Transcriptome Analyses of Myometrium from Fibroid Patients Reveals Phenotypic Differences Compared to Non-Diseased Myometrium
Abstract
1. Introduction
2. Results
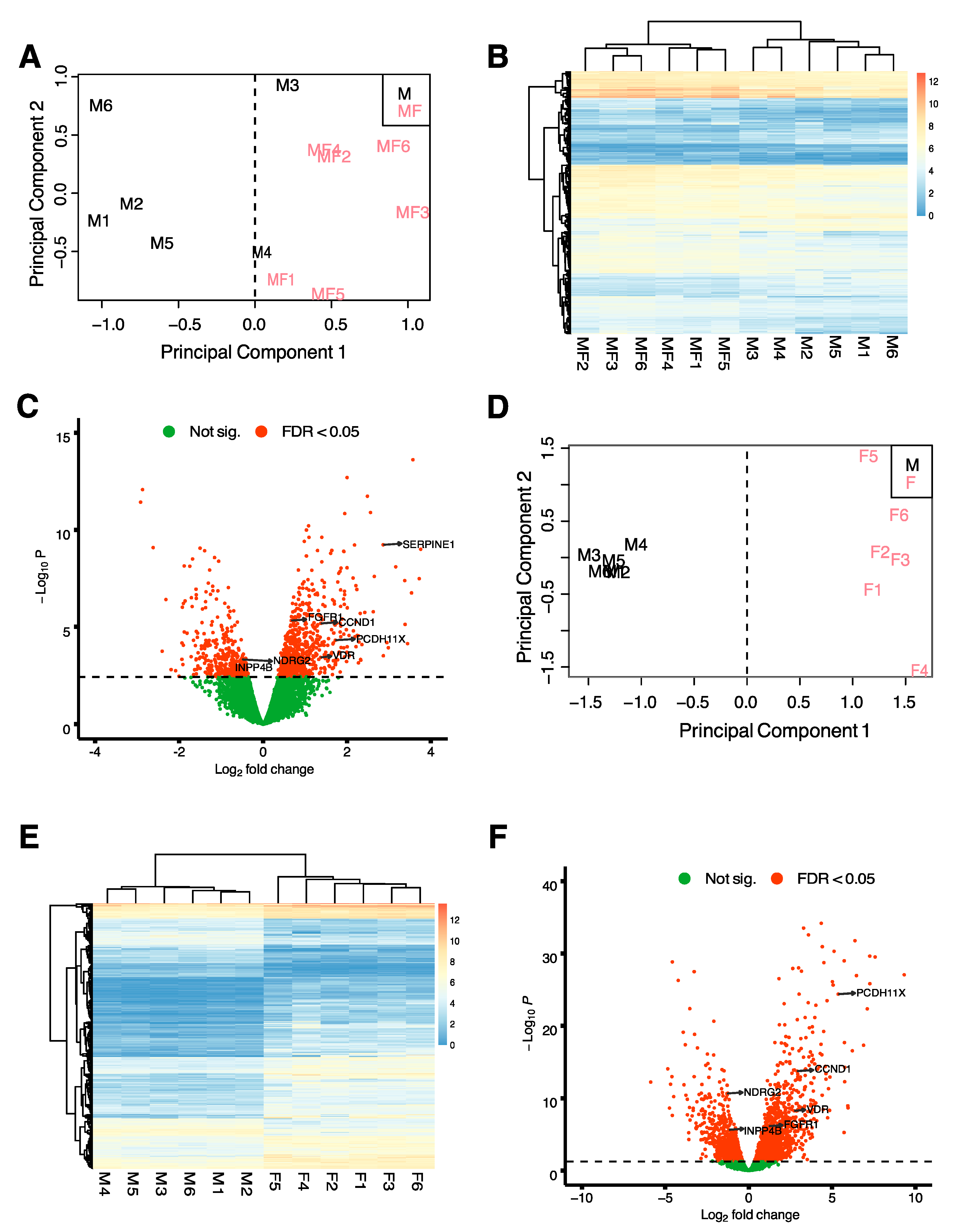
2.1. Myometrial Samples from Fibroid Patients Differentially Express Fibroid-Associated Genes
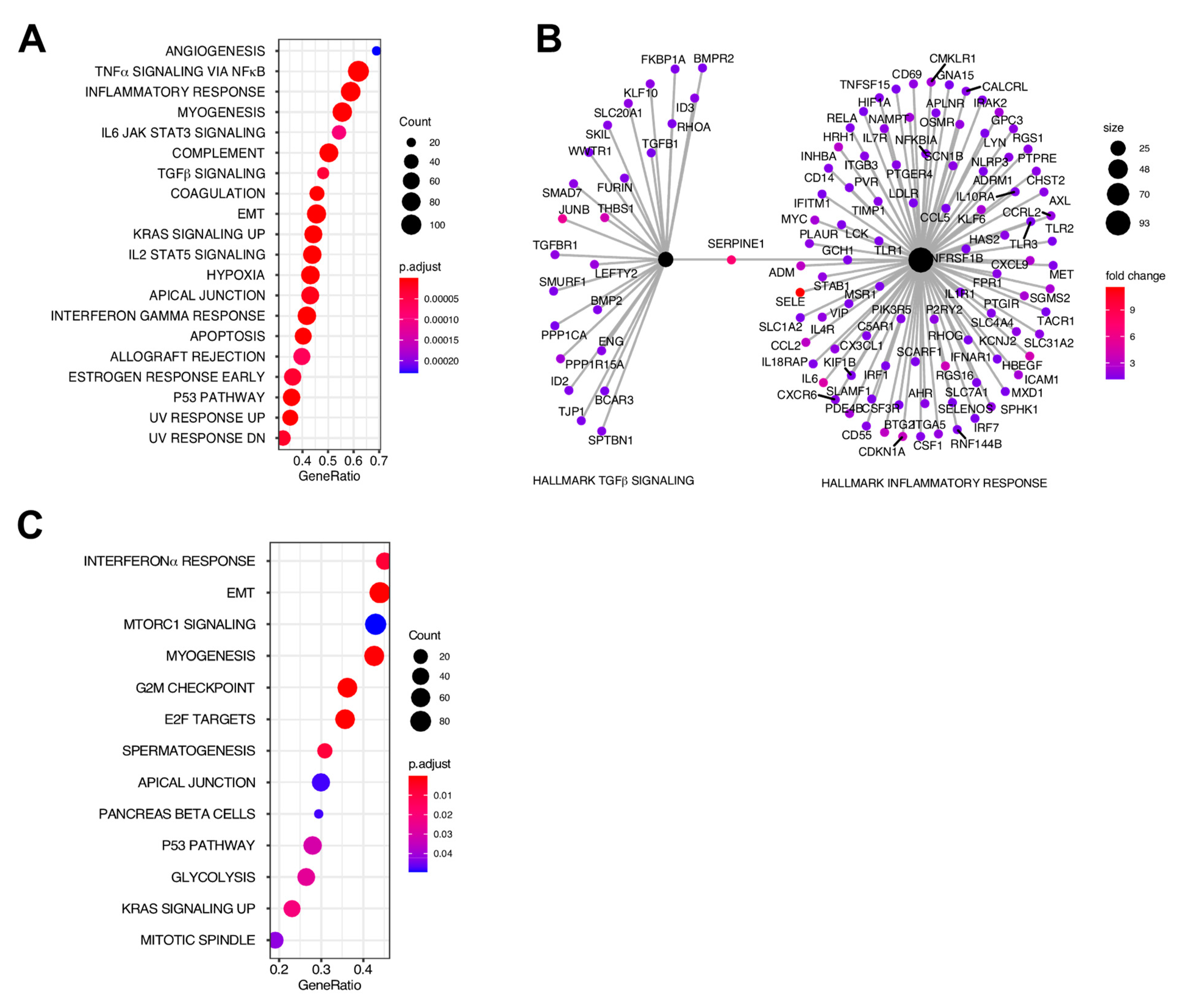
2.2. Myometrial Samples from Fibroid Patients Are Enriched for Multiple Gene Sets That May Be Involved in the Development of the Disease
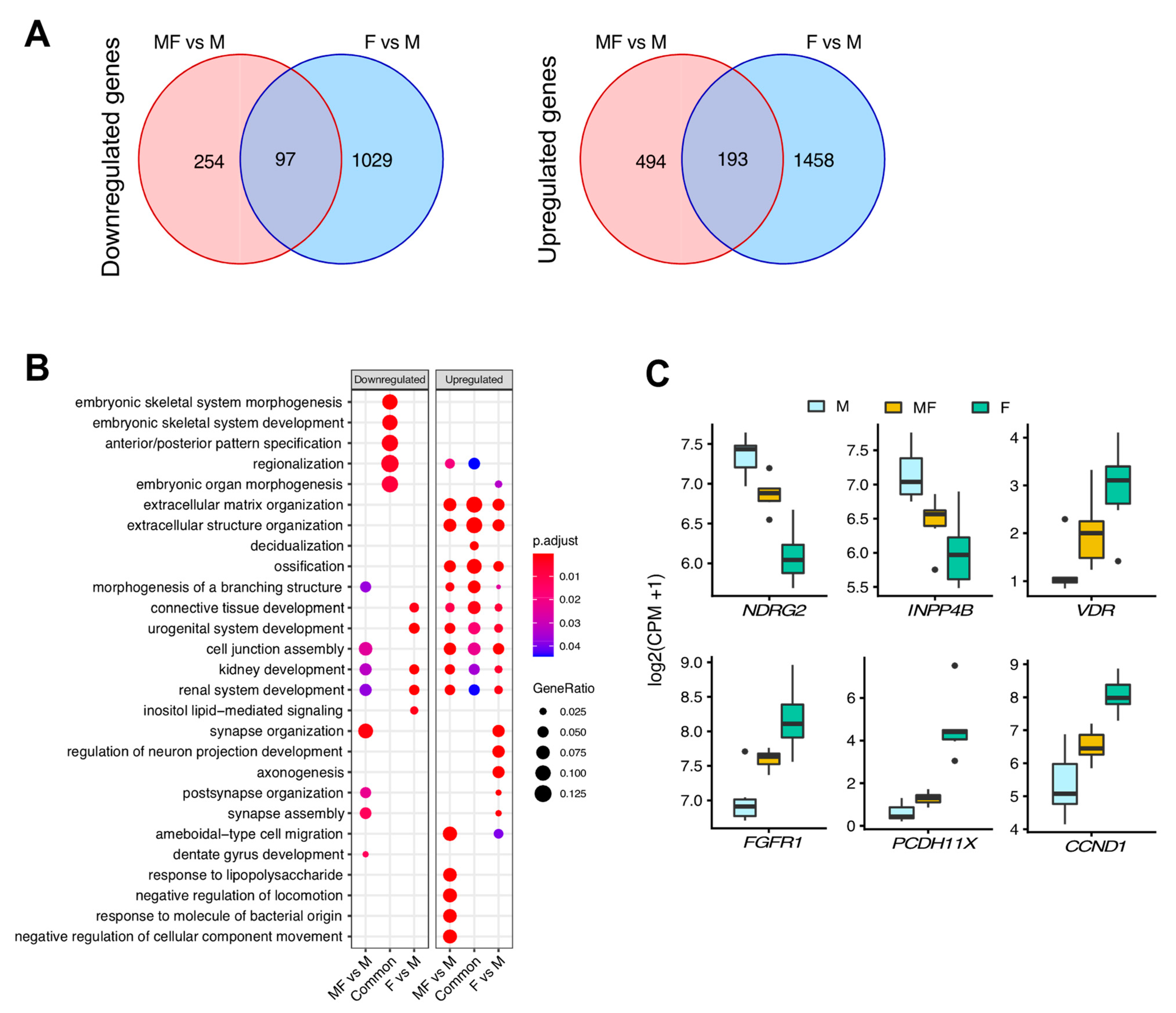
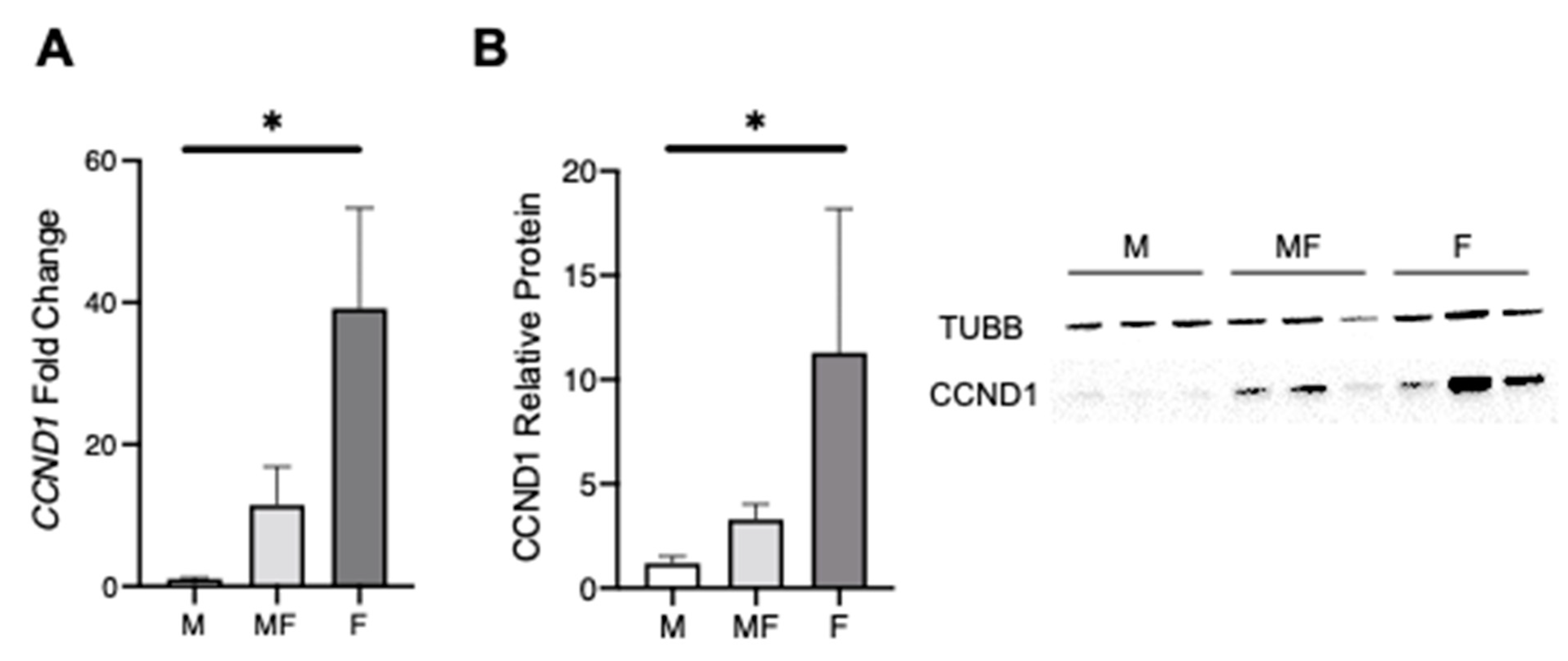
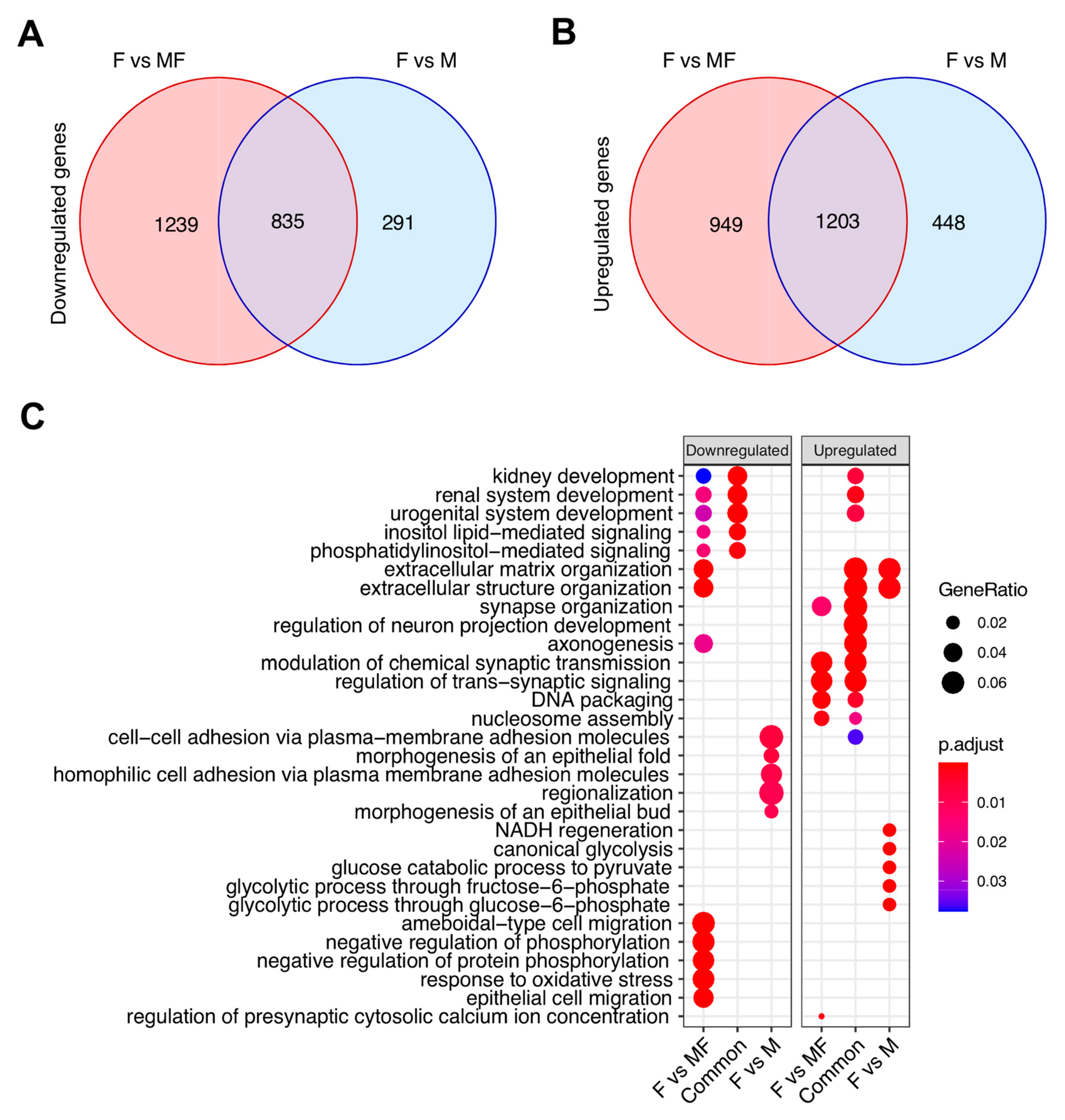
2.3. Overall Comparison of Myometrial Samples from Non-Fibroid Patients (M), Myometrial Samples from Fibroid Patients (MF), and Fibroids Samples (F)
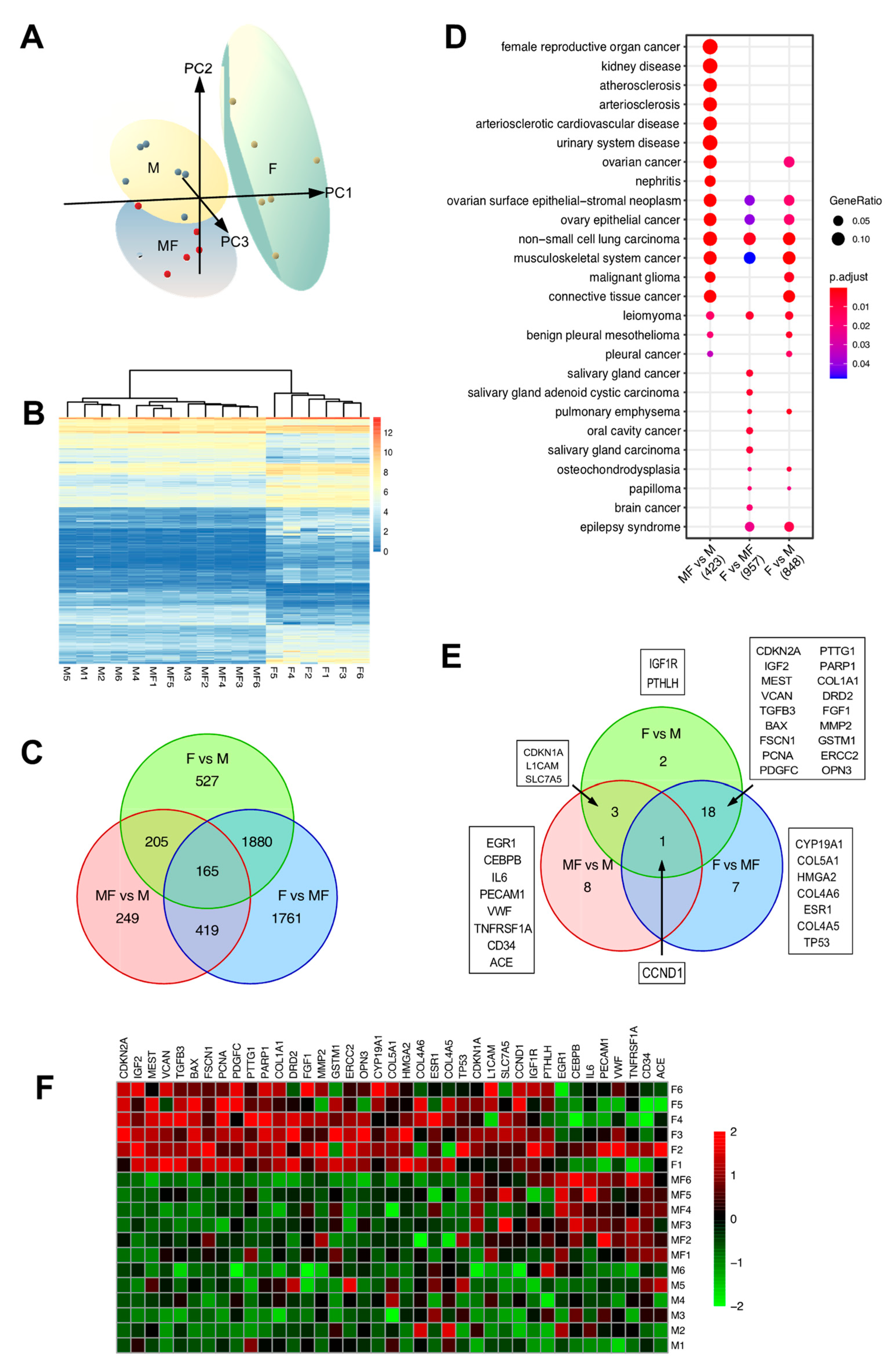
2.4. Leiomyoma Gene List Involved in Early Pathogenesis and Establish Disease
3. Discussion
4. Materials and Methods
4.1. Sample Collection
4.2. RNA Isolation
4.3. Library Preparation and Sequencing
4.4. RNA-Seq Analyses
4.5. Quantitative Real Time PCR
4.6. Western Blot
4.7. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Moravek, M.B.; Bulun, S.E. Endocrinology of uterine fibroids: Steroid hormones, stem cells, and genetic contribution. Curr. Opin. Obstet. Gynecol. 2015, 27, 276–283. [Google Scholar] [CrossRef]
- Commandeur, A.E.; Styer, A.K.; Teixeira, J.M. Epidemiological and genetic clues for molecular mechanisms involved in uterine leiomyoma development and growth. Hum. Reprod. Update 2015, 21, 593–615. [Google Scholar] [CrossRef] [PubMed]
- McWilliams, M.M.; Chennathukuzhi, V.M. Recent Advances in Uterine Fibroid Etiology. Semin. Reprod. Med. 2017, 35, 181–189. [Google Scholar] [CrossRef] [PubMed]
- De La Cruz, M.S.; Buchanan, E.M. Uterine Fibroids: Diagnosis and Treatment. Am. Fam. Physician 2017, 95, 100–107. [Google Scholar] [PubMed]
- Marsh, E.E.; Ekpo, G.E.; Cardozo, E.R.; Brocks, M.; Dune, T.; Cohen, L.S. Racial differences in fibroid prevalence and ultrasound findings in asymptomatic young women (18–30 years old): A pilot study. Fertil. Steril. 2013, 99, 1951–1957. [Google Scholar] [CrossRef]
- Sohn, G.S.; Cho, S.; Kim, Y.M.; Cho, C.H.; Kim, M.R.; Lee, S.R.; Working Group of Society of Uterine Leiomyoma. Current medical treatment of uterine fibroids. Obstet. Gynecol. Sci. 2018, 61, 192–201. [Google Scholar] [CrossRef]
- Makinen, N.; Mehine, M.; Tolvanen, J.; Kaasinen, E.; Li, Y.; Lehtonen, H.J.; Gentile, M.; Yan, J.; Enge, M.; Taipale, M.; et al. MED12, the mediator complex subunit 12 gene, is mutated at high frequency in uterine leiomyomas. Science 2011, 334, 252–255. [Google Scholar] [CrossRef]
- Markowski, D.N.; Helmke, B.M.; Bartnitzke, S.; Loning, T.; Bullerdiek, J. Uterine fibroids: Do we deal with more than one disease? Int. J. Gynecol. Pathol. 2014, 33, 568–572. [Google Scholar] [CrossRef] [PubMed]
- Mehine, M.; Makinen, N.; Heinonen, H.R.; Aaltonen, L.A.; Vahteristo, P. Genomics of uterine leiomyomas: Insights from high-throughput sequencing. Fertil. Steril. 2014, 102, 621–629. [Google Scholar] [CrossRef]
- Lee, Y.S.; Dutta, A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007, 21, 1025–1030. [Google Scholar] [CrossRef]
- George, J.W.; Fan, H.; Johnson, B.; Carpenter, T.J.; Foy, K.K.; Chatterjee, A.; Patterson, A.L.; Koeman, J.; Adams, M.; Madaj, Z.B.; et al. Integrated Epigenome, Exome, and Transcriptome Analyses Reveal Molecular Subtypes and Homeotic Transformation in Uterine Fibroids. Cell Rep. 2019, 29, 4069–4085.e4066. [Google Scholar] [CrossRef]
- Cirilo, P.D.; Marchi, F.A.; Barros Filho Mde, C.; Rocha, R.M.; Domingues, M.A.; Jurisica, I.; Pontes, A.; Rogatto, S.R. An integrative genomic and transcriptomic analysis reveals potential targets associated with cell proliferation in uterine leiomyomas. PLoS ONE 2013, 8, e57901. [Google Scholar] [CrossRef]
- Lochhead, P.; Chan, A.T.; Nishihara, R.; Fuchs, C.S.; Beck, A.H.; Giovannucci, E.; Ogino, S. Etiologic field effect: Reappraisal of the field effect concept in cancer predisposition and progression. Mod. Pathol. 2015, 28, 14–29. [Google Scholar] [CrossRef]
- Aran, D.; Camarda, R.; Odegaard, J.; Paik, H.; Oskotsky, B.; Krings, G.; Goga, A.; Sirota, M.; Butte, A.J. Comprehensive analysis of normal adjacent to tumor transcriptomes. Nat. Commun. 2017, 8, 1077. [Google Scholar] [CrossRef]
- Graham, K.; de las Morenas, A.; Tripathi, A.; King, C.; Kavanah, M.; Mendez, J.; Stone, M.; Slama, J.; Miller, M.; Antoine, G.; et al. Gene expression in histologically normal epithelium from breast cancer patients and from cancer-free prophylactic mastectomy patients shares a similar profile. Br. J. Cancer 2010, 102, 1284–1293. [Google Scholar] [CrossRef]
- Tripathi, A.; King, C.; de la Morenas, A.; Perry, V.K.; Burke, B.; Antoine, G.A.; Hirsch, E.F.; Kavanah, M.; Mendez, J.; Stone, M.; et al. Gene expression abnormalities in histologically normal breast epithelium of breast cancer patients. Int. J. Cancer 2008, 122, 1557–1566. [Google Scholar] [CrossRef]
- Lee, M.; Cheon, K.; Chae, B.; Hwang, H.; Kim, H.K.; Chung, Y.J.; Song, J.Y.; Cho, H.H.; Kim, J.H.; Kim, M.R. Analysis of MED12 Mutation in Multiple Uterine Leiomyomas in South Korean patients. Int. J. Med. Sci. 2018, 15, 124–128. [Google Scholar] [CrossRef] [PubMed]
- Wolanska, M.; Bankowski, E. Fibroblast growth factors (FGF) in human myometrium and uterine leiomyomas in various stages of tumour growth. Biochimie 2006, 88, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Musgrove, E.A.; Caldon, C.E.; Barraclough, J.; Stone, A.; Sutherland, R.L. Cyclin D as a therapeutic target in cancer. Nat. Rev. Cancer 2011, 11, 558–572. [Google Scholar] [CrossRef] [PubMed]
- Maekawa, R.; Yagi, S.; Ohgane, J.; Yamagata, Y.; Asada, H.; Tamura, I.; Sugino, N.; Shiota, K. Disease-dependent differently methylated regions (D-DMRs) of DNA are enriched on the X chromosome in uterine leiomyoma. J. Reprod. Dev. 2011, 57, 604–612. [Google Scholar] [CrossRef] [PubMed]
- Feng, L.; Jayes, F.L.; Jung, S.H.; Leppert, P.C. Vitamin D receptor (VDR) is over-expressed in the center of uterine fibroids. Fertil. Steril. 2010, 94, S75. [Google Scholar] [CrossRef]
- Hu, W.; Fan, C.; Jiang, P.; Ma, Z.; Yan, X.; Di, S.; Jiang, S.; Li, T.; Cheng, Y.; Yang, Y. Emerging role of N-myc downstream-regulated gene 2 (NDRG2) in cancer. Oncotarget 2016, 7, 209–223. [Google Scholar] [CrossRef] [PubMed]
- Gewinner, C.; Wang, Z.C.; Richardson, A.; Teruya-Feldstein, J.; Etemadmoghadam, D.; Bowtell, D.; Barretina, J.; Lin, W.M.; Rameh, L.; Salmena, L.; et al. Evidence that inositol polyphosphate 4-phosphatase type II is a tumor suppressor that inhibits PI3K signaling. Cancer Cell 2009, 16, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Liberzon, A.; Birger, C.; Thorvaldsdottir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.G.; Yan, G.R.; He, Q.Y. DOSE: An R/Bioconductor package for disease ontology semantic and enrichment analysis. Bioinformatics 2015, 31, 608–609. [Google Scholar] [CrossRef] [PubMed]
- Tinelli, A.; Kosmas, I.P.; Mynbaev, O.A.; Malvasi, A.; Sparic, R.; Vergara, D. The Biological Impact of Ulipristal Acetate on Cellular Networks Regulating Uterine Leiomyoma Growth. Curr. Pharm. Des. 2020, 26, 310–317. [Google Scholar] [CrossRef]
- Ono, M.; Yin, P.; Navarro, A.; Moravek, M.B.; Coon, J.S.T.; Druschitz, S.A.; Serna, V.A.; Qiang, W.; Brooks, D.C.; Malpani, S.S.; et al. Paracrine activation of WNT/beta-catenin pathway in uterine leiomyoma stem cells promotes tumor growth. Proc. Natl. Acad. Sci. USA 2013, 110, 17053–17058. [Google Scholar] [CrossRef]
- Ko, Y.A.; Jamaluddin, M.F.B.; Adebayo, M.; Bajwa, P.; Scott, R.J.; Dharmarajan, A.M.; Nahar, P.; Tanwar, P.S. Extracellular matrix (ECM) activates beta-catenin signaling in uterine fibroids. Reproduction 2018, 155, 61–71. [Google Scholar] [CrossRef]
- Yu, L.; Saile, K.; Swartz, C.D.; He, H.; Zheng, X.; Kissling, G.E.; Di, X.; Lucas, S.; Robboy, S.J.; Dixon, D. Differential expression of receptor tyrosine kinases (RTKs) and IGF-I pathway activation in human uterine leiomyomas. Mol. Med. 2008, 14, 264–275. [Google Scholar] [CrossRef]
- Acevedo, V.D.; Ittmann, M.; Spencer, D.M. Paths of FGFR-driven tumorigenesis. Cell Cycle 2009, 8, 580–588. [Google Scholar] [CrossRef]
- Tashiro, E.; Maruki, H.; Minato, Y.; Doki, Y.; Weinstein, I.B.; Imoto, M. Overexpression of cyclin D1 contributes to malignancy by up-regulation of fibroblast growth factor receptor 1 via the pRB/E2F pathway. Cancer Res. 2003, 63, 424–431. [Google Scholar]
- Fu, M.; Wang, C.; Li, Z.; Sakamaki, T.; Pestell, R.G. Minireview: Cyclin D1: Normal and abnormal functions. Endocrinology 2004, 145, 5439–5447. [Google Scholar] [CrossRef] [PubMed]
- Kovacs, K.A.; Oszter, A.; Gocze, P.M.; Kornyei, J.L.; Szabo, I. Comparative analysis of cyclin D1 and oestrogen receptor (alpha and beta) levels in human leiomyoma and adjacent myometrium. Mol. Hum. Reprod. 2001, 7, 1085–1091. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Wang, Y.; Liu, J.; Chen, Q.; Pang, D.; Jiang, Y. Effects of FGFR1 Gene Polymorphisms on the Risk of Breast Cancer and FGFR1 Protein Expression. Cell. Physiol. Biochem. 2018, 47, 2569–2578. [Google Scholar] [CrossRef]
- Tanwar, P.S.; Lee, H.J.; Zhang, L.; Zukerberg, L.R.; Taketo, M.M.; Rueda, B.R.; Teixeira, J.M. Constitutive activation of Beta-catenin in uterine stroma and smooth muscle leads to the development of mesenchymal tumors in mice. Biol. Reprod. 2009, 81, 545–552. [Google Scholar] [CrossRef]
- Zhang, D.; Liu, E.; Tian, W.; Zhang, Z.; Wang, L.; Li, J. MiR-93 blocks cell cycle progression and promotes apoptosis in uterine leiomyoma cells by targeting CCND1. Anat. Rec. 2020, 303, 2372–2381. [Google Scholar] [CrossRef]
- Xu, F.; Li, F.; Li, L.; Lin, D.; Hu, H.; Shi, Q. Vitamin D as a risk factor for the presence of asymptomatic uterine fibroids in premenopausal Han Chinese women. Fertil. Steril. 2021. [Google Scholar] [CrossRef]
- Ciebiera, M.; Ali, M.; Zgliczynska, M.; Skrzypczak, M.; Al-Hendy, A. Vitamins and Uterine Fibroids: Current Data on Pathophysiology and Possible Clinical Relevance. Int. J. Mol. Sci. 2020, 21, 5528. [Google Scholar] [CrossRef]
- Ali, M.; Shahin, S.M.; Sabri, N.A.; Al-Hendy, A.; Yang, Q. 1,25 Dihydroxyvitamin D3 Enhances the Antifibroid Effects of Ulipristal Acetate in Human Uterine Fibroids. Reprod. Sci. 2019, 26, 812–828. [Google Scholar] [CrossRef]
- Halder, S.; Al-Hendy, A. Hypovitaminosis D and high serum transforming growth factor beta-3: Important biomarkers for uterine fibroids risk. Fertil. Steril. 2016, 106, 1648–1649. [Google Scholar] [CrossRef] [PubMed]
- Halder, S.K.; Osteen, K.G.; Al-Hendy, A. 1,25-dihydroxyvitamin d3 reduces extracellular matrix-associated protein expression in human uterine fibroid cells. Biol. Reprod. 2013, 89, 150. [Google Scholar] [CrossRef] [PubMed]
- Vergara, D.; Catherino, W.H.; Trojano, G.; Tinelli, A. Vitamin D: Mechanism of Action and Biological Effects in Uterine Fibroids. Nutrients 2021, 13, 597. [Google Scholar] [CrossRef] [PubMed]
- Al-Hendy, A.; Lukes, A.S.; Poindexter, A.N., 3rd; Venturella, R.; Villarroel, C.; Critchley, H.O.D.; Li, Y.; McKain, L.; Arjona Ferreira, J.C.; Langenberg, A.G.M.; et al. Treatment of Uterine Fibroid Symptoms with Relugolix Combination Therapy. N. Engl. J. Med. 2021, 384, 630–642. [Google Scholar] [CrossRef]
- Lee, B.S.; Nowak, R.A. Human leiomyoma smooth muscle cells show increased expression of transforming growth factor-beta 3 (TGF beta 3) and altered responses to the antiproliferative effects of TGF beta. J. Clin. Endocrinol. Metab. 2001, 86, 913–920. [Google Scholar] [CrossRef]
- Islam, M.S.; Akhtar, M.M.; Ciavattini, A.; Giannubilo, S.R.; Protic, O.; Janjusevic, M.; Procopio, A.D.; Segars, J.H.; Castellucci, M.; Ciarmela, P. Use of dietary phytochemicals to target inflammation, fibrosis, proliferation, and angiogenesis in uterine tissues: Promising options for prevention and treatment of uterine fibroids? Mol. Nutr. Food Res. 2014, 58, 1667–1684. [Google Scholar] [CrossRef]
- Hoffman, P.J.; Milliken, D.B.; Gregg, L.C.; Davis, R.R.; Gregg, J.P. Molecular characterization of uterine fibroids and its implication for underlying mechanisms of pathogenesis. Fertil. Steril. 2004, 82, 639–649. [Google Scholar] [CrossRef]
- Zhang, Q.; Lei, L.; Jing, D. Knockdown of SERPINE1 reverses resistance of triplenegative breast cancer to paclitaxel via suppression of VEGFA. Oncol. Rep. 2020, 44, 1875–1884. [Google Scholar] [CrossRef]
- Greenman, C.; Stephens, P.; Smith, R.; Dalgliesh, G.L.; Hunter, C.; Bignell, G.; Davies, H.; Teague, J.; Butler, A.; Stevens, C.; et al. Patterns of somatic mutation in human cancer genomes. Nature 2007, 446, 153–158. [Google Scholar] [CrossRef]
- Sigurgeirsson, B.; Amark, H.; Jemt, A.; Ujvari, D.; Westgren, M.; Lundeberg, J.; Gidlof, S. Comprehensive RNA sequencing of healthy human endometrium at two time points of the menstrual cycle. Biol. Reprod. 2017, 96, 24–33. [Google Scholar] [CrossRef] [PubMed]
- Heiner, J.S.; Cai, L.; Ding, H.; Rutgers, J.K. Myometrial expression of mRNA encoding epidermal growth factor receptor (EGFR) throughout the menstrual cycle. Am. J. Reprod. Immunol. 1994, 32, 152–156. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10. [Google Scholar] [CrossRef]
- Zhou, X.; Lindsay, H.; Robinson, M.D. Robustly detecting differential expression in RNA sequencing data using observation weights. Nucleic Acids Res. 2014, 42, e91. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Mootha, V.K.; Lindgren, C.M.; Eriksson, K.F.; Subramanian, A.; Sihag, S.; Lehar, J.; Puigserver, P.; Carlsson, E.; Ridderstrale, M.; Laurila, E.; et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003, 34, 267–273. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paul, E.N.; Burns, G.W.; Carpenter, T.J.; Grey, J.A.; Fazleabas, A.T.; Teixeira, J.M. Transcriptome Analyses of Myometrium from Fibroid Patients Reveals Phenotypic Differences Compared to Non-Diseased Myometrium. Int. J. Mol. Sci. 2021, 22, 3618. https://doi.org/10.3390/ijms22073618
Paul EN, Burns GW, Carpenter TJ, Grey JA, Fazleabas AT, Teixeira JM. Transcriptome Analyses of Myometrium from Fibroid Patients Reveals Phenotypic Differences Compared to Non-Diseased Myometrium. International Journal of Molecular Sciences. 2021; 22(7):3618. https://doi.org/10.3390/ijms22073618
Chicago/Turabian StylePaul, Emmanuel N., Gregory W. Burns, Tyler J. Carpenter, Joshua A. Grey, Asgerally T. Fazleabas, and Jose M. Teixeira. 2021. "Transcriptome Analyses of Myometrium from Fibroid Patients Reveals Phenotypic Differences Compared to Non-Diseased Myometrium" International Journal of Molecular Sciences 22, no. 7: 3618. https://doi.org/10.3390/ijms22073618
APA StylePaul, E. N., Burns, G. W., Carpenter, T. J., Grey, J. A., Fazleabas, A. T., & Teixeira, J. M. (2021). Transcriptome Analyses of Myometrium from Fibroid Patients Reveals Phenotypic Differences Compared to Non-Diseased Myometrium. International Journal of Molecular Sciences, 22(7), 3618. https://doi.org/10.3390/ijms22073618





