A Bit Stickier, a Bit Slower, a Lot Stiffer: Specific vs. Nonspecific Binding of Gal4 to DNA
Abstract
1. Introduction
2. Results
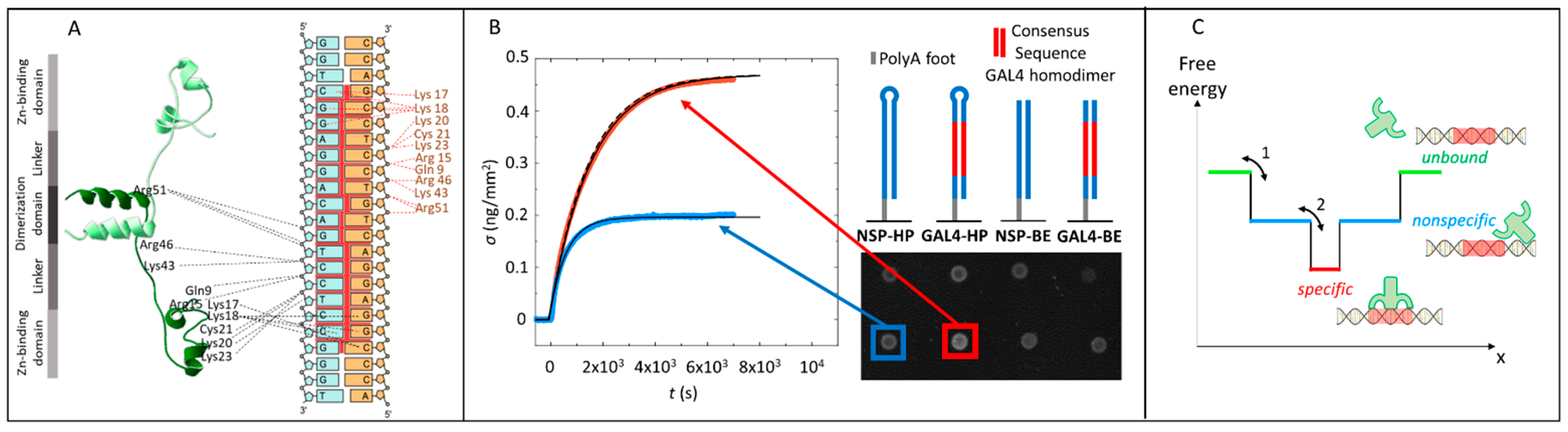
2.1. Gal4 Binding to Specific and Nonspecific DNA Sequences
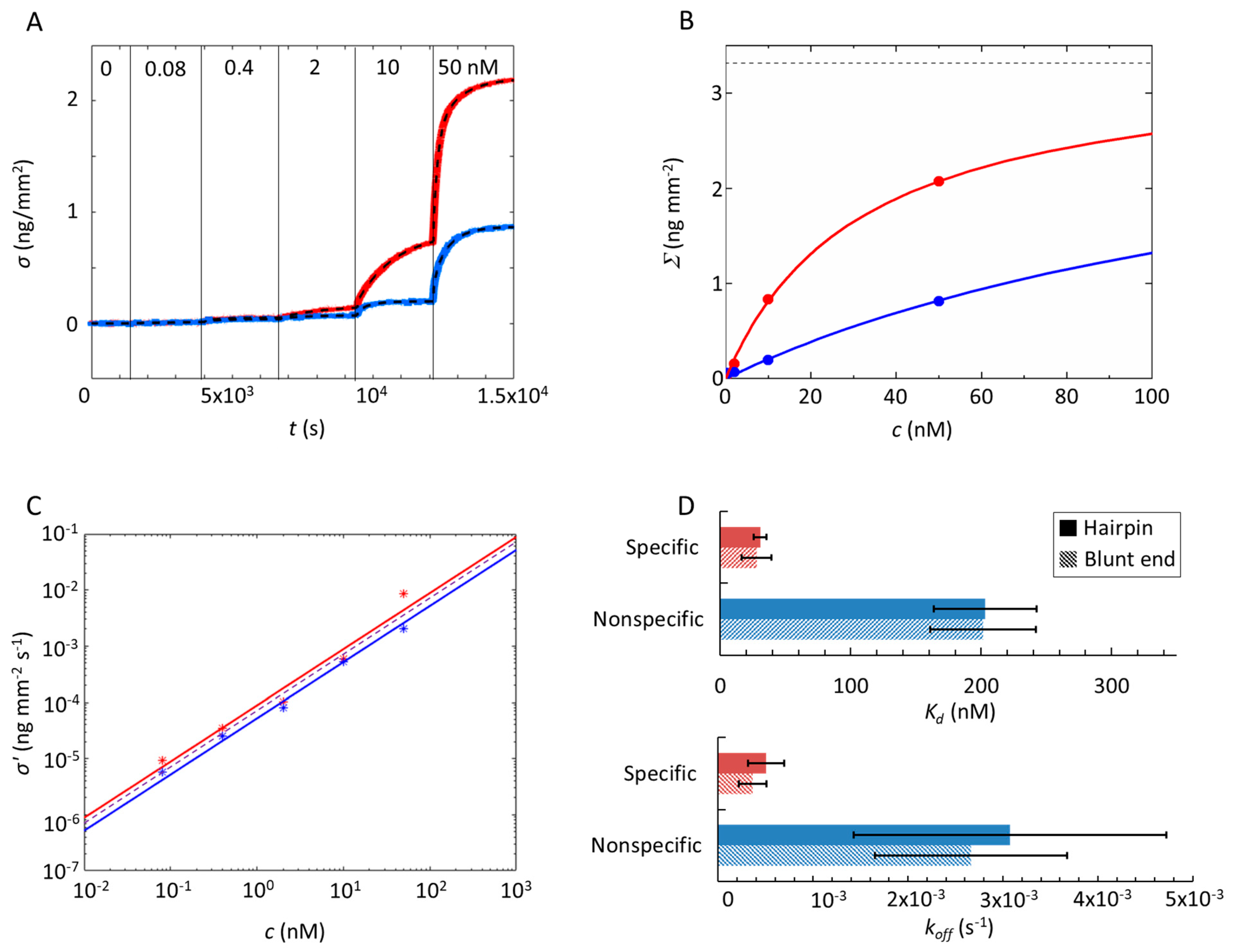
2.2. Analysis of Equilibrium and Kinetics through a Nested–Well Binding Model
2.3. Entropy–Enthalpy Compensation
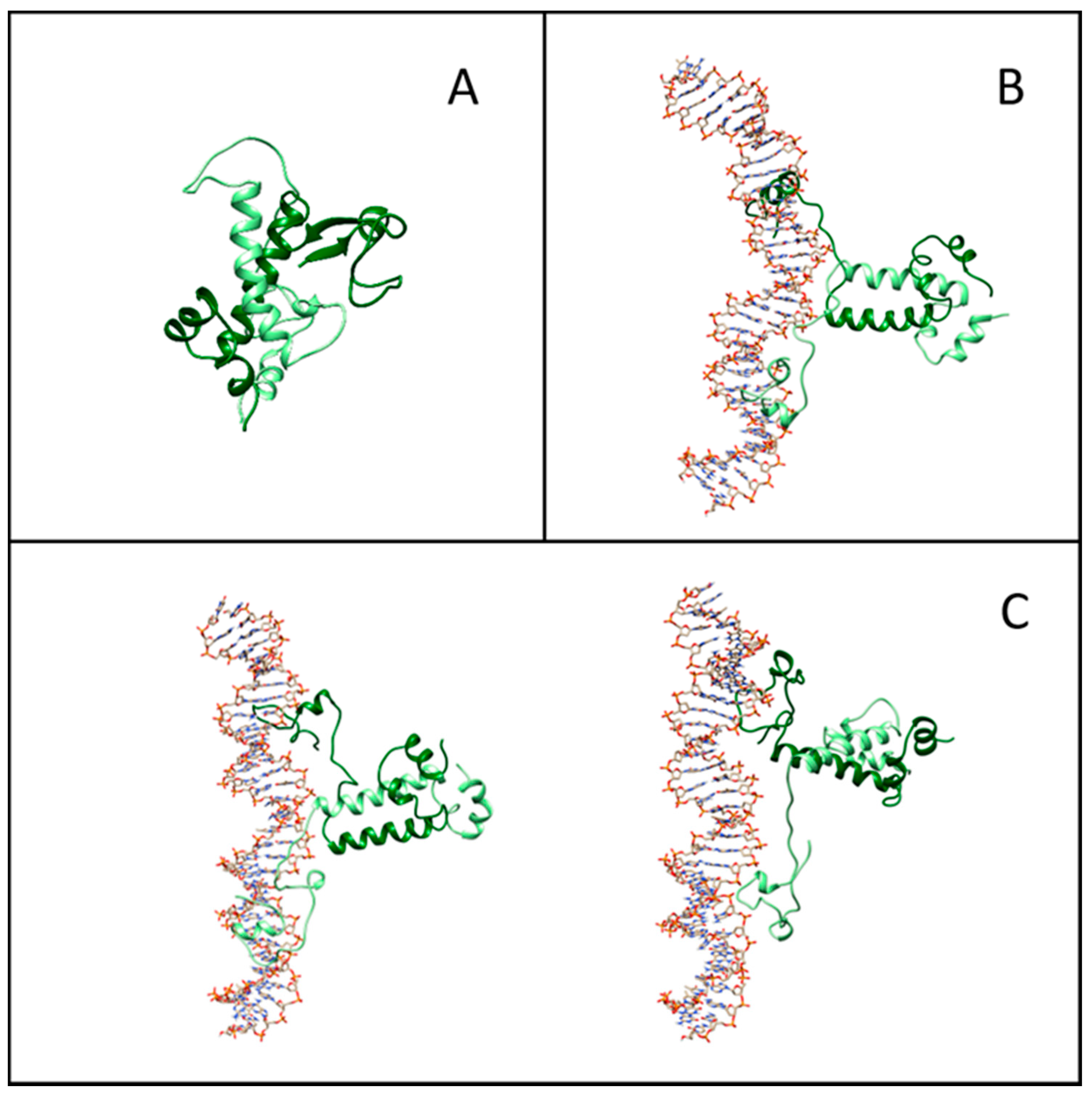
2.4. Structure and Interface of the DNA–Gal4 Complex: A Molecular Dynamics Study
2.5. Entropy–Enthalpy Compensation upon Binding from the Molecular Dynamics Study
3. Discussion and Conclusions
4. Materials and Methods
4.1. Biomolecules and Reagents
4.2. RPI Measurements
4.3. All-Atom Molecular Dynamics Simulations and Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Williams, M.C.; Maher, L.J. Introduction to Biophysics of DNA–Protein Interactions: From Single Molecules to Biological Systems; Springer: Berlin/Heidelberg, Germany, 2010; pp. 3–8. [Google Scholar]
- Zakrzewska, K.; Lavery, R. Towards a molecular view of transcriptional control. Curr. Opin. Struct. Biol. 2012, 22, 160–167. [Google Scholar] [CrossRef] [PubMed]
- Slutsky, M.; Mirny, L.A. Kinetics of Protein-DNA Interaction: Facilitated Target Location in Sequence-Dependent Potential. Biophys. J. 2004, 87, 4021–4035. [Google Scholar] [CrossRef]
- Kalodimos, C.G.; Biris, N.; Bonvin, A.M.J.J.; Levandoski, M.M.; Guennuegues, M.; Boelens, R.; Kaptein, R. Structure and Flexibility Adaptation in Nonspecific and Specific Protein-DNA Complexes. Science 2004, 305, 386–389. [Google Scholar] [CrossRef]
- Furini, S.; Barbini, P.; Domene, C. DNA-recognition process described by MD simulations of the lactose repressor protein on a specific and a non-specific DNA sequence. Nucleic Acids Res. 2013, 41, 3963–3972. [Google Scholar] [CrossRef] [PubMed]
- Wunderlich, Z.; Mirny, L.A. Different gene regulation strategies revealed by analysis of binding motifs. Trends Genet. 2009, 25, 434–440. [Google Scholar] [CrossRef] [PubMed]
- Badis, G.; Berger, M.F.; Philippakis, A.A.; Talukder, S.; Gehrke, A.R.; Jaeger, S.A.; Chan, E.T.; Metzler, G.; Vedenko, A.; Chen, X.; et al. Diversity and Complexity in DNA Recognition by Transcription Factors. Science 2009, 324, 1720–1723. [Google Scholar] [CrossRef] [PubMed]
- Siggers, T.; Gordân, R. Protein–DNA binding: Complexities and multi-protein codes. Nucleic Acids Res. 2014, 42, 2099–2111. [Google Scholar] [CrossRef] [PubMed]
- Rosanova, A.; Colliva, A.; Osella, M.; Caselle, M. Modelling the evolution of transcription factor binding preferences in complex eukaryotes. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef]
- Yoo, J.; Winogradoff, D.; Aksimentiev, A. Molecular dynamics simulations of DNA–DNA and DNA–protein interactions. Curr. Opin. Struct. Biol. 2020, 64, 88–96. [Google Scholar] [CrossRef]
- Afek, A.; Schipper, J.L.; Horton, J.; Gordân, R.; Lukatsky, D.B. Protein−DNA binding in the absence of specific base-pair recognition. Proc. Natl. Acad. Sci. USA 2014, 111, 17140–17145. [Google Scholar] [CrossRef]
- Zhang, X.; Daaboul, G.G.; Spuhler, P.S.; Dröge, P.; Ünlü, M.S. Quantitative characterization of conformational-specific protein–DNA binding using a dual-spectral interferometric imaging biosensor. Nanoscale 2016, 8, 5587–5598. [Google Scholar] [CrossRef]
- Hellman, L.M.; Fried, M.G. Electrophoretic mobility shift assay (EMSA) for detecting protein–nucleic acid interactions. Nat. Protoc. 2007, 2, 1849–1861. [Google Scholar] [CrossRef]
- Weirauch, M.T.; Yang, A.; Albu, M.; Cote, A.G.; Montenegro-Montero, A.; Drewe, P.; Najafabadi, H.S.; Lambert, S.A.; Mann, I.; Cook, K.; et al. Determination and Inference of Eukaryotic Transcription Factor Sequence Specificity. Cell 2014, 158, 1431–1443. [Google Scholar] [CrossRef] [PubMed]
- Bulyk, M.L.; Huang, X.; Choo, Y.; Church, G.M. Exploring the DNA-binding specificities of zinc fingers with DNA microarrays. Proc. Natl. Acad. Sci. USA 2001, 98, 7158–7163. [Google Scholar] [CrossRef]
- Zanchetta, G.; Lanfranco, R.; Giavazzi, F.; Bellini, T.; Buscaglia, M. Emerging applications of label-free optical biosensors. Nanophotonics 2017, 6, 627–645. [Google Scholar] [CrossRef]
- Giavazzi, F.; Salina, M.; Cerbino, R.; Bassi, M.; Prosperi, D.; Ceccarello, E.; Damin, F.; Sola, L.; Rusnati, M.; Chiari, M.; et al. Multispot, label-free biodetection at a phantom plastic–water interface. Proc. Natl. Acad. Sci. USA 2013, 110, 9350–9355. [Google Scholar] [CrossRef] [PubMed]
- Salina, M.; Giavazzi, F.; Lanfranco, R.; Ceccarello, E.; Sola, L.; Chiari, M.; Chini, B.; Cerbino, R.; Bellini, T.; Buscaglia, M. Multi-spot, label-free immunoassay on reflectionless glass. Biosens. Bioelectron. 2015, 74, 539–545. [Google Scholar] [CrossRef]
- Nava, G.; Ceccarello, E.; Giavazzi, F.; Salina, M.; Damin, F.; Buscaglia, M.; Chiari, M.; Bellini, T.; Zanchetta, G. Label-free detection of DNA single-base mismatches using a simple reflectance-based optical technique. Phys. Chem. Chem. Phys. 2016, 18, 13395–13402. [Google Scholar] [CrossRef]
- Vanjur, L.; Carzaniga, T.; Casiraghi, L.; Chiari, M.; Zanchetta, G.; Buscaglia, M. Non-Langmuir Kinetics of DNA Surface Hybridization. Biophys. J. 2020, 119, 989–1001. [Google Scholar] [CrossRef]
- Marmorstein, R.; Carey, M.; Ptashne, M.; Harrison, S.C. DNA recognition by GAL4: Structure of a protein-DNA complex. Nat. Cell Biol. 1992, 356, 408–414. [Google Scholar] [CrossRef]
- Hong, M.; Fitzgerald, M.X.; Harper, S.; Luo, C.; Speicher, D.W.; Marmorstein, R. Structural Basis for Dimerization in DNA Recognition by Gal4. Structure 2008, 16, 1019–1026. [Google Scholar] [CrossRef] [PubMed]
- Corton, J.C.; Moreno, E.; Johnston, S.A. Alterations in the GAL4 DNA-binding Domain Can Affect Transcriptional Activation Independent of DNA Binding. J. Biol. Chem. 1998, 273, 13776–13780. [Google Scholar] [CrossRef] [PubMed]
- Shumaker-Parry, J.S.; Aebersold, R.; Campbell, C.T. Parallel, Quantitative Measurement of Protein Binding to a 120-Element Double-Stranded DNA Array in Real Time Using Surface Plasmon Resonance Microscopy. Anal. Chem. 2004, 76, 2071–2082. [Google Scholar] [CrossRef] [PubMed]
- MacPherson, S.; LaRochelle, M.; Turcotte, B. A Fungal Family of Transcriptional Regulators: The Zinc Cluster Proteins. Microbiol. Mol. Biol. Rev. 2006, 70, 583–604. [Google Scholar] [CrossRef]
- Reece, R.; Ptashne, M. Determinants of binding-site specificity among yeast C6 zinc cluster proteins. Science 1993, 261, 909–911. [Google Scholar] [CrossRef]
- Liang, S.D.; Marmorstein, R.; Harrison, S.C.; Ptashine, M. DNA sequence preferences of GAL4 and PPR1: How a subset of Zn2 Cys6 binuclear cluster proteins recognizes DNA. Mol. Cell. Biol. 1996, 16, 3773–3780. [Google Scholar] [CrossRef]
- Li, M.; Hada, A.; Sen, P.; Olufemi, L.; Hall, M.A.; Smith, B.Y.; Forth, S.; McKnight, J.N.; Patel, A.; Bowman, G.D.; et al. Dynamic regulation of transcription factors by nucleosome remodeling. eLife 2015, 4, e06249. [Google Scholar] [CrossRef]
- Bintu, L.; Buchler, N.E.; Garcia, H.G.; Gerland, U.; Hwa, T.; Kondev, J.; Phillips, R. Transcriptional regulation by the numbers: Models. Curr. Opin. Genet. Dev. 2005, 15, 116–124. [Google Scholar] [CrossRef]
- Bintu, L.; Buchler, N.E.; Garcia, H.G.; Gerland, U.; Hwa, T.; Kondev, J.; Kuhlman, T.; Phillips, R. Transcriptional regulation by the numbers: Applications. Curr. Opin. Genet. Dev. 2005, 15, 125–135. [Google Scholar] [CrossRef]
- Luo, Y.; North, J.A.; Poirier, M.G. Single Molecule Fluorescence Methodologies for Investigating Transcription Factor Binding Kinetics to Nucleosomes and DNA. Methods 2014, 70, 108–118. [Google Scholar] [CrossRef]
- Zhu, C.; Byers, K.J.; Mccord, R.P.; Shi, Z.; Berger, M.F.; Newburger, D.E.; Saulrieta, K.; Smith, Z.; Shah, M.V.; Radhakrishnan, M.; et al. High-resolution DNA-binding specificity analysis of yeast transcription factors. Genome Res. 2009, 19, 556–566. [Google Scholar] [CrossRef]
- Drake, A.W.; Myszka, D.G.; Klakamp, S.L. Characterizing high-affinity antigen/antibody complexes by kinetic- and equilibrium-based methods. Anal. Biochem. 2004, 328, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Privalov, P.L.; Dragan, A.I.; Crane-Robinson, C.; Breslauer, K.J.; Remeta, D.P.; Minetti, C.A. What Drives Proteins into the Major or Minor Grooves of DNA? J. Mol. Biol. 2007, 365, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Privalov, P.L.; Dragan, A.I.; Crane-Robinson, C. Interpreting protein/DNA interactions: Distinguishing specific from non-specific and electrostatic from non-electrostatic components. Nucleic Acids Res. 2010, 39, 2483–2491. [Google Scholar] [CrossRef] [PubMed]
- Gerland, U.; Moroz, J.D.; Hwa, T. Physical constraints and functional characteristics of transcription factor-DNA interaction. Proc. Natl. Acad. Sci. USA 2002, 99, 12015–12020. [Google Scholar] [CrossRef] [PubMed]
- Esadze, A.; Kemme, C.A.; Kolomeisky, A.B.; Iwahara, J. Positive and negative impacts of nonspecific sites during target location by a sequence-specific DNA-binding protein: Origin of the optimal search at physiological ionic strength. Nucleic Acids Res. 2014, 42, 7039–7046. [Google Scholar] [CrossRef]
- Cencini, M.; Pigolotti, S. Energetic funnel facilitates facilitated diffusion. Nucleic Acids Res. 2017, 46, 558–567. [Google Scholar] [CrossRef]
- Pace, C.N.; Shirley, B.A.; McNutt, M.; Gajiwala, K. Forces contributing to the conformational stability of proteins. FASEB J. 1996, 10, 75–83. [Google Scholar] [CrossRef]
- Fersht, A.R. The hydrogen bond in molecular recognition. Trends Biochem. Sci. 1987, 12, 301–304. [Google Scholar] [CrossRef]
- Smith, J.D.; Cappa, C.D.; Wilson, K.R.; Messer, B.M.; Cohen, R.C.; Saykally, R.J. Energetics of Hydrogen Bond Network Rearrangements in Liquid Water. Science 2004, 306, 851–853. [Google Scholar] [CrossRef] [PubMed]
- Traven, A.; Jelicic, B.; Sopta, M. Yeast Gal4: A transcriptional paradigm revisited. EMBO Rep. 2006, 7, 496–499. [Google Scholar] [CrossRef]
- Huggins, D.J. Quantifying the Entropy of Binding for Water Molecules in Protein Cavities by Computing Correlations. Biophys. J. 2015, 108, 928–936. [Google Scholar] [CrossRef][Green Version]
- Hahn, S.; Young, E.T. Transcriptional Regulation in Saccharomyces cerevisiae: Transcription Factor Regulation and Function, Mechanisms of Initiation, and Roles of Activators and Coactivators. Genetics 2011, 189, 705–736. [Google Scholar] [CrossRef] [PubMed]
- Meijsing, S.H.; Pufall, M.A.; So, A.Y.; Bates, D.L.; Chen, L.; Yamamoto, K.R. DNA Binding Site Sequence Directs Glucocorticoid Receptor Structure and Activity. Science 2009, 324, 407–410. [Google Scholar] [CrossRef] [PubMed]
- Schöne, S.; Jurk, M.; Helabad, M.B.; Dror, I.; Lebars, I.; Kieffer, B.; Imhof, P.; Rohs, R.; Vingron, M.; Thomas-Chollier, M.; et al. Sequences flanking the core-binding site modulate glucocorticoid receptor structure and activity. Nat. Commun. 2016, 7, 12621. [Google Scholar] [CrossRef] [PubMed]
- Carey, M.; Kakidani, H.; Leatherwood, J.; Mostashari, F.; Ptashne, M. An amino-terminal fragment of GAL4 binds DNA as a dimer. J. Mol. Biol. 1989, 209, 423–432. [Google Scholar] [CrossRef]
- Lavery, R.; Zakrzewska, K.; Sklenar, H. JUMNA (junction minimisation of nucleic acids). Comput. Phys. Commun. 1995, 91, 135–158. [Google Scholar] [CrossRef]
- Pronk, S.; Páll, S.; Schulz, R.; Larsson, P.; Bjelkmar, P.; Apostolov, R.; Shirts, M.R.; Smith, J.C.; Kasson, P.M.; van der Spoel, D.; et al. GROMACS 4.5: A high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 2013, 29, 845–854. [Google Scholar] [CrossRef]
- Lavery, R.; Moakher, M.; Maddocks, J.H.; Petkeviciute, D.; Zakrzewska, K. Conformational analysis of nucleic acids revisited: Curves+. Nucleic Acids Res. 2009, 37, 5917–5929. [Google Scholar] [CrossRef] [PubMed]



| # | Name | Length | Sequence |
|---|---|---|---|
| 1 | GAL4-HP | 106 | /5AmMC6/AAA AAA AAA ATG AAA TGT TGG AAG GGT CGG AGG ACA GTC CTC CGG GTG GTA TAG TCT CCT ACC TAT ACC ACC CGG AGG ACT GTC CTC CGA CCC TTC CAA CAT TTC A |
| 2 | NSP-HP | 106 | /5AmMC6/AAA AAA AAA ATG AAA TGT TGG TTG CGT CTC TCC TAT GTT GCG TCG GTG GTA TAG TCT CCT ACC TAT ACC ACC GAC GCA ACA TAG GAG AGA CGC AAC CAA CAT TTC A |
| 3 | GAL4-BE | 54 | /5AmMC6/AAA AAA AAA ATG AAA TGT TGG AAG GGT CGG AGG ACA GTC CTC CGG GTG GTA TAG |
| 4 | NSP-BE | 54 | /5AmMC6/AAA AAA AAA ATG AAA TGT TGG TTG CGT CTC TCC TAT GTT GCG TCG GTG GTA TAG |
| 5 | GAL4-BE-C | 44 | CTA TAC CAC CCG GAG GAC TGT CCT CCG ACC CTT CCA ACA TTT CA |
| 6 | NSP-BE-C | 44 | CTA TAC CAC CGA CGC AAC ATA GGA GAG ACG CAA CCA ACA TTT CA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carzaniga, T.; Zanchetta, G.; Frezza, E.; Casiraghi, L.; Vanjur, L.; Nava, G.; Tagliabue, G.; Dieci, G.; Buscaglia, M.; Bellini, T. A Bit Stickier, a Bit Slower, a Lot Stiffer: Specific vs. Nonspecific Binding of Gal4 to DNA. Int. J. Mol. Sci. 2021, 22, 3813. https://doi.org/10.3390/ijms22083813
Carzaniga T, Zanchetta G, Frezza E, Casiraghi L, Vanjur L, Nava G, Tagliabue G, Dieci G, Buscaglia M, Bellini T. A Bit Stickier, a Bit Slower, a Lot Stiffer: Specific vs. Nonspecific Binding of Gal4 to DNA. International Journal of Molecular Sciences. 2021; 22(8):3813. https://doi.org/10.3390/ijms22083813
Chicago/Turabian StyleCarzaniga, Thomas, Giuliano Zanchetta, Elisa Frezza, Luca Casiraghi, Luka Vanjur, Giovanni Nava, Giovanni Tagliabue, Giorgio Dieci, Marco Buscaglia, and Tommaso Bellini. 2021. "A Bit Stickier, a Bit Slower, a Lot Stiffer: Specific vs. Nonspecific Binding of Gal4 to DNA" International Journal of Molecular Sciences 22, no. 8: 3813. https://doi.org/10.3390/ijms22083813
APA StyleCarzaniga, T., Zanchetta, G., Frezza, E., Casiraghi, L., Vanjur, L., Nava, G., Tagliabue, G., Dieci, G., Buscaglia, M., & Bellini, T. (2021). A Bit Stickier, a Bit Slower, a Lot Stiffer: Specific vs. Nonspecific Binding of Gal4 to DNA. International Journal of Molecular Sciences, 22(8), 3813. https://doi.org/10.3390/ijms22083813








