Abstract
All molecular systems, from small molecules to macromolecules, exhibit specific characteristics for a specific environment and time. In order to gain an accurate understanding of the functions of all types of molecules, studies of their structure and dynamics are essential. Through dynamic studies, using techniques such as spectroscopy, structure determination, and computer analysis, it is possible to collect functional information on molecules at specific times and in specific environments. Such information not only reveals the properties and mechanisms of action of molecules but also provides insights that can be applied to various industries, such as the development of new materials and drugs. Herein, I discuss the importance of molecular dynamics studies, present the time scale of molecular motion, and review techniques for analyzing molecular dynamics.
Imagine this example of why molecular dynamics research is important. There is an advertisement for a new vehicle in the newspaper. It is built primarily for driving on the road like a car, but it can fly like a light aircraft and can be sailed like a ship on rivers or seas. Of course, it is equipped with fully autonomous driving capability, and powered by sustainable energy. Below the introduction to each of these versatile features is a picture of the vehicle engaged in each mode of transport (ground, air, and sea). We can extrapolate a lot from these three pictures by imagination, but some key questions remain unanswered. How will this vehicle transform from one mode of transport to another (i.e., car to plane to ship)? How long will each transformation take? How long can this transport run without replacing its energy source? How perfect/safe is its autonomous driving? How does it handle traffic conditions on the road, sky, and sea? We cannot say anything with confidence with just three pictures. Each mode of transport will have its own operating characteristics, and these characteristics will also change in adaptation to changes in operating environment. We will need tons of pictures and/or intelligently produced videos to accurately understand the vehicle’s characteristics under all conditions we may encounter in each mode of transportation. With continuous information (as is provided by videos), we can accurately understand the vehicle’s performance, whereas fragmented information (such as that from pictures) is insufficient to understand the new vehicle.
This transportation-based analogy illustrates the importance of continuous observation, like movies, for the understanding of complex processes such as those studied in the sciences. In nature, there are many molecules (ranging from small molecules to macromolecules) with interesting and important molecular properties that can be elucidated using various scientific techniques.
Among these, techniques for obtaining high-resolution structural information are essential as they provide intuitive insights into important structural and functional characteristics of molecules that play important roles in life. Currently, the three dimensional structures of small molecules can be obtained from repositories such as the Cambridge Structural Database (CSD) [1], which contains 1,123,962 small-molecule organic or metal-organic structures obtained using X-ray, neutron diffraction, or microcrystal electron diffraction (MicroED) techniques [2]. The three dimensional structure of macromolecules can be obtained from repositories such as the Protein Data Bank (PDB) [3] and Electron Microscopy Data Bank (EMDB), containing 175,759 and 14,493 macromolecular structures (protein, DNA, RNA, virus, or protein-nucleic acid complexes), respectively, determined using X-ray, nuclear magnetic resonance (NMR), cryogenic electron microscopy (Cryo-EM), or MicroED techniques. These structures are very useful for describing molecular functions and features and are applied as inputs for computational analysis models such as molecular dynamics (MD) simulation [4]. However, since most of this structural information mainly deals with static information in a specific state, it cannot provide accurate information on structural changes caused by external environments or stimuli. Interpretation of a molecule’s function using static information cannot explain the complete underlying mechanism and can be misleading [5]. In order to understand the exact function of a molecule, it is necessary to obtain dynamic information about the molecule in a spatiotemporal context [6], as the molecular functions are intimately related to structure and dynamics [7]. In order to observe the dynamics of a molecule, it is important to understand the time scale of the motion of the target molecule (Figure 1). The time scales of molecular motion such as electronic motion, photodissociation, photoionization, bond vibrations, molecular collisions, vibration relaxation, solvation, proton transfer, temperature jump Raman, fluorescence, molecular rotations, large molecule relaxation, molecular diffusion, and phosphorescence motion varies between a few seconds to femtoseconds [8,9,10]. When observing the dynamics of a molecule, the movement time scale of the molecule must be considered and an appropriate experimental technique must be applied to observe it.
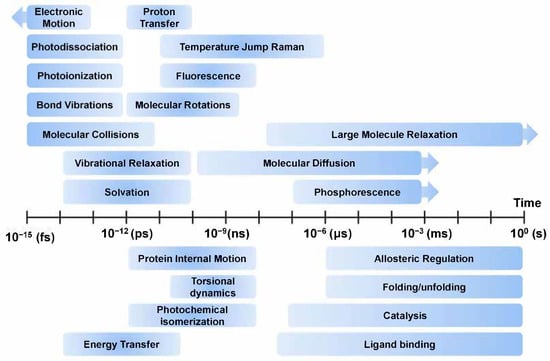
Figure 1.
Overview of time scales of molecular motions. The contents of this figure were excerpted from the contents of the study mentioned in [8,9,10].
Various techniques such as spectroscopy, structure determination and computer analysis are applied to the study of molecular dynamics (Figure 2). In order to observe the dynamics of a molecule at a specific time, it is necessary to observe the molecule at much shorter time intervals. In this context, ultrafast lasers have a shorter pulse duration than most characteristic relaxation times of the condensed phase, making it possible to carry out detailed characterization of the temporal, spatial, and spectral properties of materials [11]. The development of femtosecond spectroscopy ushered in the era of real-time investigation of the motion of atoms in solid, liquid, and gaseous state molecules [12,13]. Furthermore, advances in time-resolved spectroscopy allows us to visualize the dynamics of the various types of atoms and molecules at dynamic time units (up to tens of femtoseconds) [12]. Time-resolved vibrational spectroscopies, such as infrared absorption spectroscopy (time-resolved IR spectroscopy) and Raman scattering (time-resolved Raman spectroscopy), have been widely applied as a solution for studying molecular dynamics [14].
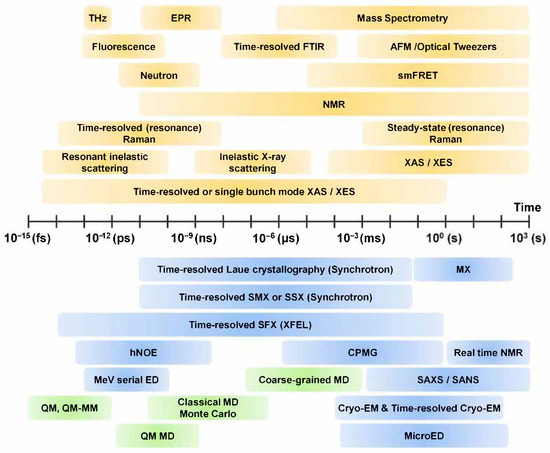
Figure 2.
Techniques for structure and dynamics. THz: terahertz spectroscopy, EPR: electron paramagnetic resonance spectroscopy, FTIR: Fourier-transform infrared spectroscopy, smFRET: single molecule fluorescence resonance energy transfer, SMX: serial millisecond crystallography, SSX: serial synchrotron crystallography, SFX: serial femtosecond crystallography, hNOE: heteronuclear nuclear Overhauser effect, CPMG: Carr–Purcell–Meiboom–Gill, AFM: atomic force microscopy, ED: electron diffraction, SAXS: small angle X-ray scattering, SANS: small angle neutron scattering, MD: molecular dynamics, QM: quantum mechanics, MM: molecular mechanics. The contents of this figure were excerpted from the contents of the study mentioned in [15,16,17,18,19].
Meanwhile, techniques such as X-ray diffraction/scattering, NMR, Cryo-EM, MicroED, and neutron scattering/diffraction are widely applied to study the mechanism of action by revealing the structure of a molecule [2,15,20]. These techniques enable the study of the flexibility of a molecule within a single static structure, and they provide dynamic information by collecting data regarding external stimuli/environment or other static data in a complex. With the development of X-ray technology, an X-ray free electron laser (XFEL) with ultrashort pulse width has been created, which generates more intense X-rays than does the existing synchrotron radiation [21,22,23]. The serial femtosecond crystallography (SFX) or spectroscopic techniques using XFEL enables the determination of the molecular structures at room temperature without radiation damage [24,25,26]. Additionally, time-resolved SFX studies using an optical laser or mix-and-inject technique contribute to the making movies with high resolution molecular dynamics [15,27]. Moreover, due to advances in X-ray-focusing technology and detectors, time-resolved serial millisecond crystallography (SMX) studies as well as general serial synchrotron crystallography (SSX) are being performed at the existing synchrotron facilities [28,29,30]. Recently, the analysis of large complex structures using Cryo-EM technology has been in the spotlight [31], and the time-resolved EM technique is expected to be useful in the future for obtaining molecular dynamics information [32].
Additionally, computational molecular dynamics analysis by density-functional theory (DFT) or MD simulation provides an understanding of the functions of molecules and insights into their industrial or medical applications, such as in drug design [33,34,35,36,37].
Comprehensive molecular dynamics information for pinpointing the functions and properties of molecules can be inferred from information gleaned from multiple snapshots obtained using traditional techniques and the most advanced techniques. Examples of these include temporal characterization based on autocorrelation and pump probe technology combined with microscopy, spatial evolution analysis using X-ray and electron diffraction techniques, and monitoring of the temporal and spatial evolution of materials using nonlinear time-resolved spectroscopy [11].
In this special issue, we deal with the topic of molecular dynamics—those of small molecules to macromolecules. The issue includes a collection of comprehensive articles and reviews of research findings on a wide range of subjects related to molecular dynamics and the development of the technologies that makes its study possible. The results from this broad range of studies not only provide important insights about the technology but also facilitate a broader understanding of molecular dynamics studies across the various sciences.
Funding
This work was funded by the National Research Foundation of Korea (NRF-2017M3A9F6029736).
Conflicts of Interest
The authors declare that they have no competing interests.
References
- Groom, C.R.; Bruno, I.J.; Lightfoot, M.P.; Ward, S.C. The Cambridge Structural Database. Acta Cryst. Sect. B 2016, 72, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Jones, C.G.; Martynowycz, M.W.; Hattne, J.; Fulton, T.J.; Stoltz, B.M.; Rodriguez, J.A.; Nelson, H.M.; Gonen, T. The CryoEM Method MicroED as a Powerful Tool for Small Molecule Structure Determination. ACS Cent. Sci. 2018, 4, 1587–1592. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Hollingsworth, S.A.; Dror, R.O. Molecular Dynamics Simulation for All. Neuron 2018, 99, 1129–1143. [Google Scholar] [CrossRef] [PubMed]
- Pearson, A.R.; Mehrabi, P. Serial synchrotron crystallography for time-resolved structural biology. Curr. Opin. Struct. Biol. 2020, 65, 168–174. [Google Scholar] [CrossRef] [PubMed]
- Di Rienzo, C.; Gratton, E.; Beltram, F.; Cardarelli, F. Spatiotemporal Fluctuation Analysis: A Powerful Tool for the Future Nanoscopy of Molecular Processes. Biophys. J. 2016, 111, 679–685. [Google Scholar] [CrossRef] [PubMed]
- Šponer, J.; Bussi, G.; Krepl, M.; Banáš, P.; Bottaro, S.; Cunha, R.A.; Gil-Ley, A.; Pinamonti, G.; Poblete, S.; Jurečka, P.; et al. RNA Structural Dynamics As Captured by Molecular Simulations: A Comprehensive Overview. Chem. Rev. 2018, 118, 4177–4338. [Google Scholar] [CrossRef]
- Farr, E.P.; Quintana, J.C.; Reynoso, V.; Ruberry, J.D.; Shin, W.R.; Swartz, K.R. Introduction to Time-Resolved Spectroscopy: Nanosecond Transient Absorption and Time-Resolved Fluorescence of Eosin B. J. Chem. Educ. 2018, 95, 864–871. [Google Scholar] [CrossRef]
- Sahoo, S.K.; Umapathy, S.; Parker, A.W. Time-Resolved Resonance Raman Spectroscopy: Exploring Reactive Intermediates. Appl. Spectrosc. 2011, 65, 1087–1115. [Google Scholar] [CrossRef]
- Fisette, O.; Lagüe, P.; Gagné, S.; Morin, S. Synergistic Applications of MD and NMR for the Study of Biological Systems. J. Biomed. Biotechnol. 2012, 2012, 1–12. [Google Scholar] [CrossRef]
- Haglund, R.F. Time and Space-Resolved Spectroscopy. In Laser Ablation and Its Applications; Springer: Boston, MA, USA, 2007; pp. 185–213. [Google Scholar]
- Stolow, A.; Bragg, A.E.; Neumark, D.M. Femtosecond Time-Resolved Photoelectron Spectroscopy. Chem. Rev. 2004, 104, 1719–1758. [Google Scholar] [CrossRef]
- Chergui, M. Editorial: Welcome to Structural Dynamics—A new open-access journal co-published by AIP Publishing and the American Crystallographic Association. Struct. Dyn. 2014, 1, 010401. [Google Scholar] [CrossRef]
- Hwan Kim, K.; Kim, J.; Hyuk Lee, J.; Ihee, H. Topical Review: Molecular reaction and solvation visualized by time-resolved X-ray solution scattering: Structure, dynamics, and their solvent dependence. Struct. Dyn. 2014, 1, 011301. [Google Scholar] [CrossRef]
- Orville, A.M. Recent results in time resolved serial femtosecond crystallography at XFELs. Curr. Opin. Struct. Biol. 2020, 65, 193–208. [Google Scholar] [CrossRef]
- Ode, H.; Nakashima, M.; Kitamura, S.; Sugiura, W.; Sato, H. Molecular dynamics simulation in virus research. Front. Microbiol. 2012, 3, 258. [Google Scholar] [CrossRef]
- García-Guevara, F.; Avelar, M.; Ayala, M.; Segovia, L. Computational Tools Applied to Enzyme Design—A review. Biocatalysis 2016, 1, 109–117. [Google Scholar] [CrossRef][Green Version]
- Hsu, C.C.; Buehler, M.J.; Tarakanova, A. The Order-Disorder Continuum: Linking Predictions of Protein Structure and Disorder through Molecular Simulation. Sci. Rep. 2020, 10, 2068. [Google Scholar] [CrossRef]
- Kovermann, M.; Rogne, P.; Wolf-Watz, M. Protein dynamics and function from solution state NMR spectroscopy. Q. Rev. Biophys. 2016, 49. [Google Scholar] [CrossRef]
- Yee, A.A.; Savchenko, A.; Ignachenko, A.; Lukin, J.; Xu, X.; Skarina, T.; Evdokimova, E.; Liu, C.S.; Semesi, A.; Guido, V.; et al. NMR and X-ray Crystallography, Complementary Tools in Structural Proteomics of Small Proteins. J. Am. Chem. Soc. 2005, 127, 16512–16517. [Google Scholar] [CrossRef]
- McNeil, B.W.J.; Thompson, N.R. X-ray free-electron lasers. Nat. Photonics 2010, 4, 814–821. [Google Scholar] [CrossRef]
- Kang, H.-S.; Min, C.-K.; Heo, H.; Kim, C.; Yang, H.; Kim, G.; Nam, I.; Baek, S.Y.; Choi, H.-J.; Mun, G.; et al. Hard X-ray free-electron laser with femtosecond-scale timing jitter. Nat. Photonics 2017, 11, 708–713. [Google Scholar] [CrossRef]
- Kim, J.; Kim, H.Y.; Park, J.; Kim, S.; Kim, S.; Rah, S.; Lim, J.; Nam, K.H. Focusing X-ray free-electron laser pulses using Kirkpatrick-Baez mirrors at the NCI hutch of the PAL-XFEL. J. Synchrotron Radiat. 2018, 25, 289–292. [Google Scholar] [CrossRef]
- Boutet, S.; Lomb, L.; Williams, G.J.; Barends, T.R.M.; Aquila, A.; Doak, R.B.; Weierstall, U.; DePonte, D.P.; Steinbrener, J.; Shoeman, R.L.; et al. High-Resolution Protein Structure Determination by Serial Femtosecond Crystallography. Science 2012, 337, 362–364. [Google Scholar] [CrossRef]
- Nam, K.H. Sample Delivery Media for Serial Crystallography. Int. J. Mol. Sci. 2019, 20, 1094. [Google Scholar] [CrossRef]
- Nam, K.H. Approach of Serial Crystallography. Crystals 2020, 10, 854. [Google Scholar] [CrossRef]
- Schmidt, M. Time-Resolved Macromolecular Crystallography at Pulsed X-ray Sources. Int. J. Mol. Sci. 2019, 20, 1401. [Google Scholar] [CrossRef]
- Diederichs, K.; Wang, M. Serial Synchrotron X-Ray Crystallography (SSX). In Protein Crystallography; Humana Press: New York, NY, USA, 2017; pp. 239–272. [Google Scholar]
- Park, S.Y.; Nam, K.H. Sample delivery using viscous media, a syringe and a syringe pump for serial crystallography. J. Synchrotron Radiat. 2019, 26, 1815–1819. [Google Scholar] [CrossRef]
- Park, S.-Y.; Choi, H.; Eo, C.; Cho, Y.; Nam, K.H. Fixed-Target Serial Synchrotron Crystallography Using Nylon Mesh and Enclosed Film-Based Sample Holder. Crystals 2020, 10, 803. [Google Scholar] [CrossRef]
- Bai, X.-C.; McMullan, G.; Scheres, S.H.W. How cryo-EM is revolutionizing structural biology. Trends Biochem. Sci. 2015, 40, 49–57. [Google Scholar] [CrossRef]
- Frank, J. Time-resolved cryo-electron microscopy: Recent progress. J. Struct. Biol. 2017, 200, 303–306. [Google Scholar] [CrossRef]
- Jones, R.O. Density functional theory: Its origins, rise to prominence, and future. Rev. Mod. Phys. 2015, 87, 897–923. [Google Scholar] [CrossRef]
- Kapil, J.; Shukla, P.; Pathak, A. Review Article on Density Functional Theory. In Recent Trends in Materials and Devices, Proceedings of the International Conference on Recent Trends in Materials and Devices (ICRTMD 2019), Noida, India, 18–19 December 2019; Springer: Singapore, 2020; pp. 211–220. [Google Scholar]
- Borhani, D.W.; Shaw, D.E. The future of molecular dynamics simulations in drug discovery. J. Comput. Aided Mol. Des. 2011, 26, 15–26. [Google Scholar] [CrossRef] [PubMed]
- Durrant, J.D.; McCammon, J.A. Molecular dynamics simulations and drug discovery. BMC Biol. 2011, 9, 71. [Google Scholar] [CrossRef] [PubMed]
- Gelpi, J.; Hospital, A.; Goñi, R.; Orozco, M. Molecular dynamics simulations: Advances and applications. Adv. Appl. Bioinform. Chem. 2015, 8, 37–47. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).