The Mitogen-Activated Protein Kinase PlMAPK2 Is Involved in Zoosporogenesis and Pathogenicity of Peronophythoralitchii
Abstract
1. Introduction
2. Results
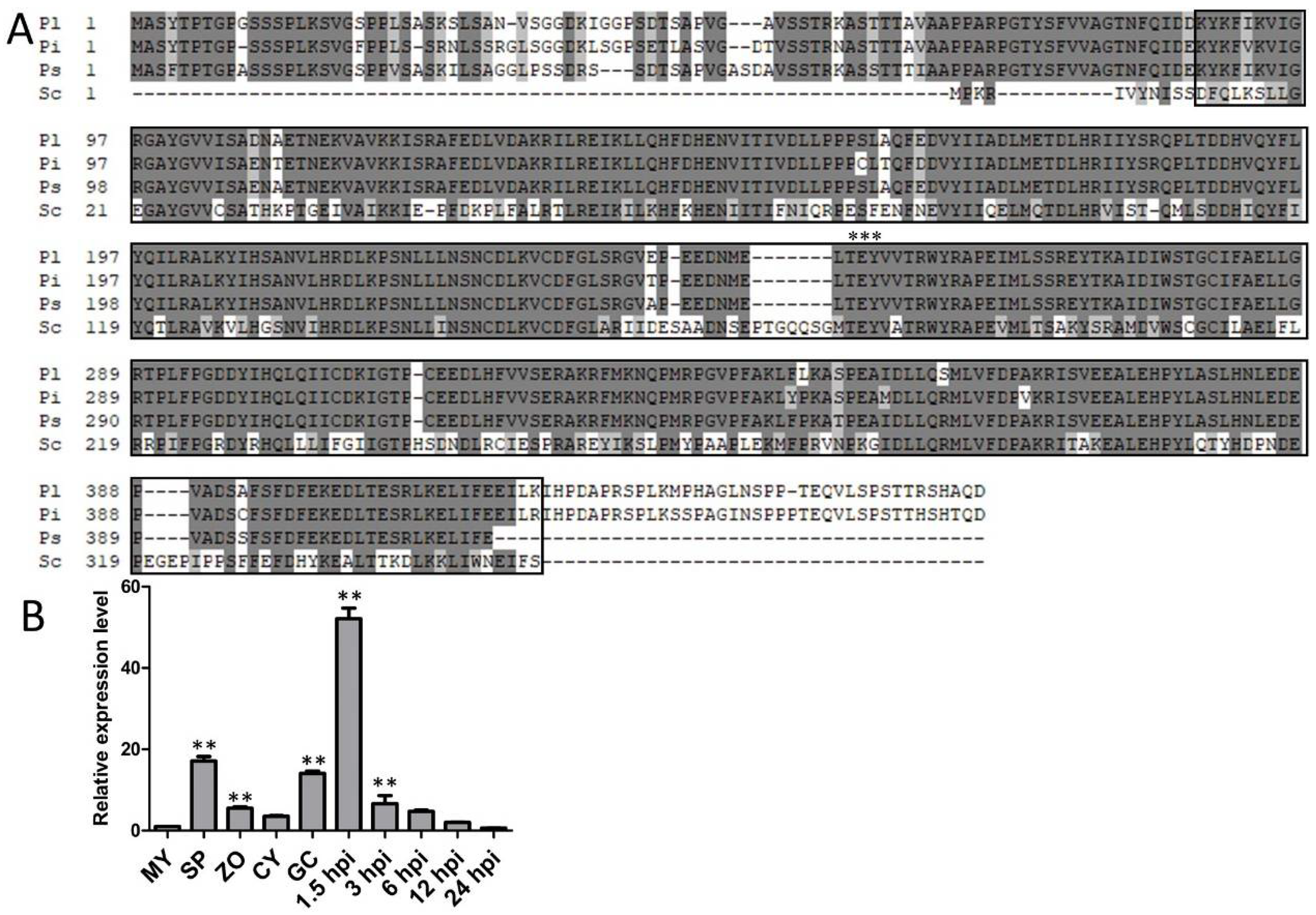
2.1. PlMAPK2 Is Conserved in P. litchii and Phytophthora Species and Up-Regulated in Asexual Development and Early Stage of Infection
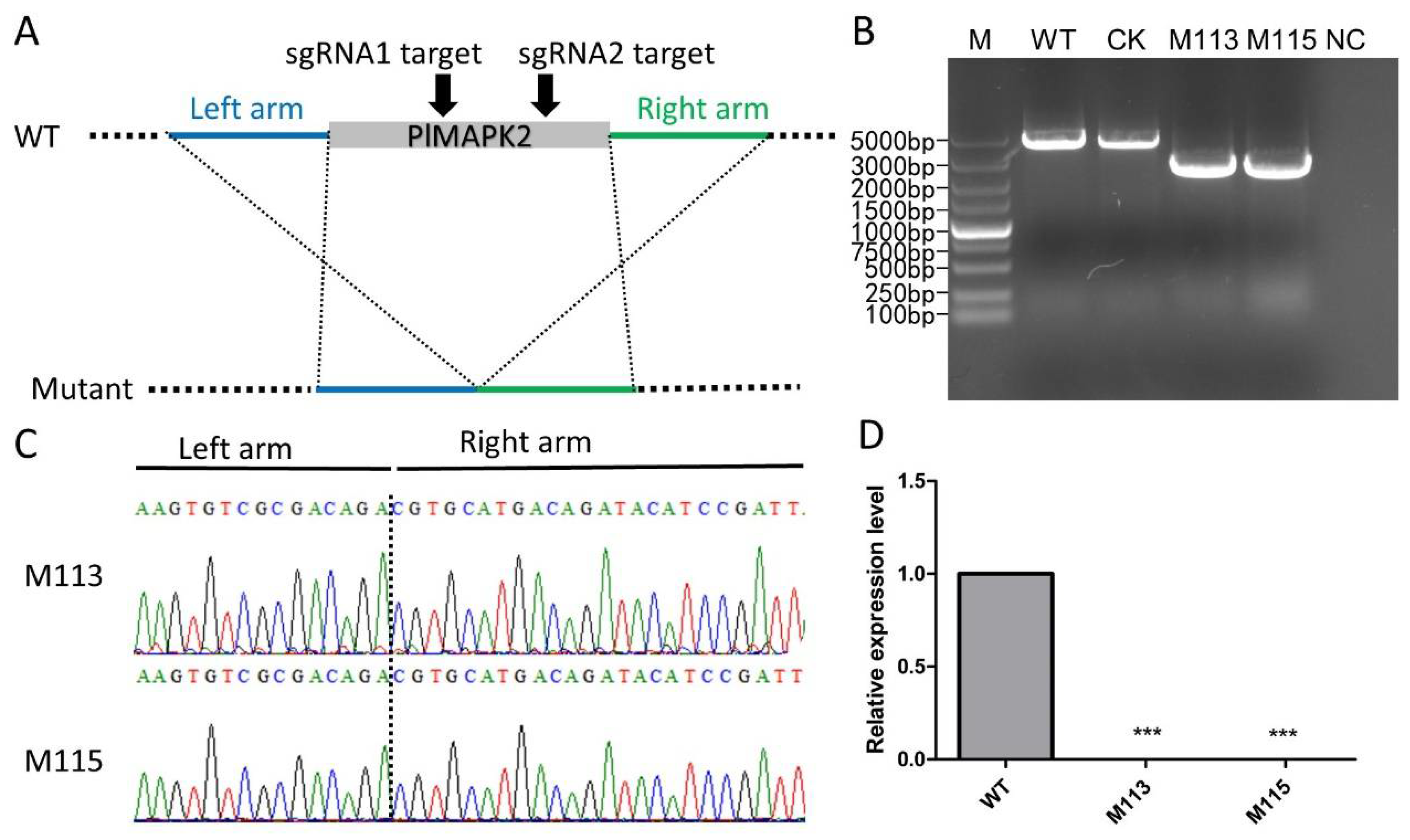
2.2. Generation of PlMAPK2 Knockout Mutants Using CRISPR/Cas9 Genome Editing Technology
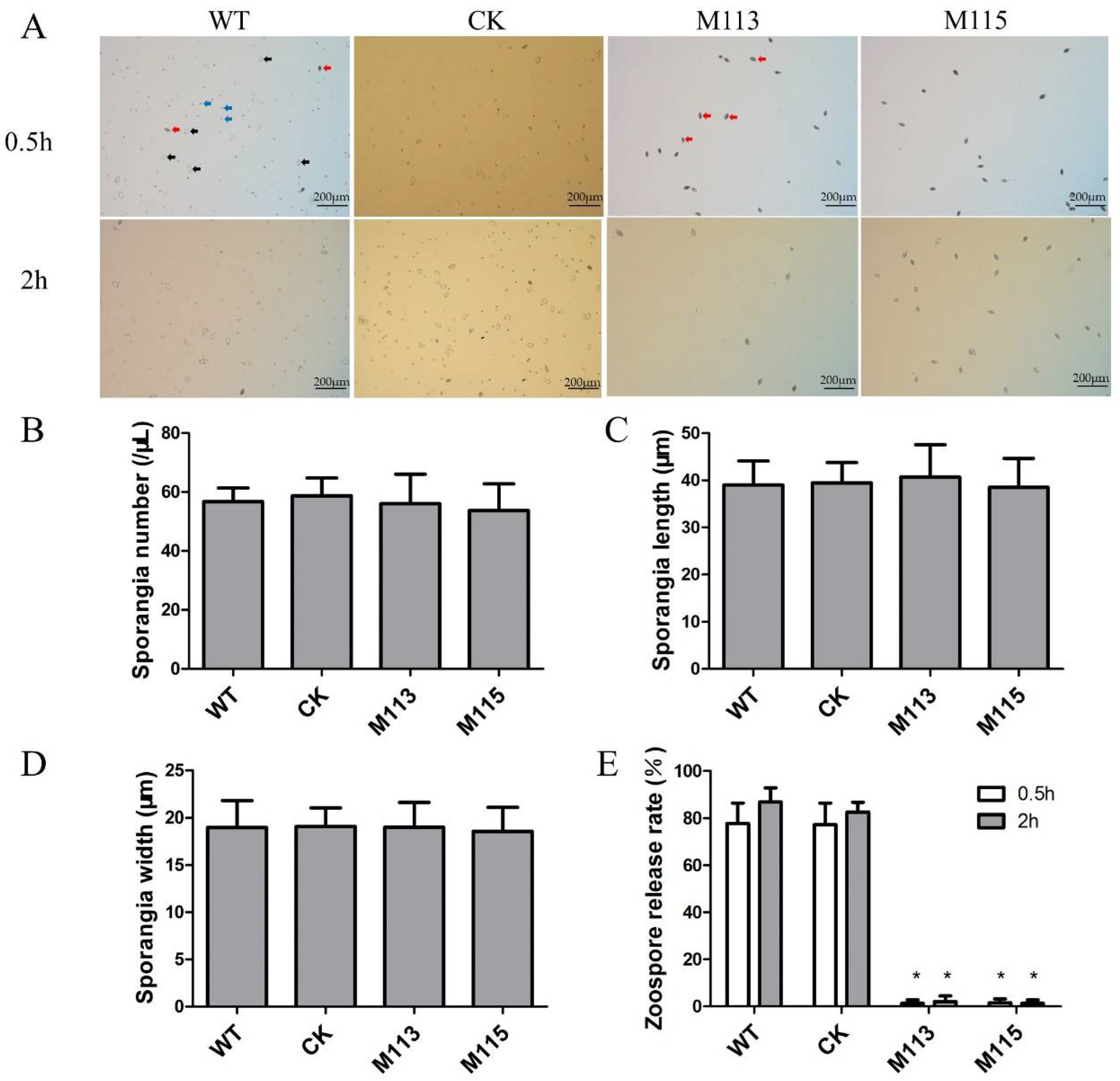
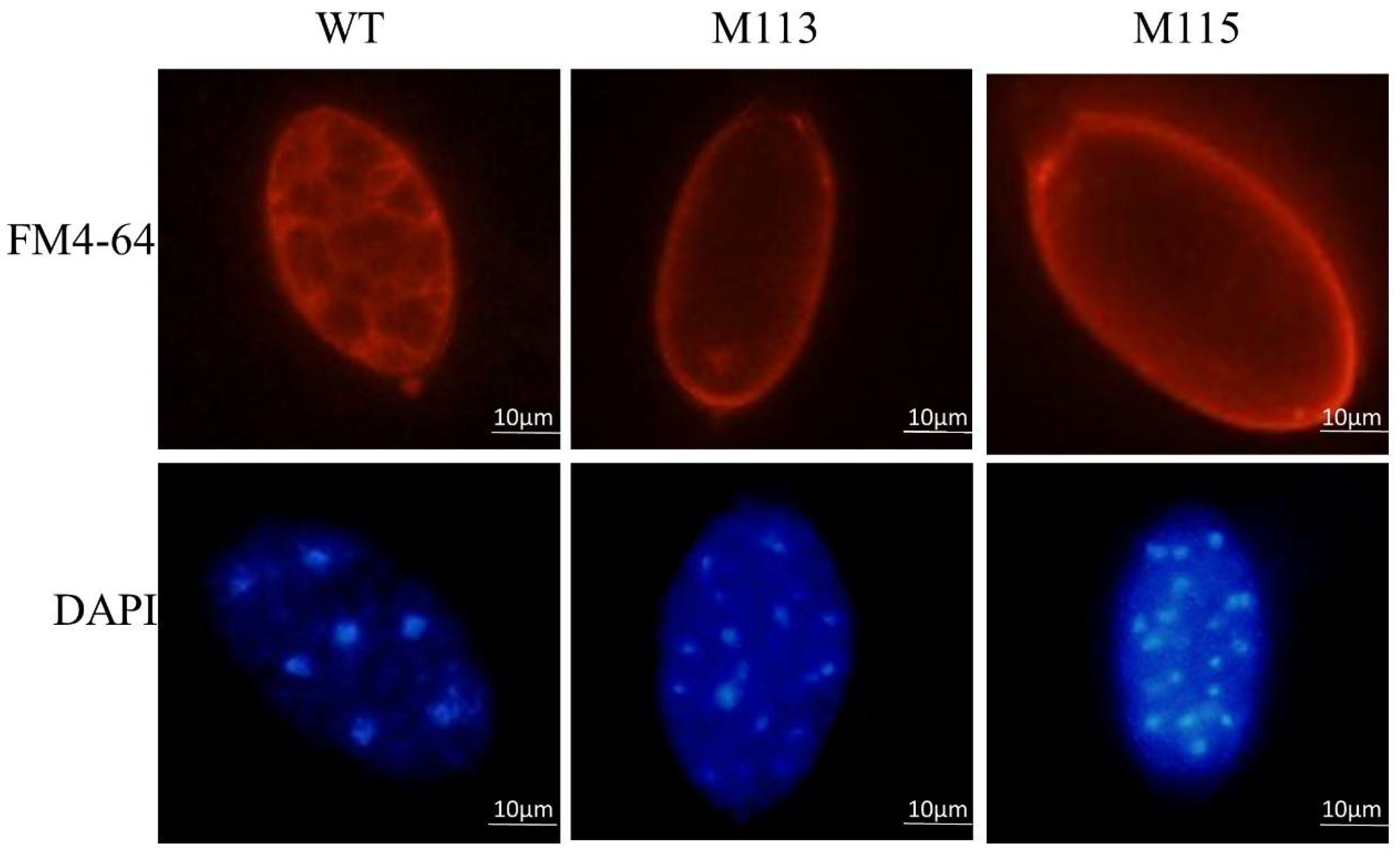
2.3. Knockout of PlMAPK2 Affects Zoosporogenesis of P. litchii
2.4. PlMAPK2 Is Required for Sporangial Cleavage during Zoospore Development
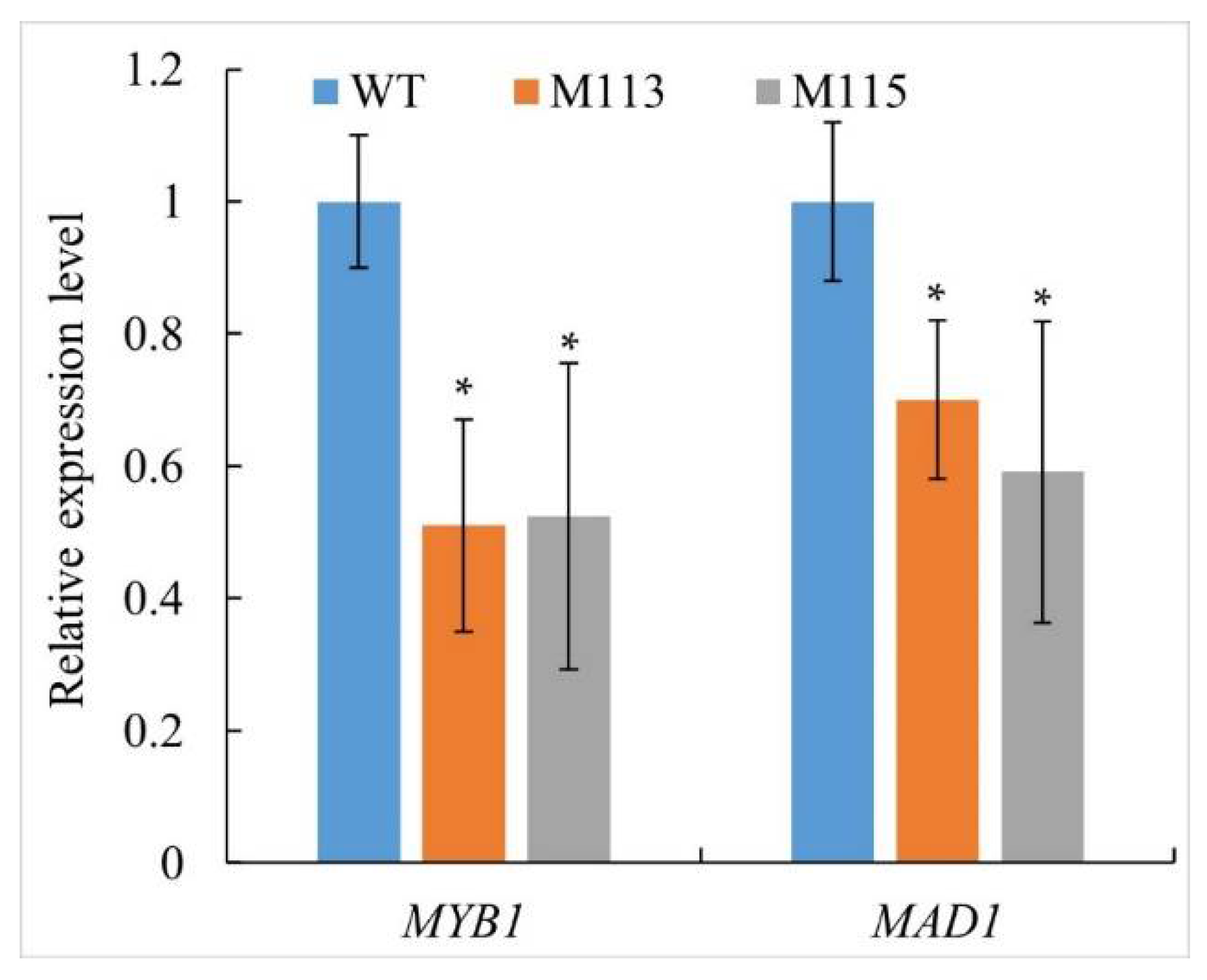
2.5. PlMAD1 and PlMYB1 Were Down-Regulated in PlMAPK2 Mutants
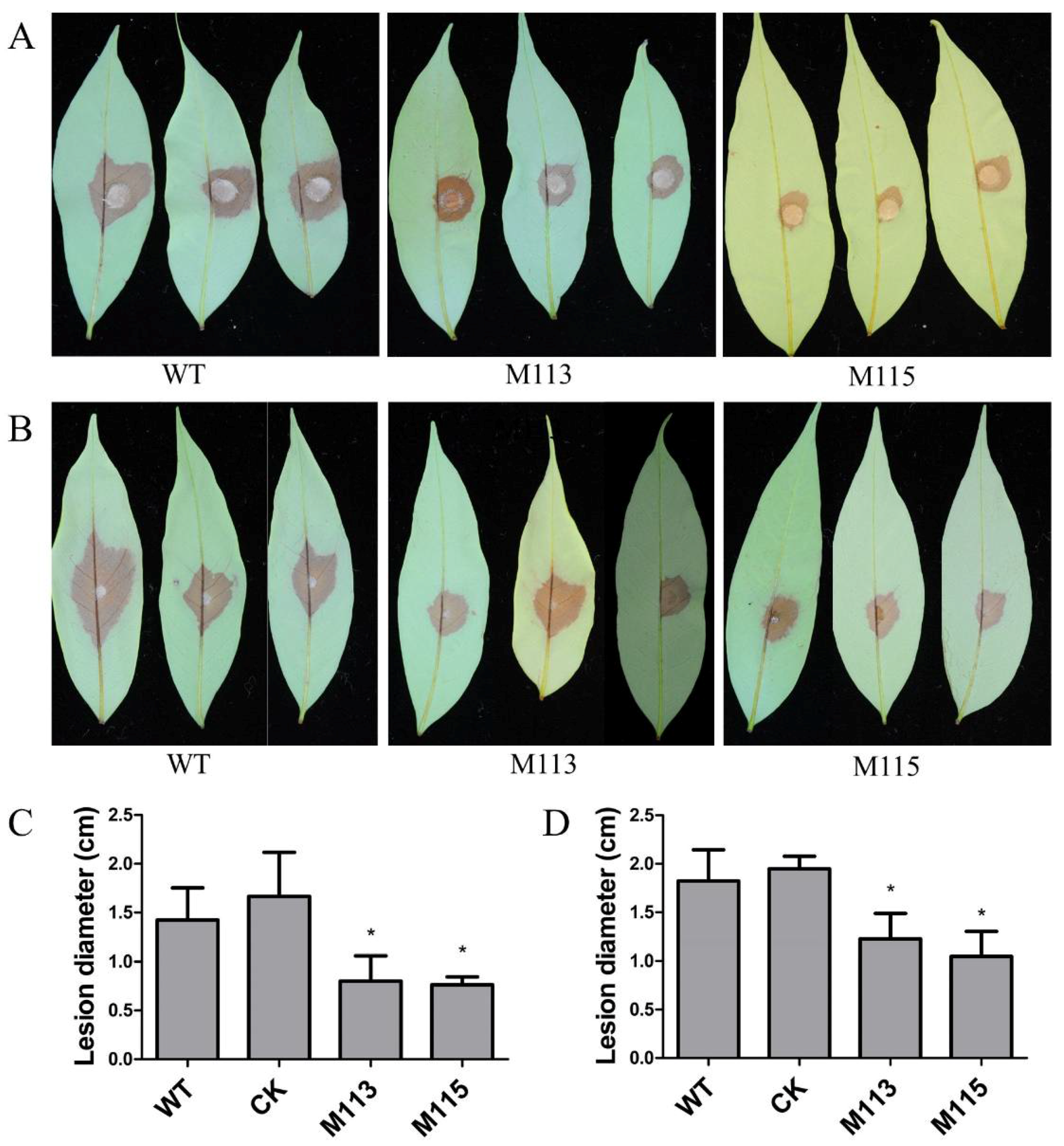
2.6. PlMAPK2 Is Required for Full Virulence and Regulates Laccase Activity in P. litchii
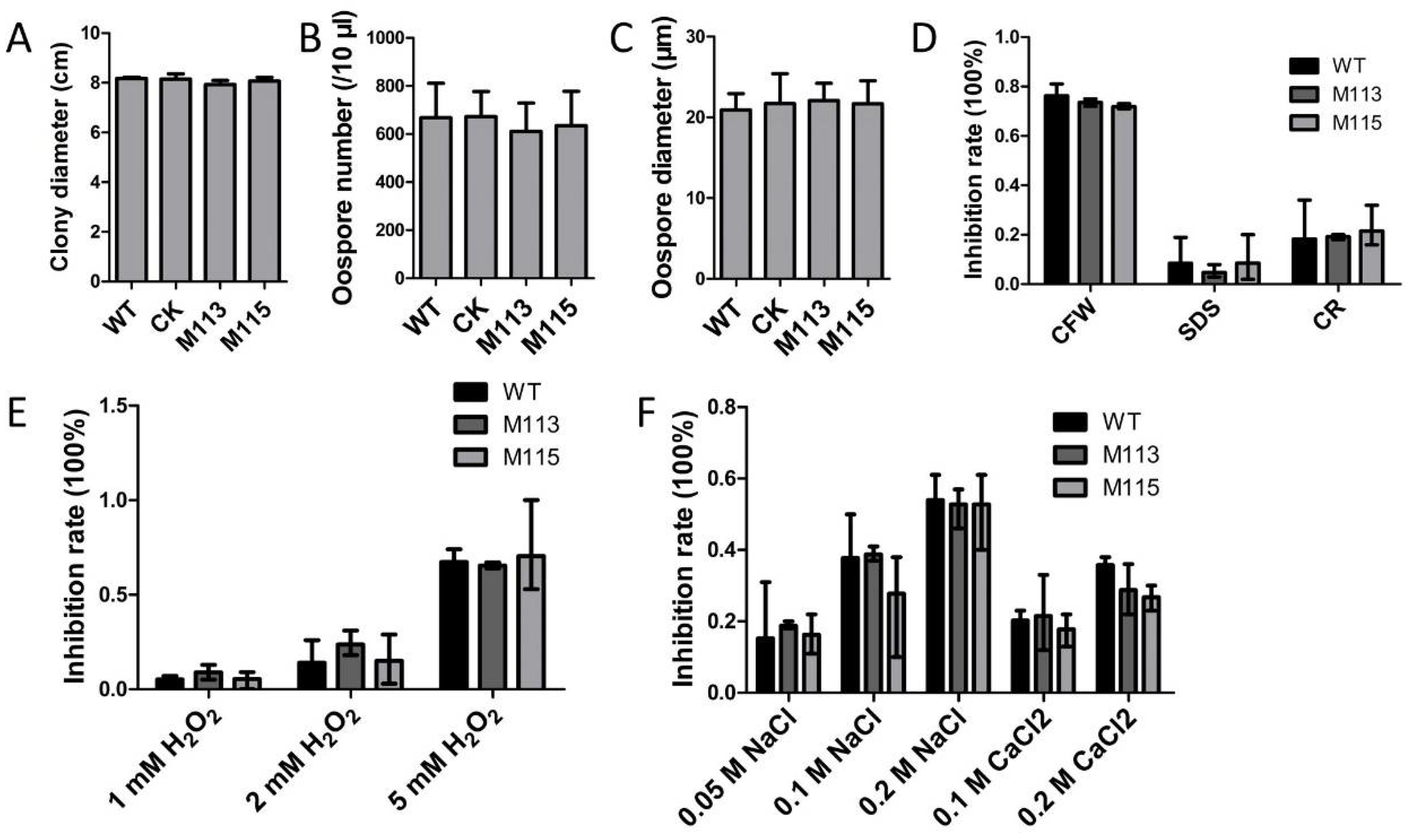
2.7. Assessment of Growth and Stress Response in the PlMAPK2 Mutants
3. Discussion
4. Materials and Methods
4.1. Sequences and Phylogenetic Analysis
4.2. Strains and Culture Conditions
4.3. Transcriptional Analysis
4.4. CRISPR/Cas9 Editing for PlMAPK2
4.5. Analysis of Sporangium, Zoospore and Oospore Development
4.6. Chemical Dyes with FM4-64 and DAPI (4’,6-Diamidino-2-Phenylindole, Dilactate)
4.7. Pathogenicity Test
4.8. Laccase Activity Assays
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cavalier-Smith, T.; Chao, E.E.; Lewis, R. Multigene phylogeny and cell evolution of chromist infrakingdom Rhizaria: Contrasting cell organisation of sister phyla Cercozoa and Retaria. Protoplasma 2018, 255, 1517–1574. [Google Scholar] [CrossRef]
- Tyler, B.M.; Tripathy, S.; Zhang, X.; Dehal, P.; Jiang, R.H.; Aerts, A.; Arredondo, F.D.; Baxter, L.; Bensasson, D.; Beynon, J.L.; et al. Phytophthora genome sequences uncover evolutionary origins and mechanisms of Pathogenesis. Science 2006, 313, 1261–1266. [Google Scholar] [CrossRef] [PubMed]
- Kamoun, S.; Furzer, O.; Jones, J.D.G.; Judelson, H.S.; Ali, G.S.; Dalio, R.J.D.; Roy, S.G.; Schena, L.; Zambounis, A.; Panabières, F.; et al. The Top 10 oomycete pathogens in molecular plant pathology. Mol. Plant Pathol. 2015, 16, 413–434. [Google Scholar] [CrossRef] [PubMed]
- Kong, G.; Li, T.; Huang, W.; Li, M.; Shen, W.; Jiang, L.; Hsiang, T.; Jiang, Z.; Xi, P. Detection of Peronophythora litchii on lychee by loop-mediated isothermal amplification assay. Crop. Prot. 2021, 139, 105370. [Google Scholar] [CrossRef]
- Kong, G.; Chen, Y.; Deng, Y.; Feng, D.; Jiang, L.; Wan, L.; Li, M.; Jiang, Z.; Xi, P. The Basic Leucine Zipper Transcription Factor PlBZP32 Associated with the Oxidative Stress Response Is Critical for Pathogenicity of the Lychee Downy Blight Oomycete Peronophythora litchii. mSphere 2020, 5, e00261–e00320. [Google Scholar] [CrossRef]
- Hamel, L.-P.; Nicole, M.-C.; Duplessis, S.; Ellis, B.E. Mitogen-Activated Protein Kinase Signaling in Plant-Interacting Fungi: Distinct Messages from Conserved Messengers. Plant Cell 2012, 24, 1327–1351. [Google Scholar] [CrossRef]
- Widmann, C.; Gibson, S.; Jarpe, M.B.; Johnson, G.L. Mitogen-Activated Protein Kinase: Conservation of a Three-Kinase Module from Yeast to Human. Physiol. Rev. 1999, 79, 143–180. [Google Scholar] [CrossRef]
- Jiang, C.; Zhang, X.; Liu, H.; Xu, J.-R. Mitogen-activated protein kinase signaling in plant pathogenic fungi. PLoS Pathog. 2018, 14, e1006875. [Google Scholar] [CrossRef] [PubMed]
- González-Rubio, G.; Fernández-Acero, T.; Martín, H.; Molina, M. Mitogen-Activated Protein Kinase Phosphatases (MKPs) in Fungal Signaling: Conservation, Function, and Regulation. Int. J. Mol. Sci. 2019, 20, 1709. [Google Scholar] [CrossRef]
- Jiang, L.; Situ, J.; Deng, Y.Z.; Wan, L.; Xu, D.; Chen, Y.; Xi, P.; Jiang, Z. PlMAPK10, a Mitogen-Activated Protein Kinase (MAPK) in Peronophythora litchii, Is Required for Mycelial Growth, Sporulation, Laccase Activity, and Plant Infection. Front. Microbiol. 2018, 9, 426. [Google Scholar] [CrossRef]
- Li, A.; Zhang, M.; Wang, Y.; Li, D.; Liu, X.; Tao, K.; Ye, W.; Wang, Y. PsMPK1, an SLT2-type mitogen-activated protein kinase, is required for hyphal growth, zoosporogenesis, cell wall integrity, and pathogenicity in Phytophthora sojae. Fungal Genet. Biol. 2014, 65, 14–24. [Google Scholar] [CrossRef]
- Li, A.; Wang, Y.; Tao, K.; Dong, S.; Huang, Q.; Dai, T.; Zheng, X.; Wang, Y. PsSAK1, a Stress-Activated MAP Kinase of Phytophthora sojae, Is Required for Zoospore Viability and Infection of Soybean. Mol. Plant-Microbe Interact. 2010, 23, 1022–1031. [Google Scholar] [CrossRef]
- Gao, J.; Cao, M.; Ye, W.; Li, H.; Kong, L.; Zheng, X.; Wang, Y. PsMPK7, a stress-associated mitogen-activated protein kinase (MAPK) in Phytophthora sojae, is required for stress tolerance, reactive oxygenated species detoxification, cyst germination, sexual reproduction and infection of soybean. Mol. Plant Pathol. 2014, 16, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Judelson, H.S.; Blanco, F.A. The spores of Phytophthora: Weapons of the plant destroyer. Nat. Rev. Genet. 2005, 3, 47–58. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Ye, W.; Wu, J.; Xuan, M.; Li, Y.; Gao, J.; Wang, Y.; Wang, Y.; Dong, S.; Wang, Y. The MADS-box Transcription Factor PsMAD1 Is Involved in Zoosporogenesis and Pathogenesis of Phytophthora sojae. Front. Microbiol. 2018, 9, 2259. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Lu, J.; Tao, K.; Ye, W.; Li, A.; Liu, X.; Kong, L.; Dong, S.; Zheng, X.; Wang, Y. A Myb Transcription Factor of Phytophthora sojae, Regulated by MAP Kinase PsSAK1, Is Required for Zoospore Development. PLoS ONE 2012, 7, e40246. [Google Scholar] [CrossRef]
- Cázares-García, S.V.; Vázquez-Garcidueñas, M.S.; Vázquez-Marrufo, G. Structural and Phylogenetic Analysis of Laccases from Trichoderma: A Bioinformatic Approach. PLoS ONE 2013, 8, e55295. [Google Scholar] [CrossRef]
- Sheng, Y.; Wang, Y.; Meijer, H.J.; Yang, X.; Hua, C.; Ye, W.; Tao, K.; Liu, X.; Govers, F.; Wang, Y. The heat shock transcription factor PsHSF1 of Phytophthora sojae is required for oxidative stress tolerance and detoxifying the plant oxidative burst. Environ. Microbiol. 2015, 17, 1351–1364. [Google Scholar] [CrossRef]
- Fong, A.M.V.A.; Judelson, H.S. Cell cycle regulator Cdc14 is expressed during sporulation but not hyphal growth in the fungus-like oomycete Phytophthora infestans. Mol. Microbiol. 2003, 50, 487–494. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Yang, X.; Dong, S.; Sheng, Y.; Wang, Y.; Zheng, X. Transient silencing mediated by in vitro synthesized double-stranded RNA indicates that PsCdc14 is required for sporangial development in a soybean root rot pathogen. Sci. China Life Sci. 2011, 54, 1143–1150. [Google Scholar] [CrossRef]
- Guo, M.; Chen, Y.; Du, Y.; Dong, Y.; Guo, W.; Zhai, S.; Zhang, H.; Dong, S.; Zhang, Z.; Wang, Y.; et al. The bZIP Transcription Factor MoAP1 Mediates the Oxidative Stress Response and Is Critical for Pathogenicity of the Rice Blast Fungus Magnaporthe oryzae. PLoS Pathog. 2011, 7, e1001302. [Google Scholar] [CrossRef]
- Ye, W.; Wang, Y.; Shen, D.; Li, D.; Pu, T.; Jiang, Z.; Zhang, Z.; Zheng, X.; Tyler, B.M.; Wang, Y. Sequencing of the Litchi Downy Blight Pathogen Reveals It Is a Phytophthora Species with Downy Mildew-Like Characteristics. Mol. Plant-Microbe Interact. 2016, 29, 573–583. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Ye, W.; Situ, J.; Chen, Y.; Yang, X.; Kong, G.; Liu, Y.; Tinashe, R.J.; Xi, P.; Wang, Y.; et al. A Puf RNA-binding protein encoding gene PlM90 regulates the sexual and asexual life stages of the litchi downy blight pathogen Peronophythora litchii. Fungal Genet. Biol. 2017, 98, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Fang, Y.; Tyler, B.M. Efficient disruption and replacement of an effector gene in the oomycete Phytophthora sojae using CRISPR/Cas9. Mol. Plant Pathol. 2016, 17, 127–139. [Google Scholar] [CrossRef]
- Situ, J.; Jiang, L.; Fan, X.; Yang, W.; Li, W.; Xi, P.; Deng, Y.; Kong, G.; Jiang, Z. An RXLR effector PlAvh142 from Peronophythora litchii triggers plant cell death and contributes to virulence. Mol. Plant Pathol. 2020, 21, 415–428. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, J.; Xi, P.; Deng, Y.; Huang, W.; Wang, J.; Zhao, Q.; Yang, W.; Li, W.; Situ, J.; Jiang, L.; et al. The Mitogen-Activated Protein Kinase PlMAPK2 Is Involved in Zoosporogenesis and Pathogenicity of Peronophythoralitchii. Int. J. Mol. Sci. 2021, 22, 3524. https://doi.org/10.3390/ijms22073524
Huang J, Xi P, Deng Y, Huang W, Wang J, Zhao Q, Yang W, Li W, Situ J, Jiang L, et al. The Mitogen-Activated Protein Kinase PlMAPK2 Is Involved in Zoosporogenesis and Pathogenicity of Peronophythoralitchii. International Journal of Molecular Sciences. 2021; 22(7):3524. https://doi.org/10.3390/ijms22073524
Chicago/Turabian StyleHuang, Jiamin, Pinggen Xi, Yizhen Deng, Weixiong Huang, Jingrui Wang, Qingqing Zhao, Wensheng Yang, Wen Li, Junjian Situ, Liqun Jiang, and et al. 2021. "The Mitogen-Activated Protein Kinase PlMAPK2 Is Involved in Zoosporogenesis and Pathogenicity of Peronophythoralitchii" International Journal of Molecular Sciences 22, no. 7: 3524. https://doi.org/10.3390/ijms22073524
APA StyleHuang, J., Xi, P., Deng, Y., Huang, W., Wang, J., Zhao, Q., Yang, W., Li, W., Situ, J., Jiang, L., Guan, T., Li, M., Jiang, Z., & Kong, G. (2021). The Mitogen-Activated Protein Kinase PlMAPK2 Is Involved in Zoosporogenesis and Pathogenicity of Peronophythoralitchii. International Journal of Molecular Sciences, 22(7), 3524. https://doi.org/10.3390/ijms22073524





