M. Accumulation of long-lived mRNAs associated with germination in embryos during seed development of rice. J. Exp. Bot. 2015, 66, 4035–4046. [Google Scholar] [CrossRef]
- Marcus, A.; Feeley, J. Activation of protein synthesis in the imbibition phase of seed germination. Proc. Natl. Acad. Sci. USA 1964, 51, 1075. [Google Scholar] [CrossRef] [PubMed]
- Fleming, M.B.; Richards, C.M.; Walters, C. Decline in RNA integrity of dry-stored soybean seeds correlates with loss of germination potential. J. Exp. Bot. 2017, 68, 2219–2230. [Google Scholar] [CrossRef]
- Fleming, M.B.; Patterson, E.L.; Reeves, P.A.; Richards, C.M.; Gaines, T.A.; Walters, C. Exploring the fate of mRNA in aging seeds: Protection, destruction, or slow decay? J. Exp. Bot. 2018, 69, 4309–4321. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Wang, S.; Fu, Y.-B.; Wang, H. Arabidopsis seed stored mRNAs are degraded constantly over aging time, as revealed by new quantification methods. Front. Plant Sci. 2020, 10, 1764. [Google Scholar] [CrossRef] [PubMed]
- Martin, R.C.; Liu, P.-P.; Nonogaki, H. Simple purification of small RNAs from seeds and efficient detection of multiple microRNAs expressed in Arabidopsis thaliana and tomato (Lycopersicon esculentum) seeds. Seed Sci. Res. 2005, 15, 319–328. [Google Scholar] [CrossRef]
- Martin, R.C.; Liu, P.-P.; Nonogaki, H. microRNAs in seeds: Modified detection techniques and potential applications. Botany 2006, 84, 189–198. [Google Scholar] [CrossRef]
- Abdelmohsen, K.; Srikantan, S.; Kang, M.-J.; Gorospe, M. Regulation of senescence by microRNA biogenesis factors. Ageing Res. Rev. 2012, 11, 491–500. [Google Scholar] [CrossRef]
- Kim, J.; Kim, J.H.; Lyu, J.I.; Woo, H.R.; Lim, P.O. New insights into the regulation of leaf senescence in Arabidopsis. J. Exp. Bot. 2018, 69, 787–799. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Gan, L.; Chen, D.; Zhang, Y.; Zhang, Y.; Liu, X.; Chen, S.; Wei, Z.; Tong, L.; Song, Z. Integration of small RNA, degradome and proteome sequencing in Oryza sativa reveals a delayed senescence network in tetraploid rice seed. PLoS ONE 2020, 15, e0242260. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhou, S.; Wang, L.; Wu, D.; Cheng, H.; Du, X.; Mao, D.; Zhang, C.; Jiang, X. miR164c and miR168a regulate seed vigor in rice. J. Integr. Plant Biol. 2020, 62, 470–486. [Google Scholar] [CrossRef]
- Galleschi, L.; Capocchi, A.; Ghiringhelli, S.; Saviozzi, F.; Calucci, L.; Pinzino, C.; Zandomeneghi, M. Antioxidants, free radicals, storage proteins, and proteolytic activities in wheat (Triticum durum) seeds during accelerated aging. J. Agric. Food Chem. 2002, 50, 5450–5457. [Google Scholar] [CrossRef] [PubMed]
- Xin, X.; Lin, X.H.; Zhou, Y.C.; Chen, X.L.; Liu, X.; Lu, X.X. Proteome analysis of maize seeds: The effect of artificial ageing. Physiol. Plant. 2011, 143, 126–138. [Google Scholar] [CrossRef] [PubMed]
- Sajeev, N.; Bai, B.; Bentsink, L. Seeds: A unique system to study translational regulation. Trends Plant Sci. 2019, 24, 487–495. [Google Scholar] [CrossRef] [PubMed]
- Puchta, M.; Boczkowska, M.; Groszyk, J. Low RIN Value for RNA-Seq Library Construction from Long-Term Stored Seeds: A Case Study of Barley Seeds. Genes 2020, 11, 1190. [Google Scholar] [CrossRef]
- Aryani, A.; Denecke, B. In vitro application of ribonucleases: Comparison of the effects on mRNA and miRNA stability. BMC Res. Notes 2015, 8, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Balzano, F.; Deiana, M.; Dei Giudici, S.; Oggiano, A.; Baralla, A.; Pasella, S.; Mannu, A.; Pescatori, M.; Porcu, B.; Fanciulli, G. miRNA stability in frozen plasma samples. Molecules 2015, 20, 19030–19040. [Google Scholar] [CrossRef] [PubMed]
- Kakimoto, Y.; Tanaka, M.; Kamiguchi, H.; Ochiai, E.; Osawa, M. MicroRNA stability in FFPE tissue samples: Dependence on GC content. PLoS ONE 2016, 11, e0163125. [Google Scholar] [CrossRef] [PubMed]
- Jung, M.; Schaefer, A.; Steiner, I.; Kempkensteffen, C.; Stephan, C.; Erbersdobler, A.; Jung, K. Robust microRNA stability in degraded RNA preparations from human tissue and cell samples. Clin. Chem. 2010, 56, 998–1006. [Google Scholar] [CrossRef]
- Tang, F.; Hajkova, P.; O’Carroll, D.; Lee, C.; Tarakhovsky, A.; Lao, K.; Surani, M.A. MicroRNAs are tightly associated with RNA-induced gene silencing complexes in vivo. Biochem. Biophys. Res. Commun. 2008, 372, 24–29. [Google Scholar] [CrossRef]
- Diederichs, S.; Haber, D.A. Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell 2007, 131, 1097–1108. [Google Scholar] [CrossRef]
- Mahalingam, R. Shotgun proteomics of the barley seed proteome. BMC Genom. 2017, 18, 44. [Google Scholar] [CrossRef] [PubMed]
- Papatheodorou, I.; Moreno, P.; Manning, J.; Fuentes, A.M.-P.; George, N.; Fexova, S.; Fonseca, N.A.; Füllgrabe, A.; Green, M.; Huang, N. Expression Atlas update: From tissues to single cells. Nucleic Acids Res. 2020, 48, D77–D83. [Google Scholar] [CrossRef]
- Yu, B.; Yang, Z.; Li, J.; Minakhina, S.; Yang, M.; Padgett, R.W.; Steward, R.; Chen, X. Methylation as a crucial step in plant microRNA biogenesis. Science 2005, 307, 932–935. [Google Scholar] [CrossRef] [PubMed]
- Ruby, J.G.; Jan, C.; Player, C.; Axtell, M.J.; Lee, W.; Nusbaum, C.; Ge, H.; Bartel, D.P. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell 2006, 127, 1193–1207. [Google Scholar] [CrossRef]
- Kim, J.; Cho, I.S.; Hong, J.S.; Choi, Y.K.; Kim, H.; Lee, Y.S. Identification and characterization of new microRNAs from pig. Mamm. Genome 2008, 19, 570–580. [Google Scholar] [CrossRef]
- Lee, L.W.; Zhang, S.; Etheridge, A.; Ma, L.; Martin, D.; Galas, D.; Wang, K. Complexity of the microRNA repertoire revealed by next-generation sequencing. RNA 2010, 16, 2170–2180. [Google Scholar] [CrossRef]
- Lelandais-Brière, C.; Naya, L.; Sallet, E.; Calenge, F.; Frugier, F.; Hartmann, C.; Gouzy, J.; Crespi, M. Genome-wide Medicago truncatula small RNA analysis revealed novel microRNAs and isoforms differentially regulated in roots and nodules. Plant Cell 2009, 21, 2780–2796. [Google Scholar] [CrossRef]
- Starega-Roslan, J.; Koscianska, E.; Kozlowski, P.; Krzyzosiak, W.J. The role of the precursor structure in the biogenesis of microRNA. Cell. Mol. Life Sci. 2011, 68, 2859–2871. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yang, Z.; Yu, B.; Liu, J.; Chen, X. Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr. Biol. 2005, 15, 1501–1507. [Google Scholar] [CrossRef]
- Ramachandran, V.; Chen, X. Degradation of microRNAs by a family of exoribonucleases in Arabidopsis. Science 2008, 321, 1490–1492. [Google Scholar] [CrossRef]
- Henderson, I.R.; Zhang, X.; Lu, C.; Johnson, L.; Meyers, B.C.; Green, P.J.; Jacobsen, S.E. Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nat. Genet. 2006, 38, 721–725. [Google Scholar] [CrossRef] [PubMed]
- Pontes, O.; Li, C.F.; Nunes, P.C.; Haag, J.; Ream, T.; Vitins, A.; Jacobsen, S.E.; Pikaard, C.S. The Arabidopsis chromatin-modifying nuclear siRNA pathway involves a nucleolar RNA processing center. Cell 2006, 126, 79–92. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Johansen, L.K.; Gustafson, A.M.; Kasschau, K.D.; Lellis, A.D.; Zilberman, D.; Jacobsen, S.E.; Carrington, J.C. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004, 2, e104. [Google Scholar] [CrossRef]
- Margis, R.; Fusaro, A.F.; Smith, N.A.; Curtin, S.J.; Watson, J.M.; Finnegan, E.J.; Waterhouse, P.M. The evolution and diversification of Dicers in plants. FEBS Lett. 2006, 580, 2442–2450. [Google Scholar] [CrossRef]
- Borges, F.; Martienssen, R.A. The expanding world of small RNAs in plants. Nat. Rev. Mol. Cell Biol. 2015, 16, 727–741. [Google Scholar] [CrossRef]
- Han, B.W.; Hung, J.-H.; Weng, Z.; Zamore, P.D.; Ameres, S.L. The 3′-to-5′ exoribonuclease Nibbler shapes the 3′ ends of microRNAs bound to Drosophila Argonaute1. Curr. Biol. 2011, 21, 1878–1887. [Google Scholar] [CrossRef]
- Elkayam, E.; Kuhn, C.-D.; Tocilj, A.; Haase, A.D.; Greene, E.M.; Hannon, G.J.; Joshua-Tor, L. The structure of human argonaute-2 in complex with miR-20a. Cell 2012, 150, 100–110. [Google Scholar] [CrossRef]
- Schirle, N.T.; MacRae, I.J. The crystal structure of human Argonaute2. Science 2012, 336, 1037–1040. [Google Scholar] [CrossRef] [PubMed]
- Ameres, S.L.; Horwich, M.D.; Hung, J.-H.; Xu, J.; Ghildiyal, M.; Weng, Z.; Zamore, P.D. Target RNA–directed trimming and tailing of small silencing RNAs. Science 2010, 328, 1534–1539. [Google Scholar] [CrossRef]
- Wang, X.; Liu, S. Systematic curation of miRBase annotation using integrated small RNA high-throughput sequencing data for C. elegans and Drosophila. Front. Genet. 2011, 2, 25. [Google Scholar] [CrossRef] [PubMed]
- Mi, S.; Cai, T.; Hu, Y.; Chen, Y.; Hodges, E.; Ni, F.; Wu, L.; Li, S.; Zhou, H.; Long, C. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell 2008, 133, 116–127. [Google Scholar] [CrossRef] [PubMed]
- Fard, E.M.; Moradi, S.; Salekdeh, N.N.; Bakhshi, B.; Ghaffari, M.R.; Zeinalabedini, M.; Salekdeh, G.H. Plant isomiRs: Origins, biogenesis, and biological functions. Genomics 2020, 112, 3382–3395. [Google Scholar] [CrossRef]
- Fard, E.M.; Bakhshi, B.; Keshavarznia, R.; Nikpay, N.; Shahbazi, M.; Salekdeh, G.H. Drought responsive microRNAs in two barley cultivars differing in their level of sensitivity to drought stress. Plant Physiol. Biochem. 2017, 118, 121–129. [Google Scholar] [CrossRef]
- Chiang, H.R.; Schoenfeld, L.W.; Ruby, J.G.; Auyeung, V.C.; Spies, N.; Baek, D.; Johnston, W.K.; Russ, C.; Luo, S.; Babiarz, J.E. Mammalian microRNAs: Experimental evaluation of novel and previously annotated genes. Genes Dev. 2010, 24, 992–1009. [Google Scholar] [CrossRef] [PubMed]
- Jeong, D.-H.; Thatcher, S.R.; Brown, R.S.; Zhai, J.; Park, S.; Rymarquis, L.A.; Meyers, B.C.; Green, P.J. Comprehensive investigation of microRNAs enhanced by analysis of sequence variants, expression patterns, ARGONAUTE loading, and target cleavage. Plant Physiol. 2013, 162, 1225–1245. [Google Scholar] [CrossRef]
- Peláez, P.; Trejo, M.S.; Iñiguez, L.P.; Estrada-Navarrete, G.; Covarrubias, A.A.; Reyes, J.L.; Sanchez, F. Identification and characterization of microRNAs in Phaseolus vulgaris by high-throughput sequencing. BMC Genom. 2012, 13, 1–18. [Google Scholar] [CrossRef]
- Wu, L.; Zhou, H.; Zhang, Q.; Zhang, J.; Ni, F.; Liu, C.; Qi, Y. DNA methylation mediated by a microRNA pathway. Mol. Cell 2010, 38, 465–475. [Google Scholar] [CrossRef] [PubMed]
- Ding, D.; Wang, Y.; Han, M.; Fu, Z.; Li, W.; Liu, Z.; Hu, Y.; Tang, J. MicroRNA transcriptomic analysis of heterosis during maize seed germination. PLoS ONE 2012, 7, e39578. [Google Scholar]
- Zabala, G.; Campos, E.; Varala, K.K.; Bloomfield, S.; Jones, S.I.; Win, H.; Tuteja, J.H.; Calla, B.; Clough, S.J.; Hudson, M. Divergent patterns of endogenous small RNA populations from seed and vegetative tissues of Glycine max. BMC Plant Biol. 2012, 12, 177. [Google Scholar] [CrossRef]
- Zhang, L.; Chia, J.-M.; Kumari, S.; Stein, J.C.; Liu, Z.; Narechania, A.; Maher, C.A.; Guill, K.; McMullen, M.D.; Ware, D. A genome-wide characterization of microRNA genes in maize. PLoS Genet. 2009, 5, e1000716. [Google Scholar] [CrossRef] [PubMed]
- Finkelstein, R.; Reeves, W.; Ariizumi, T.; Steber, C. Molecular aspects of seed dormancy. Annu. Rev. Plant Biol. 2008, 59, 387–415. [Google Scholar] [CrossRef]
- Shamimuzzaman, M.; Vodkin, L. Identification of soybean seed developmental stage-specific and tissue-specific miRNA targets by degradome sequencing. BMC Genom. 2012, 13, 310. [Google Scholar] [CrossRef]
- Hertweck, K.L.; Kinney, M.S.; Stuart, S.A.; Maurin, O.; Mathews, S.; Chase, M.W.; Gandolfo, M.A.; Pires, J.C. Phylogenetics, divergence times and diversification from three genomic partitions in monocots. Bot. J. Linn. Soc. 2015, 178, 375–393. [Google Scholar] [CrossRef]
- Crepet, W.L.; Nixon, K.C.; Gandolfo, M.A. Fossil evidence and phylogeny: The age of major angiosperm clades based on mesofossil and macrofossil evidence from Cretaceous deposits. Am. J. Bot. 2004, 91, 1666–1682. [Google Scholar] [CrossRef]
- Wang, X.; Elling, A.A.; Li, X.; Li, N.; Peng, Z.; He, G.; Sun, H.; Qi, Y.; Liu, X.S.; Deng, X.W. Genome-wide and organ-specific landscapes of epigenetic modifications and their relationships to mRNA and small RNA transcriptomes in maize. Plant Cell 2009, 21, 1053–1069. [Google Scholar] [CrossRef]
- Millar, A.A.; Gubler, F. The Arabidopsis GAMYB-like genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development. Plant Cell 2005, 17, 705–721. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.-W.; Czech, B.; Weigel, D. miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 2009, 138, 738–749. [Google Scholar] [CrossRef] [PubMed]
- Xie, K.; Wu, C.; Xiong, L. Genomic organization, differential expression, and interaction of SQUAMOSA promoter-binding-like transcription factors and microRNA156 in rice. Plant Physiol. 2006, 142, 280–293. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Cui, X.; Meng, Z.; Huang, X.; Xie, Q.; Wu, H.; Jin, H.; Zhang, D.; Liang, W. Transcriptional regulation of Arabidopsis MIR168a and argonaute1 homeostasis in abscisic acid and abiotic stress responses. Plant Physiol. 2012, 158, 1279–1292. [Google Scholar] [CrossRef] [PubMed]
- Voinnet, O. Origin, biogenesis, and activity of plant microRNAs. Cell 2009, 136, 669–687. [Google Scholar] [CrossRef] [PubMed]
- Rhoades, M.W.; Reinhart, B.J.; Lim, L.P.; Burge, C.B.; Bartel, B.; Bartel, D.P. Prediction of plant microRNA targets. Cell 2002, 110, 513–520. [Google Scholar] [CrossRef]
- Gibbons, G.C. On the relative role of the scutellum and aleurone in the production of hydrolases during germination of barley. Carlsberg Res. Commun. 1981, 46, 215. [Google Scholar] [CrossRef]
- Okamoto, K.; Kitano, H.; Akazawa, T. Biosynthesis and excretion of hydrolases in germinating cereal seeds. Plant Cell Physiol. 1980, 21, 201–204. [Google Scholar]
- Ballance, G.M.; Manners, D.J. Structural analysis and ensymic solubilization of barley endosperm cell-walls. Carbohydr. Res. 1978, 61, 107–118. [Google Scholar] [CrossRef]
- Stuart, I.; Loi, L.; Fincher, G. Varietal and environmental variations in (1→ 3, 1→ 4)-β-glucan levels and (1→ 3, 1→ 4)-β-glucanase potential in barley: Relationships to malting quality. J. Cereal Sci. 1988, 7, 61–71. [Google Scholar] [CrossRef]
- White, J.; Pacey-Miller, T.; Bundock, P.; Henry, R. Differential LongSAGE tag abundance analysis in a barley seed germination time course and validation with relative real-time RT-PCR. Plant Sci. 2008, 175, 858–867. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data; Babraham Bioinformatics, Babraham Institute: Cambridge, UK, 2010. [Google Scholar]
- Stocks, M.B.; Moxon, S.; Mapleson, D.; Woolfenden, H.C.; Mohorianu, I.; Folkes, L.; Schwach, F.; Dalmay, T.; Moulton, V. The UEA sRNA workbench: A suite of tools for analysing and visualizing next generation sequencing microRNA and small RNA datasets. Bioinformatics 2012, 28, 2059–2061. [Google Scholar] [CrossRef]
- Mohorianu, I.; Stocks, M.B.; Applegate, C.S.; Folkes, L.; Moulton, V. The UEA small RNA workbench: A suite of computational tools for small RNA analysis. In MicroRNA Detection and Target Identification; Springer: Berlin/Heidelberg, Germany, 2017; pp. 193–224. [Google Scholar]
- Kersey, P.J.; Allen, J.E.; Allot, A.; Barba, M.; Boddu, S.; Bolt, B.J.; Carvalho-Silva, D.; Christensen, M.; Davis, P.; Grabmueller, C. Ensembl Genomes 2018: An integrated omics infrastructure for non-vertebrate species. Nucleic Acids Res. 2018, 46, D802–D808. [Google Scholar] [CrossRef]
- Zou, Q.; Mao, Y.; Hu, L.; Wu, Y.; Ji, Z. miRClassify: An advanced web server for miRNA family classification and annotation. Comput. Biol. Med. 2014, 45, 157–160. [Google Scholar] [CrossRef]
- Varet, H.; Brillet-Guéguen, L.; Coppée, J.-Y.; Dillies, M.-A. SARTools: A DESeq2-and edgeR-based R pipeline for comprehensive differential analysis of RNA-Seq data. PLoS ONE 2016, 11, e0157022. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: Berlin/Heidelberg, Germany, 2016. [Google Scholar]
- Cirera, S.; Busk, P.K. Quantification of miRNAs by a simple and specific qPCR method. In RNA Mapping; Springer: Berlin/Heidelberg, Germany, 2014; pp. 73–81. [Google Scholar]
- Kim, M.; Gee, M.; Loh, A.; Rachatasumrit, N. Ref-Finder: A refactoring reconstruction tool based on logic query templates. In Proceedings of the Eighteenth ACM SIGSOFT International Symposium on Foundations of Software Engineering, Santa Fe, NM, USA, 9–11 November 2010; pp. 371–372. [Google Scholar]
- Busk, P.K. A tool for design of primers for microRNA-specific quantitative RT-qPCR. BMC Bioinform. 2014, 15, 1–9. [Google Scholar] [CrossRef] [PubMed]
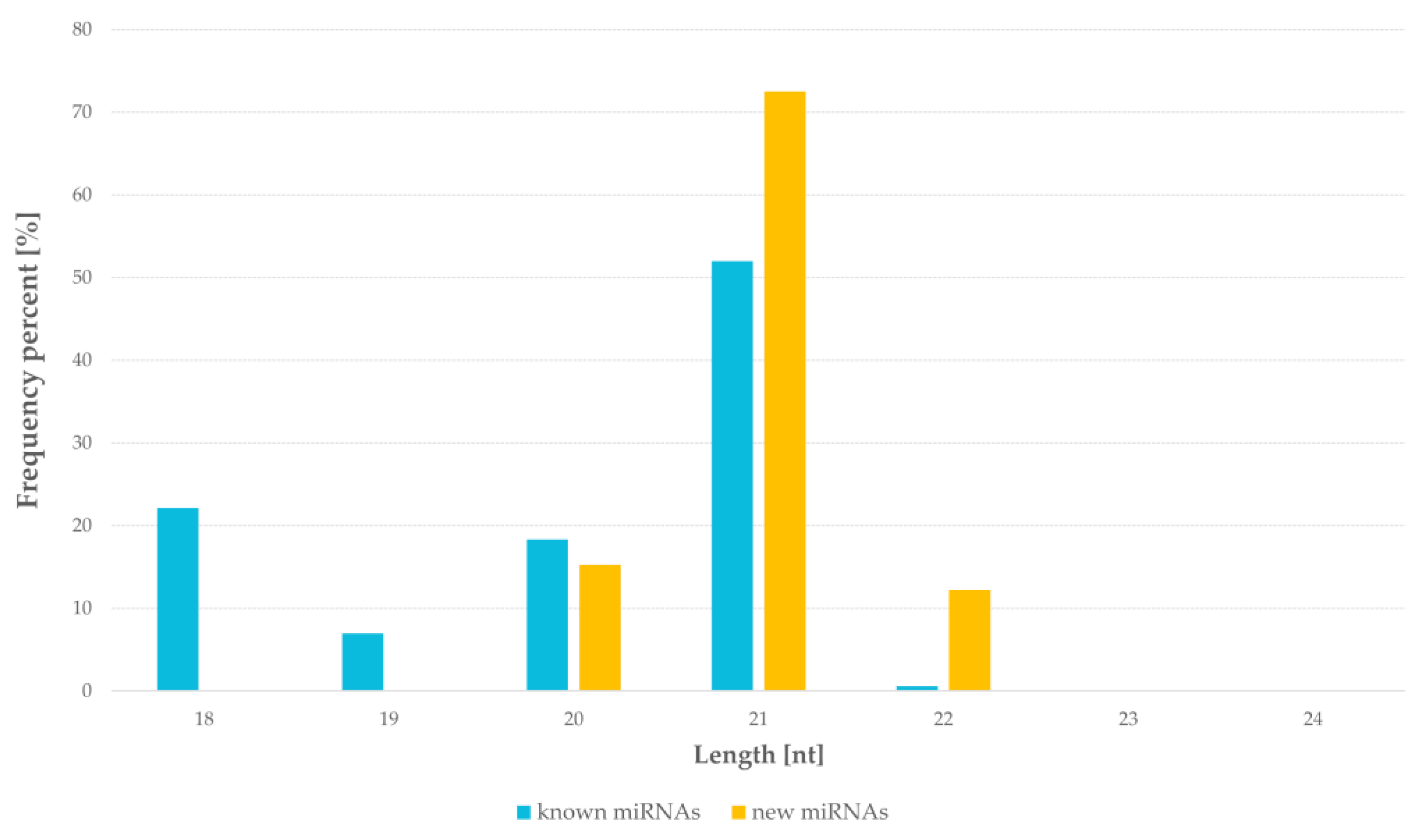
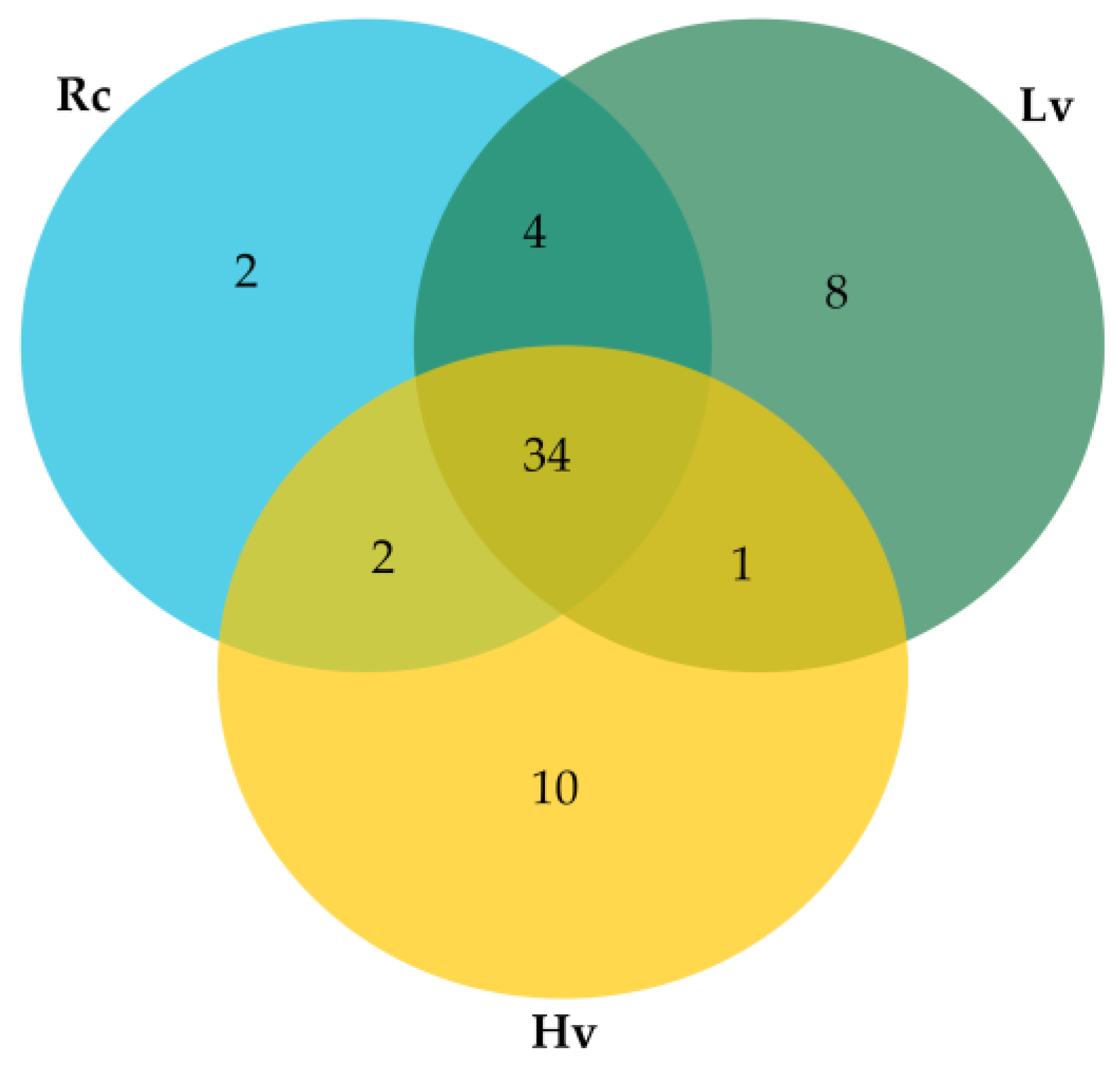
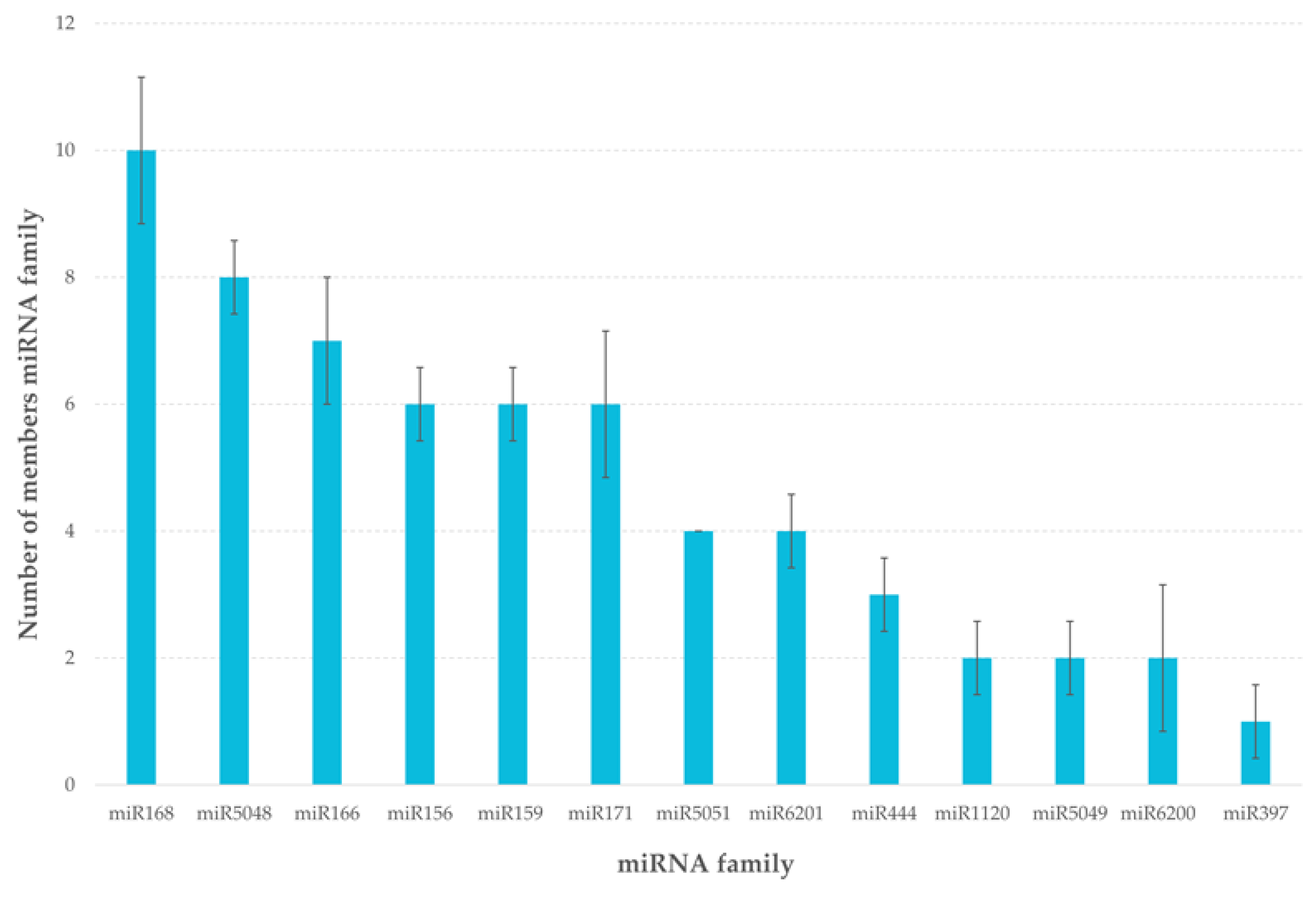

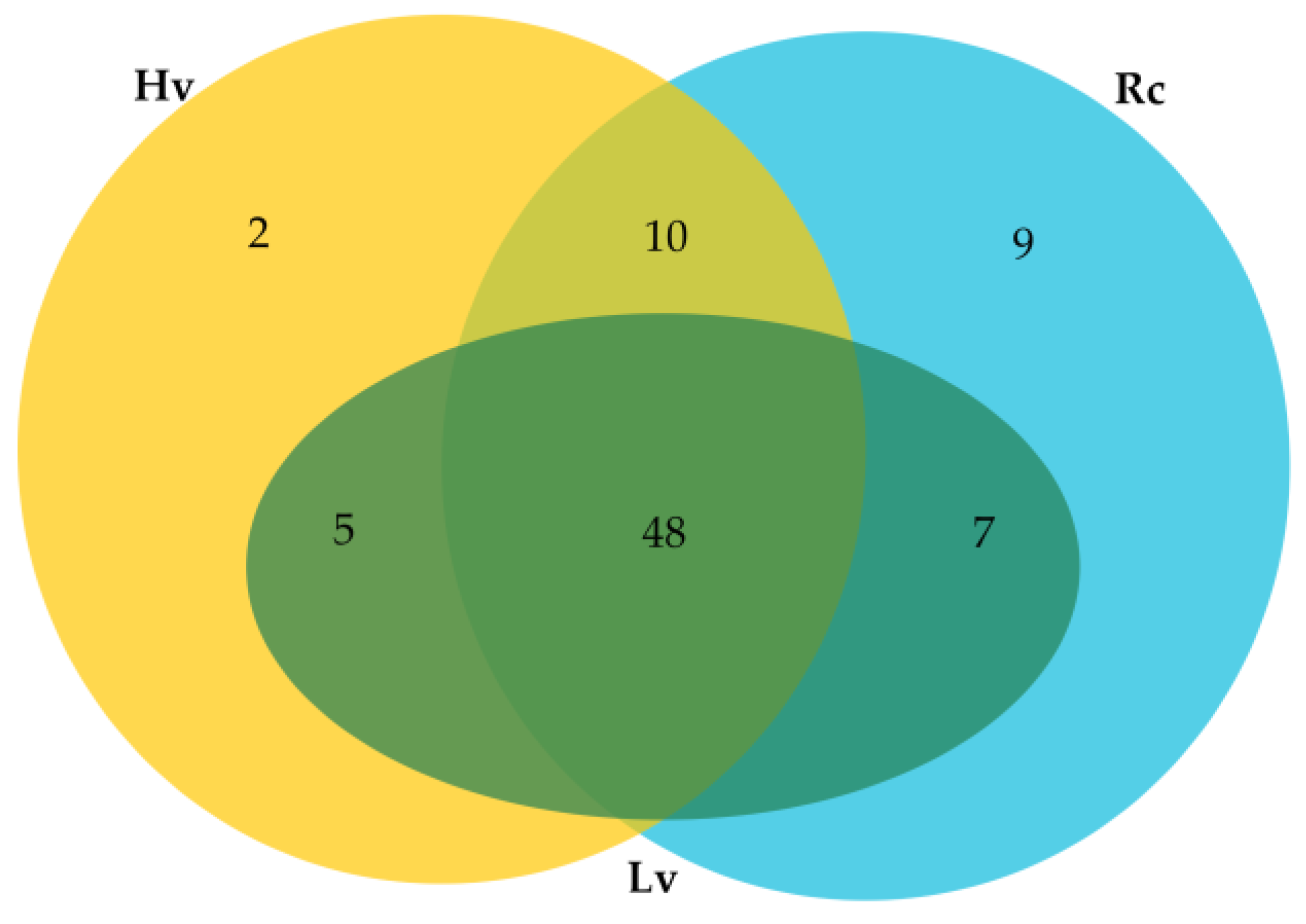
| Small RNA Data | Raw Sequencing Reads | Clean Reads | Mapped Reads | t/rRNA Matches | Match miRNA |
|---|---|---|---|---|---|
| Rc-1 | 2,496,964 | 672,701 | 239,836 (35.65%) | 54,665 (8.13%) | 874(0.13%) |
| Rc-2 | 2,593,309 | 657,323 | 240,539 (36.59%) | 66,134 (10.06%) | 1103 (0.17%) |
| Rc-3 | 5,425,372 | 1,116,857 | 438,066 (39.22%) | 40,201 (3.60%) | 1740 (0.16%) |
| Lv-1 | 2,258,988 | 511,598 | 173,417 (33.90%) | 61,253 (11.97%) | 725 (0.14%) |
| Lv-2 | 4,174,307 | 930,756 | 324,366 (34.85%) | 111,254 (11.95%) | 1444 (0.16%) |
| Lv-3 | 4,635,289 | 1,085,034 | 390,864 (36.02%) | 127,376 (11.74%) | 1553 (0.14%) |
| Hv-1 | 2,521,775 | 626,232 | 197,658 (31.56%) | 79,540 (12.70%) | 867 (0.14%) |
| Hv-2 | 3,369,389 | 821,586 | 286,502 (34.87%) | 85,666 (10.43%) | 1083 (0.13%) |
| Hv-3 | 4,932,306 | 1,162,680 | 430,005 (36.98%) | 120,497 (10.36%) | 1711 (0.15%) |
| Novel miRNA | Matched miRNA | Probability |
|---|---|---|
| hvu-new80 | miR159 | 0.77 |
| hvu-new45 | miR166 | 0.62 |
| hvu-new49 | miR167 | 0.28 |
| hvu-new50 | 0.33 | |
| hvu-new51 | 0.2 | |
| hvu-new52 | ||
| hvu-new53 | ||
| hvu-new43 | miR168 | 0.7 |
| hvu-new69 | miR396 | 0.4 |
| hvu-new71 | miR171 | 0.6 |
| hvu-new73 | miR397 | 0.5 |
| Target Accession | Expectation | Target Descriptor | Gene | Uniprot Protein Accession |
|---|---|---|---|---|
| hvu-new60 | ||||
| BF261584 | 3.5 | Precursor of CP29, core chlorophyll a/b binding (CAB) protein of photosystem II | N/A | Q40039 |
| TC240558, TC267263, TC276211, TC257011, BI953040, TC275119, BI955619, BJ482334 | 4.5 | |||
| BI951019 | 4 | Hordoindoline-B1 | HINB-1 | Q9FSI9 |
| TC265164 | 4 | Rust resistance gene ABC1041 | ABC1041 | Q2L7E7 |
| TC251527 | 5 | Cystatin Hv-CPI7 | icy7 | Q1ENE8 |
| TC238665 | 5 | NADPH-dependent thioredoxin reductase isoform 2 | NTR2 | A9LN30 |
| hvu-new41 | ||||
| TC238419, BJ468992 | 3.5 | Boron transporter | Bot1 | A9XTK3 |
| TC267583 | 4.5 | Basic helix-loop-helix protein | HvIRO2 | Q0KKX2 |
| DN185239 | 4 | Glb 1 1-3,1-4-beta-D-glucanase precursor (Lichenase precursor) | N/A | Q02345 |
| TC277443, TC239158, TC252681 | 5 | Knotted 7 | kn7 | Q717U4 |
| hvu-new2 | ||||
| TC241603 | 2.5 | Glyceraldehyde-3-phosphate dehydrogenase | GAPC | P26517 |
| TC256820 | 4 | Non-specific lipid-transfer protein 1 | LTP1 | P07597 |
| TC238588 | 3.5 | Germin-like protein 5a | GER5a | Q0GR06 |
| BI949246 | 3 | B hordein | N/A | Q40026 |
| TC278915 | 4 | Ribulose bisphosphate carboxylase large chain | rbcL | P05698 |
| TC240271 | 3.5 | Ribulose bisphosphate carboxylase/oxygenase activase B, chloroplast | RCAB | Q42450 |
| BI950805 | 2.5 | Pathogenesis-related protein PRB1-2 | N/A | P35792 |
| TC247384 | 2.5 | Sucrose-phosphatase | N/A | Q84ZX7 |
| TC273036 | 4 | Beta-amylase | Bmy1 | Q9AVJ8 |
| TC279634 | 3 | 14-3-3-like protein A | N/A | P29305 |
| TC275451 | 3 | Hordoindoline-b1 | hinb-1 | Q5IUH9 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Puchta, M.; Groszyk, J.; Małecka, M.; Koter, M.D.; Niedzielski, M.; Rakoczy-Trojanowska, M.; Boczkowska, M. Barley Seeds miRNome Stability during Long-Term Storage and Aging. Int. J. Mol. Sci. 2021, 22, 4315. https://doi.org/10.3390/ijms22094315
Puchta M, Groszyk J, Małecka M, Koter MD, Niedzielski M, Rakoczy-Trojanowska M, Boczkowska M. Barley Seeds miRNome Stability during Long-Term Storage and Aging. International Journal of Molecular Sciences. 2021; 22(9):4315. https://doi.org/10.3390/ijms22094315
Chicago/Turabian StylePuchta, Marta, Jolanta Groszyk, Magdalena Małecka, Marek D. Koter, Maciej Niedzielski, Monika Rakoczy-Trojanowska, and Maja Boczkowska. 2021. "Barley Seeds miRNome Stability during Long-Term Storage and Aging" International Journal of Molecular Sciences 22, no. 9: 4315. https://doi.org/10.3390/ijms22094315
APA StylePuchta, M., Groszyk, J., Małecka, M., Koter, M. D., Niedzielski, M., Rakoczy-Trojanowska, M., & Boczkowska, M. (2021). Barley Seeds miRNome Stability during Long-Term Storage and Aging. International Journal of Molecular Sciences, 22(9), 4315. https://doi.org/10.3390/ijms22094315







