Deficiency of NEIL3 Enhances the Chemotherapy Resistance of Prostate Cancer
Abstract
1. Introduction
2. Results
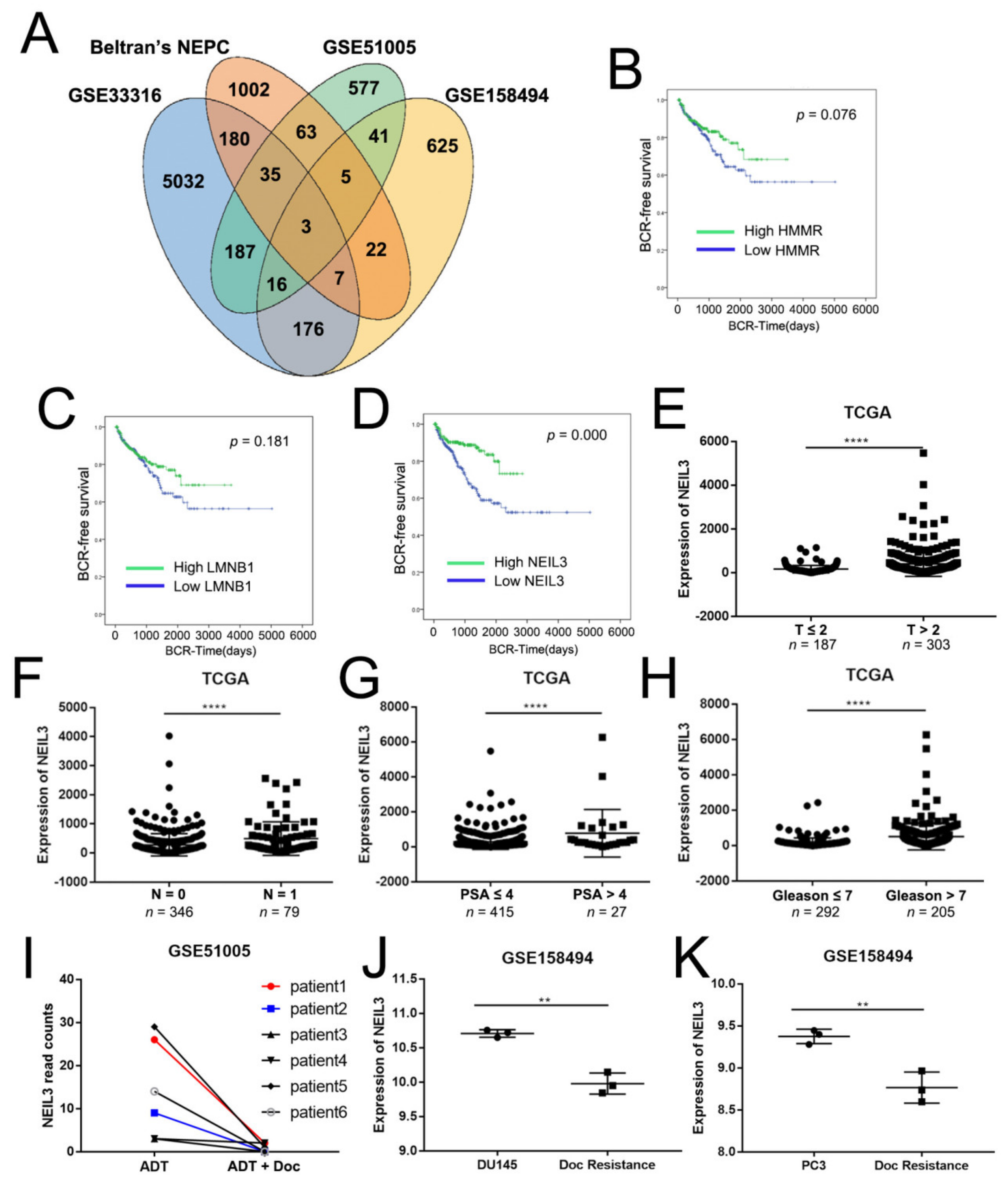
2.1. NEIL3 Was Associated with Chemotherapy Resistance in PCa
2.2. NEIL3 Showed Low Expression in Therapy-Resistant PCa
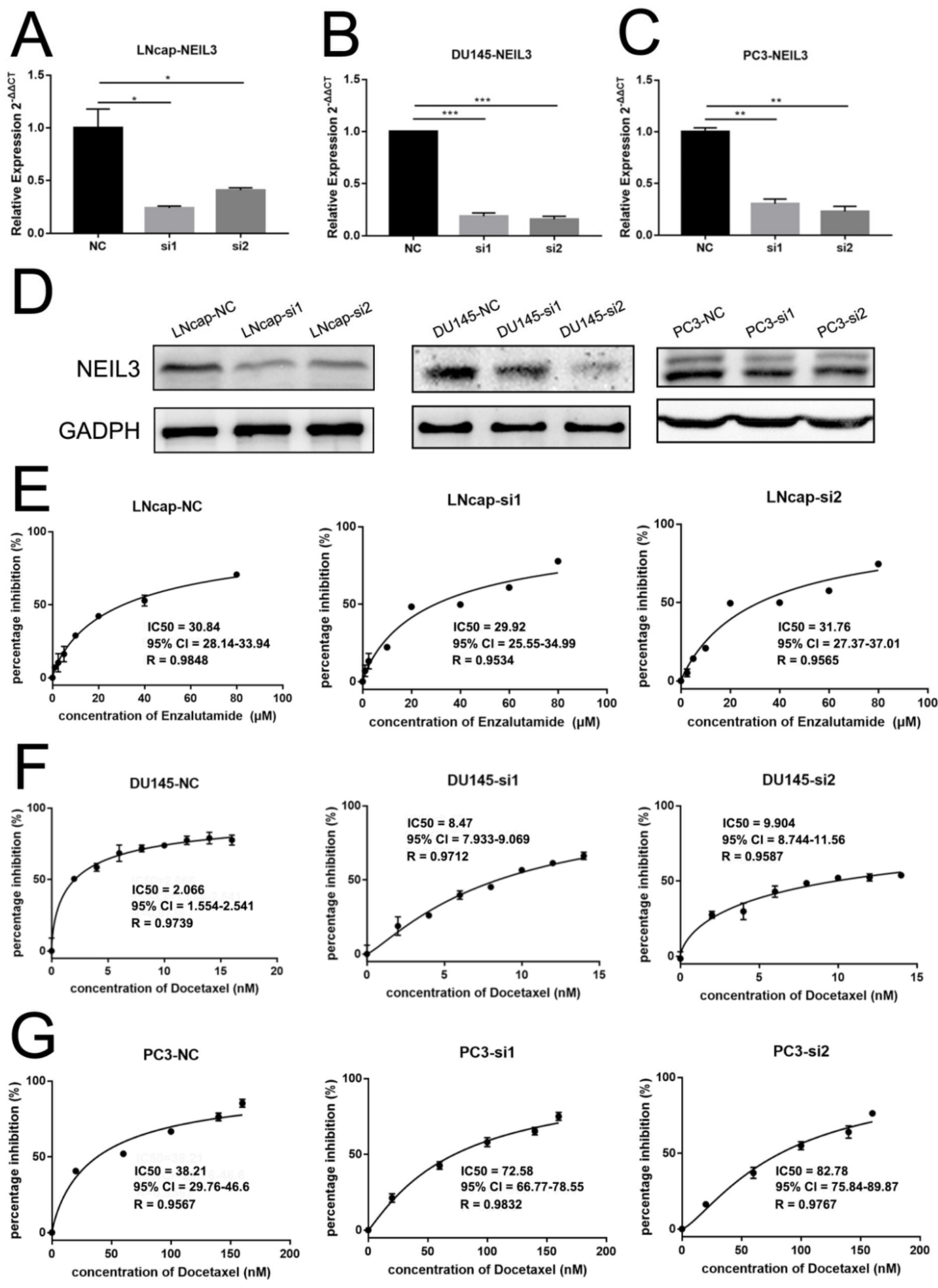
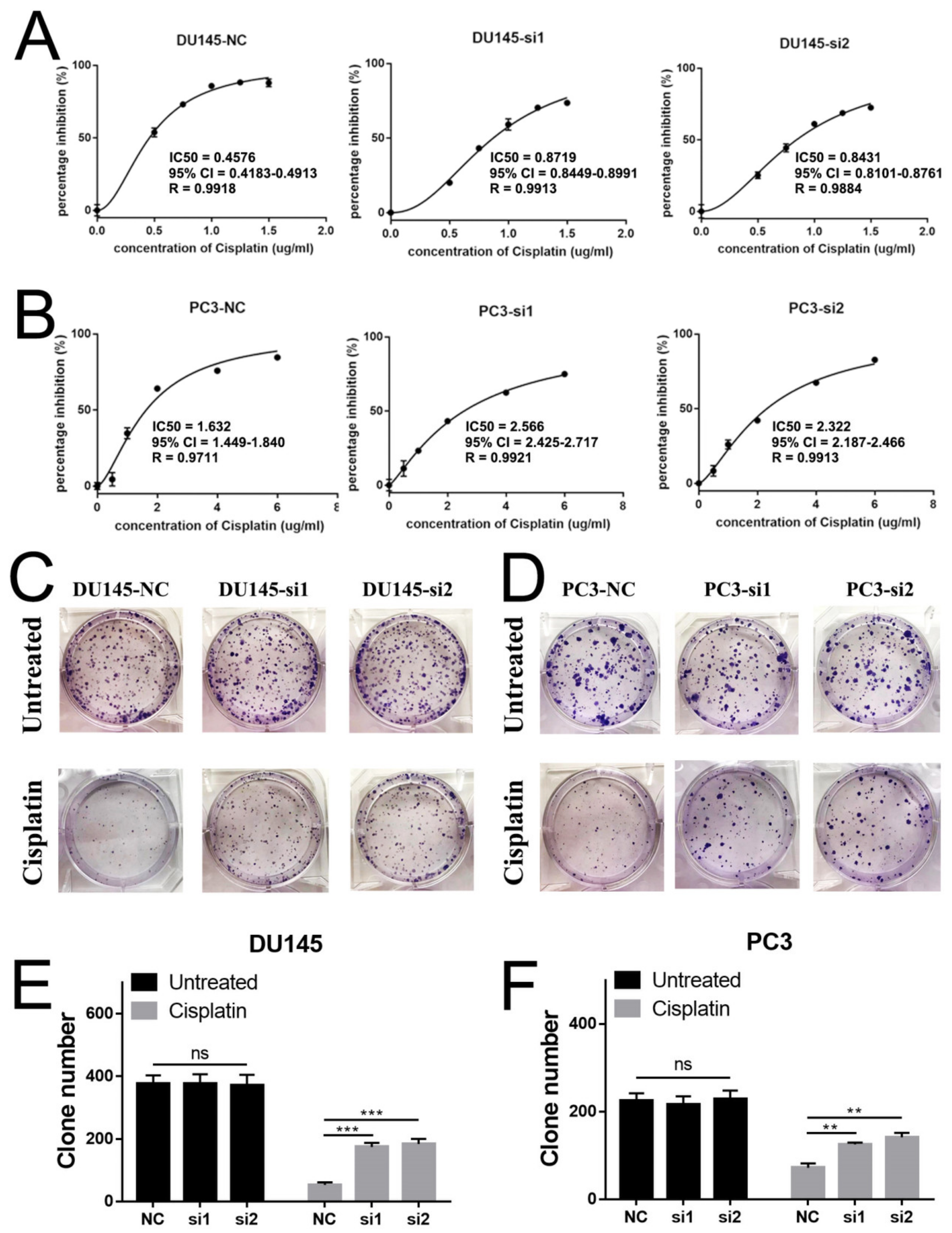
2.3. Loss of NEIL3 Facilitated Docetaxel and Cisplatin Resistance of PCa, but Could Not Promote ADT Resistance
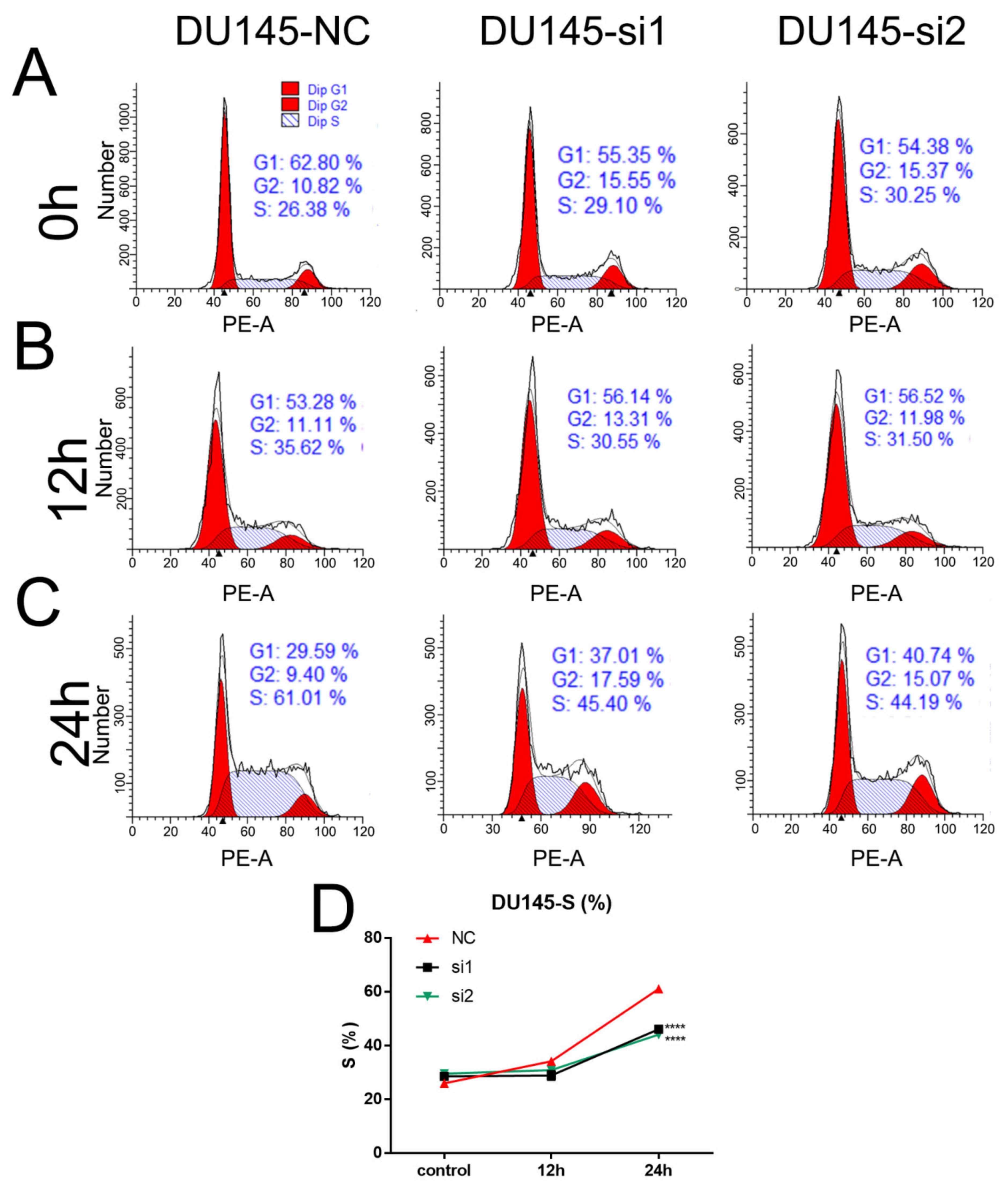
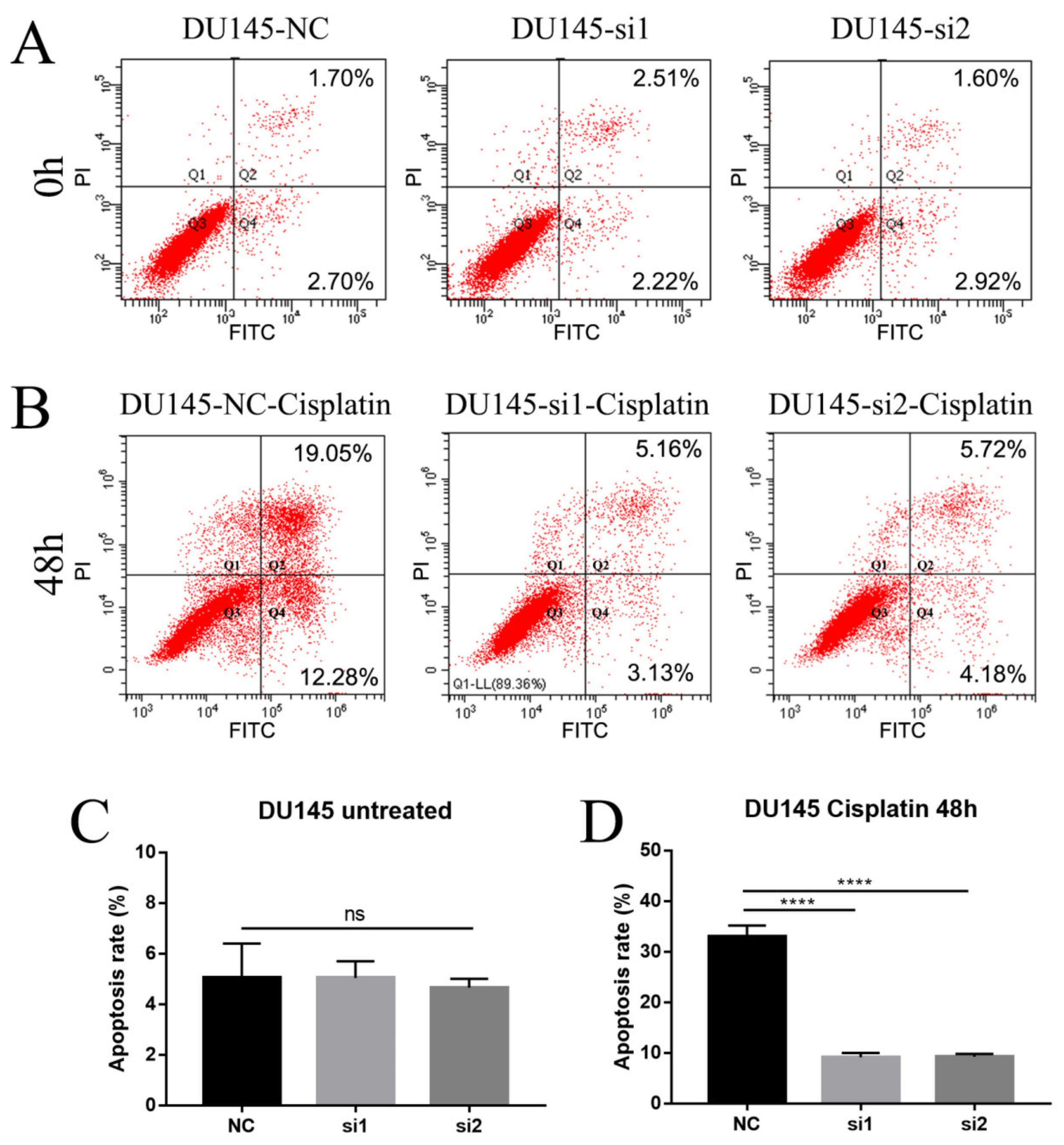
2.4. Loss of NEIL3 Facilitates Cisplatin Resistance by Regulating Apoptosis and Cell Cycle Arrest
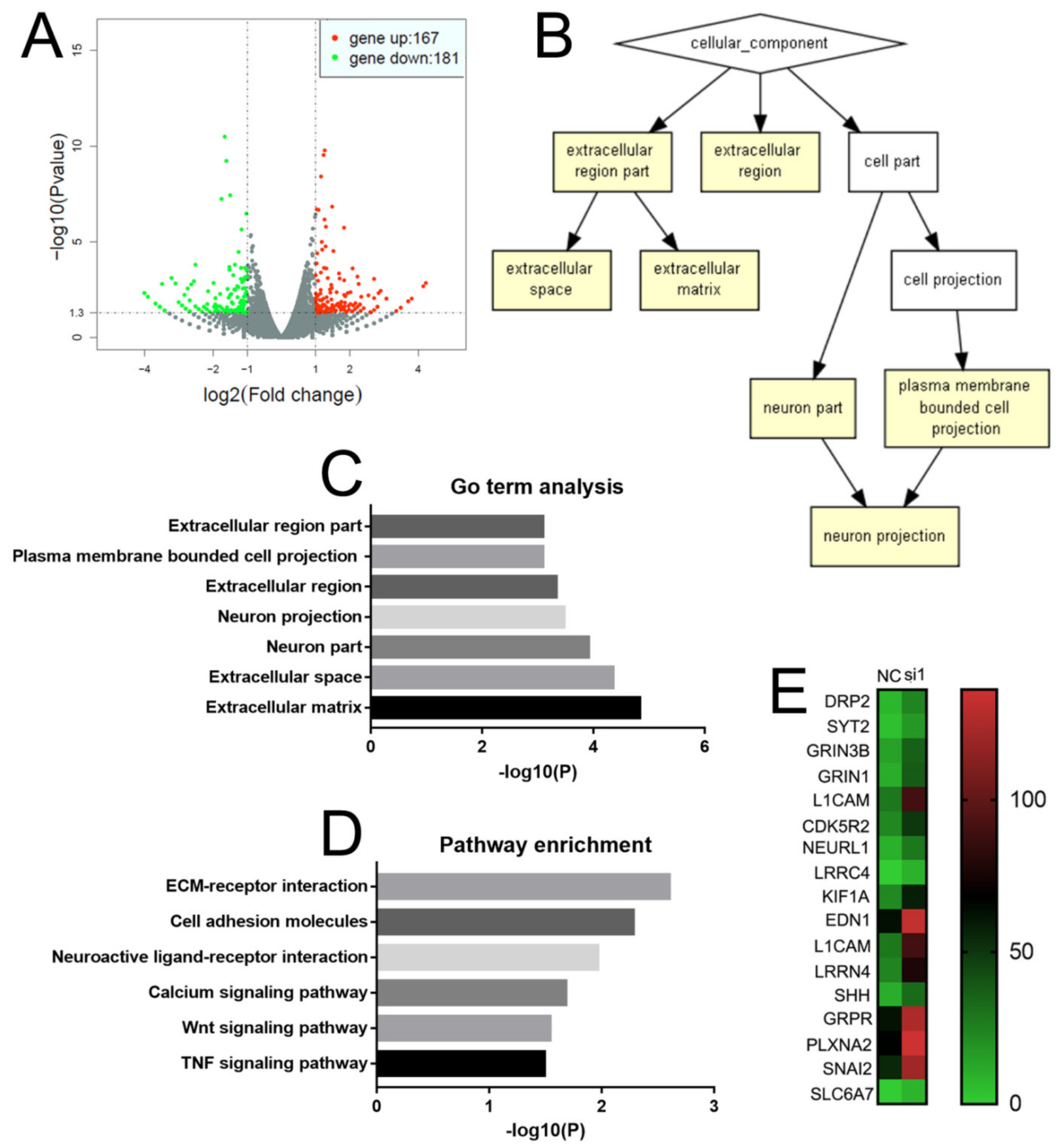
2.5. NEIL3 Affects the Level of Neuroendocrine and Invasion-Related Genes
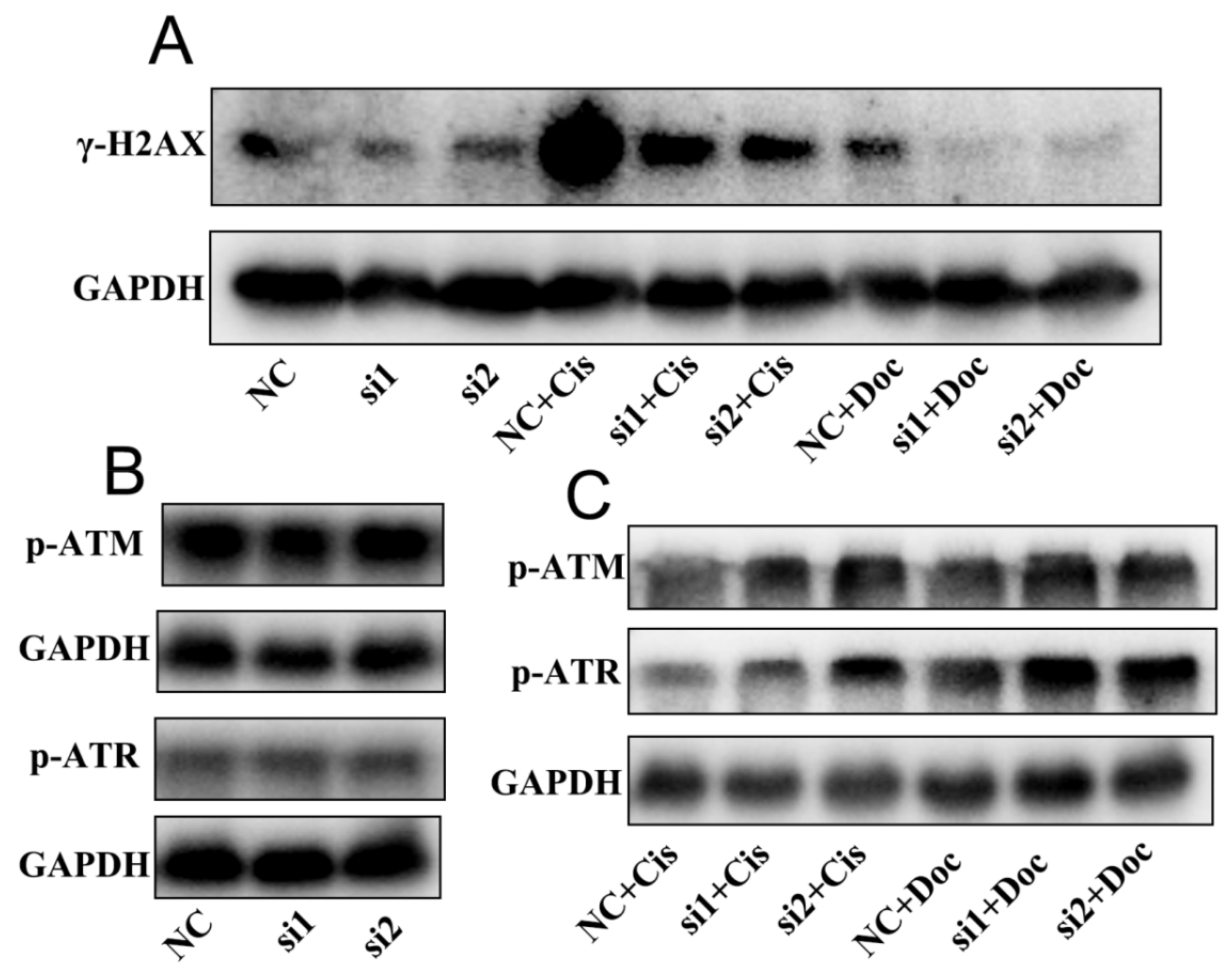
2.6. NEIL3 Regulated Chemotherapy Resistance through the ATR Serine/Threonine Kinase (ATR) and ATM Serine/Threonine Kinase (ATM) Pathways
3. Discussion
4. Materials and Methods
4.1. Database Analysis
4.2. Transient Transfection to Knock Down NEIL3 in DU145 and PC3
4.3. RNA Isolation and qPCR
4.4. Western Blotting
4.5. Cytotoxicity Assay and Colony Formation Assay
4.6. RNA Sequencing and Data Analysis
4.7. Flow Cytometry
4.8. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekar, T.; Yang, J.C.; Gao, A.C.; Evans, C.P. Mechanisms of resistance in castration-resistant prostate cancer (CRPC). Transl. Androl. Urol. 2015, 4, 365–380. [Google Scholar]
- Aparicio, A.; Logothetis, C.J.; Maity, S.N. Understanding the lethal variant of prostate cancer: Power of examining extremes. Cancer Discov. 2011, 1, 466–468. [Google Scholar] [CrossRef] [PubMed]
- Gîngu, C.; Heidenreich, A.; Andreşanu, A.; Mihancea, A.; Sinescu, I.; Baston, C. Treatment of the primary in metastatic prostate cancer. Curr. Opin. Urol. 2020, 30, 566–575. [Google Scholar] [CrossRef] [PubMed]
- Buttigliero, C.; Tucci, M.; Bertaglia, V.; Vignani, F.; Bironzo, P.; Di Maio, M.; Scagliotti, G.V. Understanding and overcoming the mechanisms of primary and acquired resistance to abiraterone and enzalutamide in castration resistant prostate cancer. Cancer Treat. Rev. 2015, 41, 884–892. [Google Scholar] [CrossRef]
- Chen, R.; Dong, X.; Gleave, M. Molecular model for neuroendocrine prostate cancer progression. BJU Int. 2018, 122, 560–570. [Google Scholar] [CrossRef]
- Carceles-Cordon, M.; Kelly, W.K.; Gomella, L.; Knudsen, K.E.; Rodriguez-Bravo, V.; Domingo-Domenech, J. Cellular rewiring in lethal prostate cancer: The architect of drug resistance. Nat. Rev. Urol. 2020, 17, 292–307. [Google Scholar] [CrossRef]
- Wade, C.A.; Kyprianou, N. Profiling Prostate Cancer Therapeutic Resistance. Int. J. Mol. Sci. 2018, 19, 904. [Google Scholar] [CrossRef]
- Conteduca, V.; Oromendia, C.; Eng, K.W.; Bareja, R.; Sigouros, M.; Molina, A.; Faltas, B.M.; Sboner, A.; Mosquera, J.M.; Elemento, O.; et al. Clinical features of neuroendocrine prostate cancer. Eur. J. Cancer 2019, 121, 7–18. [Google Scholar] [CrossRef]
- Francini, E.; Gray, K.P.; Shaw, G.K.; Evan, C.P.; Hamid, A.A.; Perry, C.E.; Kantoff, P.W.; Taplin, M.E.; Sweeney, C.J. Impact of new systemic therapies on overall survival of patients with metastatic castration-resistant prostate cancer in a hospital-based registry. Prostate Cancer Prostatic Dis. 2019, 22, 420–427. [Google Scholar] [CrossRef]
- Bandaru, V.; Sunkara, S.; Wallace, S.S.; Bond, J.P. A novel human DNA glycosylase that removes oxidative DNA damage and is homologous to Escherichia coli endonuclease VIII. DNA Repair 2002, 1, 517–529. [Google Scholar] [CrossRef]
- Li, N.; Wang, J.; Wallace, S.S.; Chen, J.; Zhou, J.; D’Andrea, A.D. Cooperation of the NEIL3 and Fanconi anemia/BRCA pathways in interstrand crosslink repair. Nucleic Acids Res. 2020, 48, 3014–3028. [Google Scholar] [CrossRef]
- Massaad, M.J.; Zhou, J.; Tsuchimoto, D.; Chou, J.; Jabara, H.; Janssen, E.; Glauzy, S.; Olson, B.G.; Morbach, H.; Ohsumi, T.K.; et al. Deficiency of base excision repair enzyme NEIL3 drives increased predisposition to autoimmunity. J. Clin. Investig. 2016, 126, 4219–4236. [Google Scholar] [CrossRef] [PubMed]
- Ha, A.; Lin, Y.; Yan, S. A non-canonical role for the DNA glycosylase NEIL3 in suppressing APE1 endonuclease-mediated ssDNA damage. J. Biol. Chem. 2020, 295, 14222–14235. [Google Scholar] [CrossRef] [PubMed]
- Beltran, H.; Rickman, D.S.; Park, K.; Chae, S.S.; Sboner, A.; MacDonald, T.Y.; Wang, Y.; Sheikh, K.L.; Terry, S.; Tagawa, S.T.; et al. Molecular characterization of neuroendocrine prostate cancer and identification of new drug targets. Cancer Discov. 2011, 1, 487–495. [Google Scholar] [CrossRef]
- Zhang, Q.; Han, Y.; Zhang, Y.; Liu, D.; Ming, J.; Huang, B.; Qiu, X. Treatment-Emergent Neuroendocrine Prostate Cancer: A Clinicopathological and Immunohistochemical Analysis of 94 Cases. Front. Oncol. 2020, 10, 571308. [Google Scholar] [CrossRef] [PubMed]
- Mylavarapu, S.; Das, A.; Roy, M. Role of BRCA Mutations in the Modulation of Response to Platinum Therapy. Front. Oncol. 2018, 8, 16. [Google Scholar] [CrossRef] [PubMed]
- Amellem, O.; Pettersen, E.O. Cell inactivation and cell cycle inhibition as induced by extreme hypoxia: The possible role of cell cycle arrest as a protection against hypoxia-induced lethal damage. Cell Prolif. 1991, 24, 127–141. [Google Scholar] [CrossRef]
- Lin, S.R.; Mokgautsi, N.; Liu, Y.N. Ras and Wnt Interaction Contribute in Prostate Cancer Bone Metastasis. Molecules 2020, 25, 2380. [Google Scholar] [CrossRef]
- Tang, H.; Jiang, L.; Zhu, C.; Liu, R.; Wu, Y.; Yan, Q.; Liu, M.; Jia, Y.; Chen, J.; Qin, Y.; et al. Loss of cell adhesion molecule L1 like promotes tumor growth and metastasis in esophageal squamous cell carcinoma. Oncogene 2019, 38, 3119–3133. [Google Scholar] [CrossRef]
- Machackova, T.; Vychytilova-Faltejskova, P.; Souckova, K.; Trachtova, K.; Brchnelova, D.; Svoboda, M.; Kiss, I.; Prochazka, V.; Kala, Z.; Slaby, O. MiR-215-5p Reduces Liver Metastasis in an Experimental Model of Colorectal Cancer through Regulation of ECM-Receptor Interactions and Focal Adhesion. Cancers 2020, 12, 3518. [Google Scholar] [CrossRef] [PubMed]
- Ren, D.; Yang, Q.; Dai, Y.; Guo, W.; Du, H.; Song, L.; Peng, X. Oncogenic miR-210-3p promotes prostate cancer cell EMT and bone metastasis via NF-κB signaling pathway. Mol. Cancer 2017, 16, 117. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; He, Y.; Butler, W.; Xu, L.; Chang, Y.; Lei, K.; Zhang, H.; Zhou, Y.; Gao, A.C.; Zhang, Q.; et al. Targeting cellular heterogeneity with CXCR2 blockade for the treatment of therapy-resistant prostate cancer. Sci. Transl. Med. 2019, 11. [Google Scholar] [CrossRef] [PubMed]
- Maréchal, A.; Zou, L. DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb. Perspect. Biol. 2013, 5. [Google Scholar] [CrossRef] [PubMed]
- Blackford, A.N.; Jackson, S.P. ATM, ATR, and DNA-PK: The Trinity at the Heart of the DNA Damage Response. Mol. Cell 2017, 66, 801–817. [Google Scholar] [CrossRef] [PubMed]
- Von Klot, C.A.; Kuczyk, M.A.; Boeker, A.; Reuter, C.; Imkamp, F.; Herrmann, T.R.; Tezval, H.; Kramer, M.W.; Perner, S.; Merseburger, A.S. Role of free testosterone levels in patients with metastatic castration-resistant prostate cancer receiving second-line therapy. Oncol. Lett. 2017, 13, 22–28. [Google Scholar] [CrossRef][Green Version]
- Niu, Y.; Guo, C.; Wen, S.; Tian, J.; Luo, J.; Wang, K.; Tian, H.; Yeh, S.; Chang, C. ADT with antiandrogens in prostate cancer induces adverse effect of increasing resistance, neuroendocrine differentiation and tumor metastasis. Cancer Lett. 2018, 439, 47–55. [Google Scholar] [CrossRef]
- Assi, T.; Rassy, E.; Farhat, F.; Kattan, C.; Kattan, J. Docetaxel Rechallenge in Patients with Metastatic Prostate Cancer: A Comprehensive Review. Oncol. Res. Treat. 2020, 43, 299–306. [Google Scholar] [CrossRef]
- Zhang, F.; Zhang, T.; Qu, Y.; Jiang, T.; Cao, Y.X.; Li, C.; Fan, L.; Mei, Q.B. Replication-dependent γ-H2AX formation is involved in docetaxel-induced apoptosis in NSCLC A549 cells. Oncol. Rep. 2010, 24, 1297–1305. [Google Scholar]
- Zhang, C.; Yan, Z.; Painter, C.L.; Zhang, Q.; Chen, E.; Arango, M.E.; Kuszpit, K.; Zasadny, K.; Hallin, M.; Hallin, J.; et al. PF-00477736 mediates checkpoint kinase 1 signaling pathway and potentiates docetaxel-induced efficacy in xenografts. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2009, 15, 4630–4640. [Google Scholar] [CrossRef]
- Ghosh, S. Cisplatin: The first metal based anticancer drug. Bioorganic Chem. 2019, 88, 102925. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Li, Z.; Yang, J.; Peng, S.; Zhou, Q.; Yao, K.; Cai, W.; Xie, Z.; Qin, F.; Li, H.; et al. Loss of NEIL3 activates radiotherapy resistance in the progression of prostate cancer. Cancer Biol. Med. 2021, in press. [Google Scholar]
- Albelazi, M.S.; Martin, P.R.; Mohammed, S.; Mutti, L.; Parsons, J.L.; Elder, R.H. The Biochemical Role of the Human NEIL1 and NEIL3 DNA Glycosylases on Model DNA Replication Forks. Genes 2019, 10, 315. [Google Scholar] [CrossRef]
- Makasheva, K.A.; Endutkin, A.V.; Zharkov, D.O. Requirements for DNA bubble structure for efficient cleavage by helix-two-turn-helix DNA glycosylases. Mutagenesis 2020, 35, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Olsen, M.B.; Hildrestrand, G.A.; Scheffler, K.; Vinge, L.E.; Alfsnes, K.; Palibrk, V.; Wang, J.; Neurauter, C.G.; Luna, L.; Johansen, J.; et al. NEIL3-Dependent Regulation of Cardiac Fibroblast Proliferation Prevents Myocardial Rupture. Cell Rep. 2017, 18, 82–92. [Google Scholar] [CrossRef]
- Shinmura, K.; Kato, H.; Kawanishi, Y.; Igarashi, H.; Goto, M.; Tao, H.; Inoue, Y.; Nakamura, S.; Misawa, K.; Mineta, H.; et al. Abnormal Expressions of DNA Glycosylase Genes NEIL1, NEIL2, and NEIL3 Are Associated with Somatic Mutation Loads in Human Cancer. Oxidative Med. Cell. Longev. 2016, 2016, 1546392. [Google Scholar] [CrossRef]
- Matsuoka, S.; Ballif, B.A.; Smogorzewska, A.; McDonald, E.R., 3rd; Hurov, K.E.; Luo, J.; Bakalarski, C.E.; Zhao, Z.; Solimini, N.; Lerenthal, Y.; et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 2007, 316, 1160–1166. [Google Scholar] [CrossRef] [PubMed]
- Klattenhoff, A.W.; Thakur, M.; Chu, C.S.; Ray, D.; Habib, S.L.; Kidane, D. Loss of NEIL3 DNA glycosylase markedly increases replication associated double strand breaks and enhances sensitivity to ATR inhibitor in glioblastoma cells. Oncotarget 2017, 8, 112942–112958. [Google Scholar] [CrossRef] [PubMed]
- Wallace, B.D.; Berman, Z.; Mueller, G.A.; Lin, Y.; Chang, T.; Andres, S.N.; Wojtaszek, J.L.; DeRose, E.F.; Appel, C.D.; London, R.E.; et al. APE2 Zf-GRF facilitates 3′-5′ resection of DNA damage following oxidative stress. Proc. Natl. Acad. Sci. USA 2017, 114, 304–309. [Google Scholar] [CrossRef]
- Nadal, R.; Schweizer, M.; Kryvenko, O.N.; Epstein, J.I.; Eisenberger, M.A. Small cell carcinoma of the prostate. Nat. Rev. Urol. 2014, 11, 213–219. [Google Scholar] [CrossRef]
- Patel, G.K.; Chugh, N.; Tripathi, M. Neuroendocrine Differentiation of Prostate Cancer-An Intriguing Example of Tumor Evolution at Play. Cancers 2019, 11, 1405. [Google Scholar] [CrossRef] [PubMed]
- Tritschler, S.; Erdelkamp, R.; Stief, C.; Hentrich, M. Neuroendocrine prostate cancer. Der. Pathol. 2018, 39, 333–343. [Google Scholar]
- Lawrence, M.G.; Obinata, D.; Sandhu, S.; Selth, L.A.; Wong, S.Q.; Porter, L.H.; Lister, N.; Pook, D.; Pezaro, C.J.; Goode, D.L.; et al. Patient-derived Models of Abiraterone- and Enzalutamide-resistant Prostate Cancer Reveal Sensitivity to Ribosome-directed Therapy. Eur. Urol. 2018, 74, 562–572. [Google Scholar] [CrossRef] [PubMed]
- Farha, N.G.; Kasi, A. Docetaxel. In StatPearls; StatPearls Publishing Copyright © 2021; StatPearls Publishing LLC.: Treasure Island, FL, USA, 2020. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Xu, L.; Shi, S.; Wu, S.; Meng, R.; Chen, H.; Jiang, Z. Deficiency of NEIL3 Enhances the Chemotherapy Resistance of Prostate Cancer. Int. J. Mol. Sci. 2021, 22, 4098. https://doi.org/10.3390/ijms22084098
Wang Y, Xu L, Shi S, Wu S, Meng R, Chen H, Jiang Z. Deficiency of NEIL3 Enhances the Chemotherapy Resistance of Prostate Cancer. International Journal of Molecular Sciences. 2021; 22(8):4098. https://doi.org/10.3390/ijms22084098
Chicago/Turabian StyleWang, Yiwei, Liuyue Xu, Shanshan Shi, Sha Wu, Ruijie Meng, Huifang Chen, and Zhenyou Jiang. 2021. "Deficiency of NEIL3 Enhances the Chemotherapy Resistance of Prostate Cancer" International Journal of Molecular Sciences 22, no. 8: 4098. https://doi.org/10.3390/ijms22084098
APA StyleWang, Y., Xu, L., Shi, S., Wu, S., Meng, R., Chen, H., & Jiang, Z. (2021). Deficiency of NEIL3 Enhances the Chemotherapy Resistance of Prostate Cancer. International Journal of Molecular Sciences, 22(8), 4098. https://doi.org/10.3390/ijms22084098




