Candidate Genes for the High-Altitude Adaptations of Two Mountain Pine Taxa
Abstract
1. Introduction
2. Results
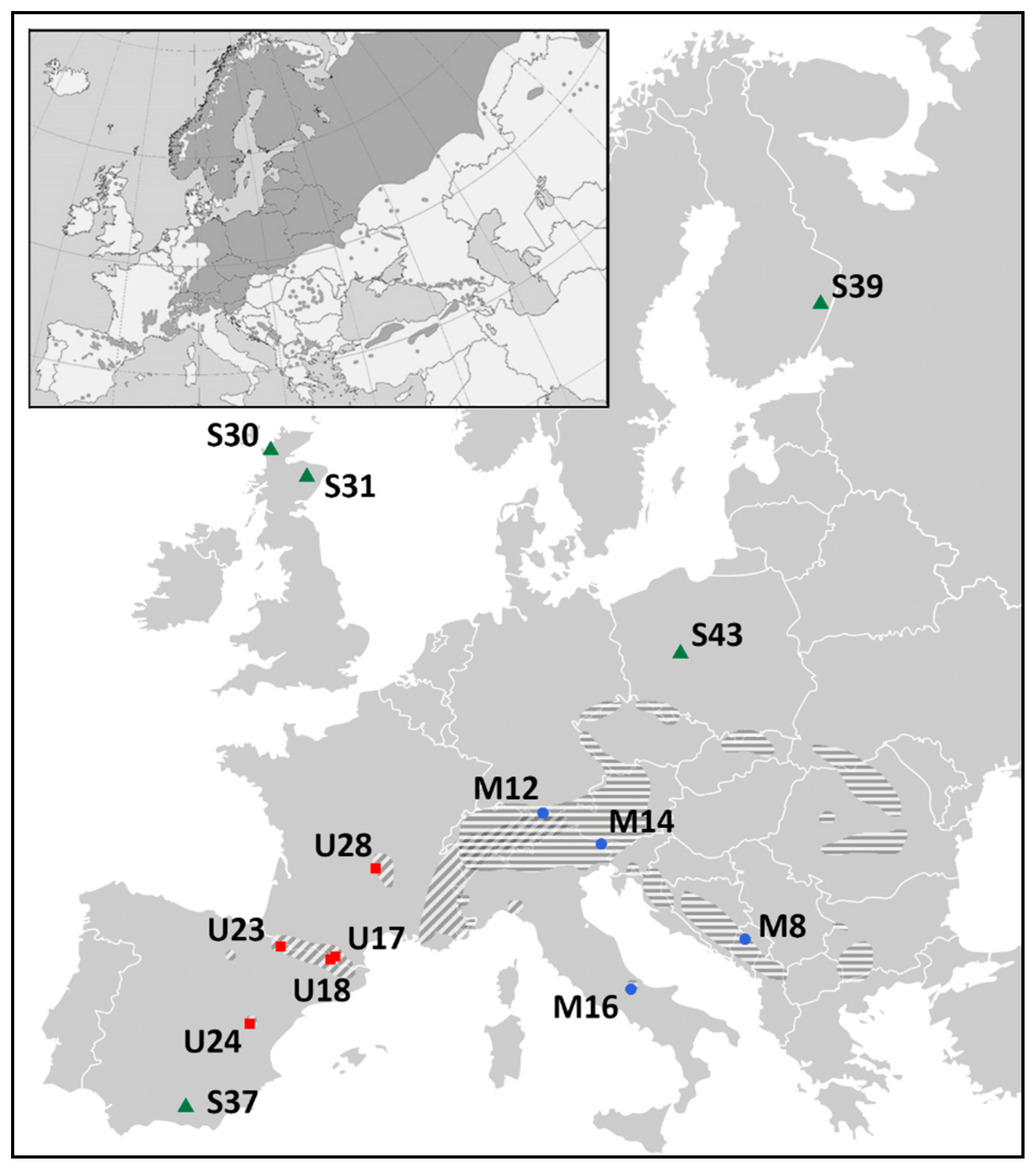
2.1. Genetic Diversity
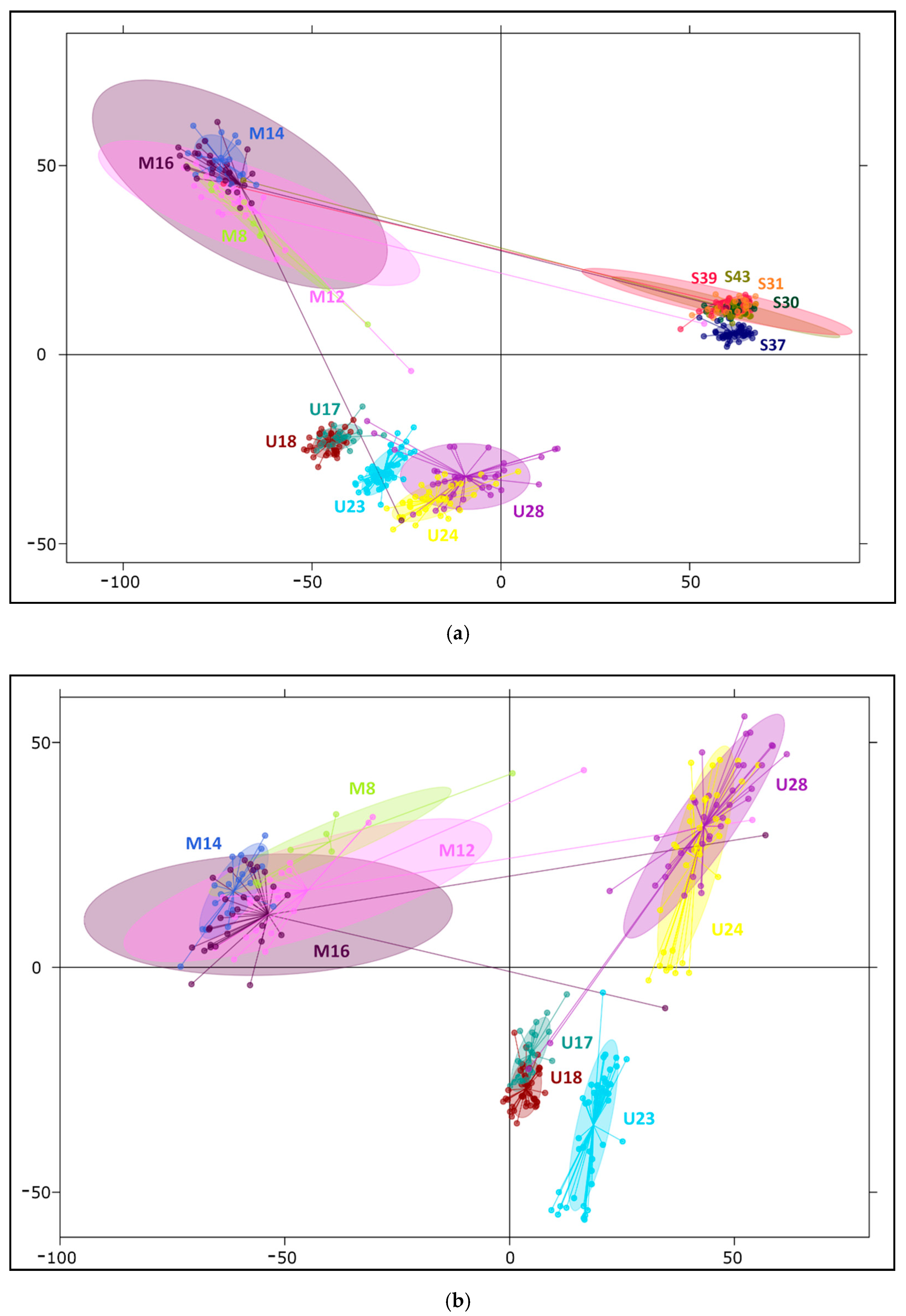
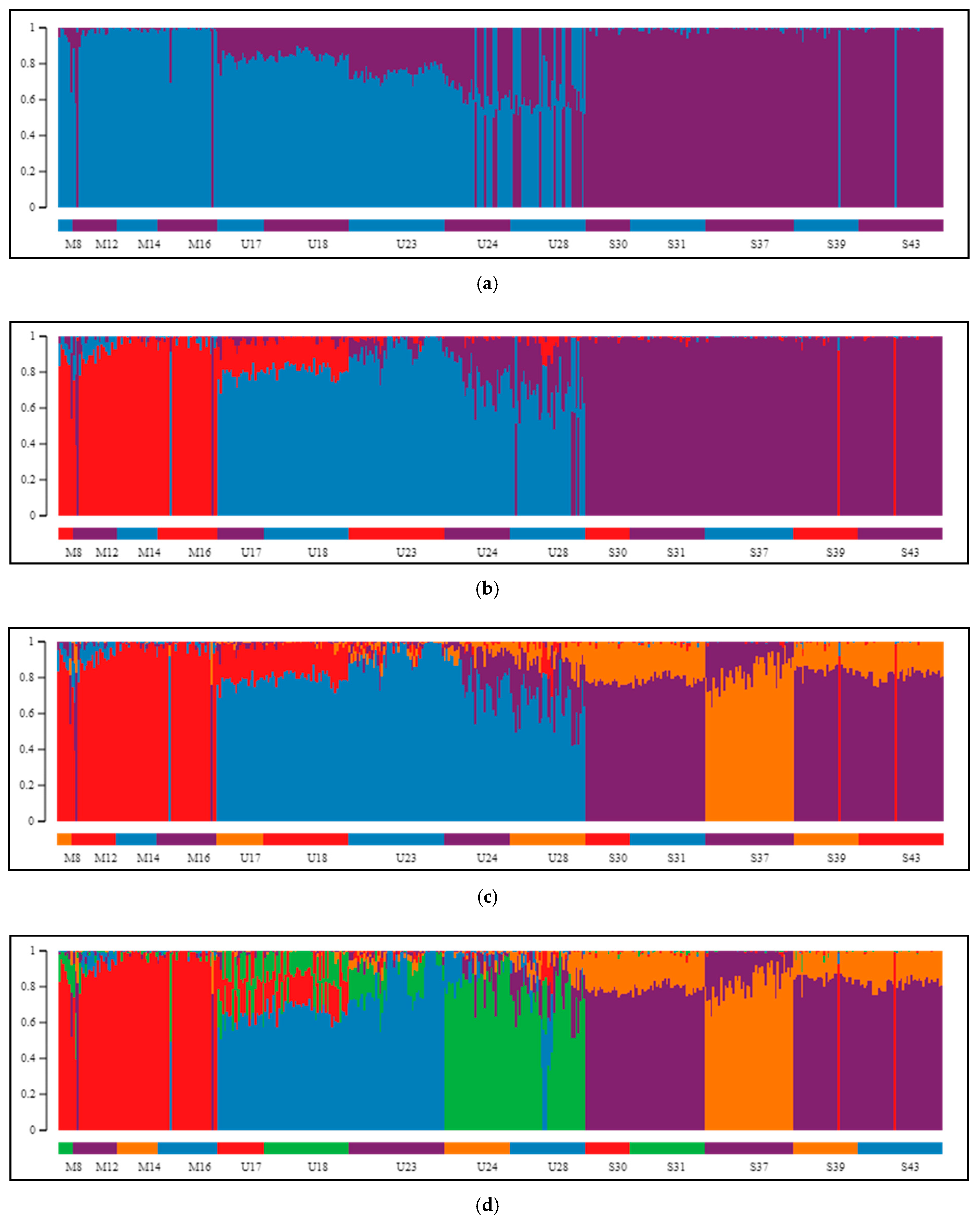
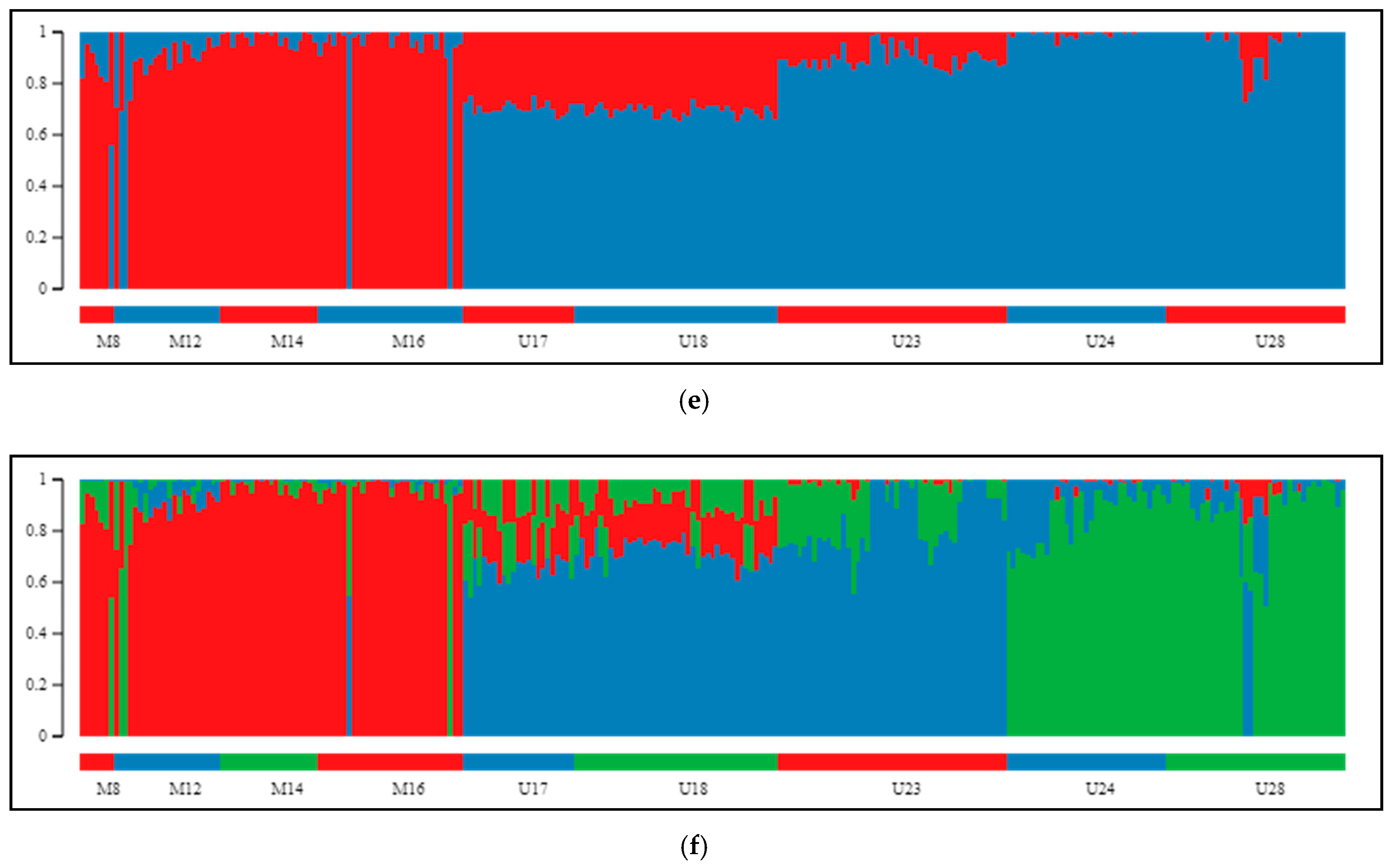
2.2. Differentiation and Grouping of Populations
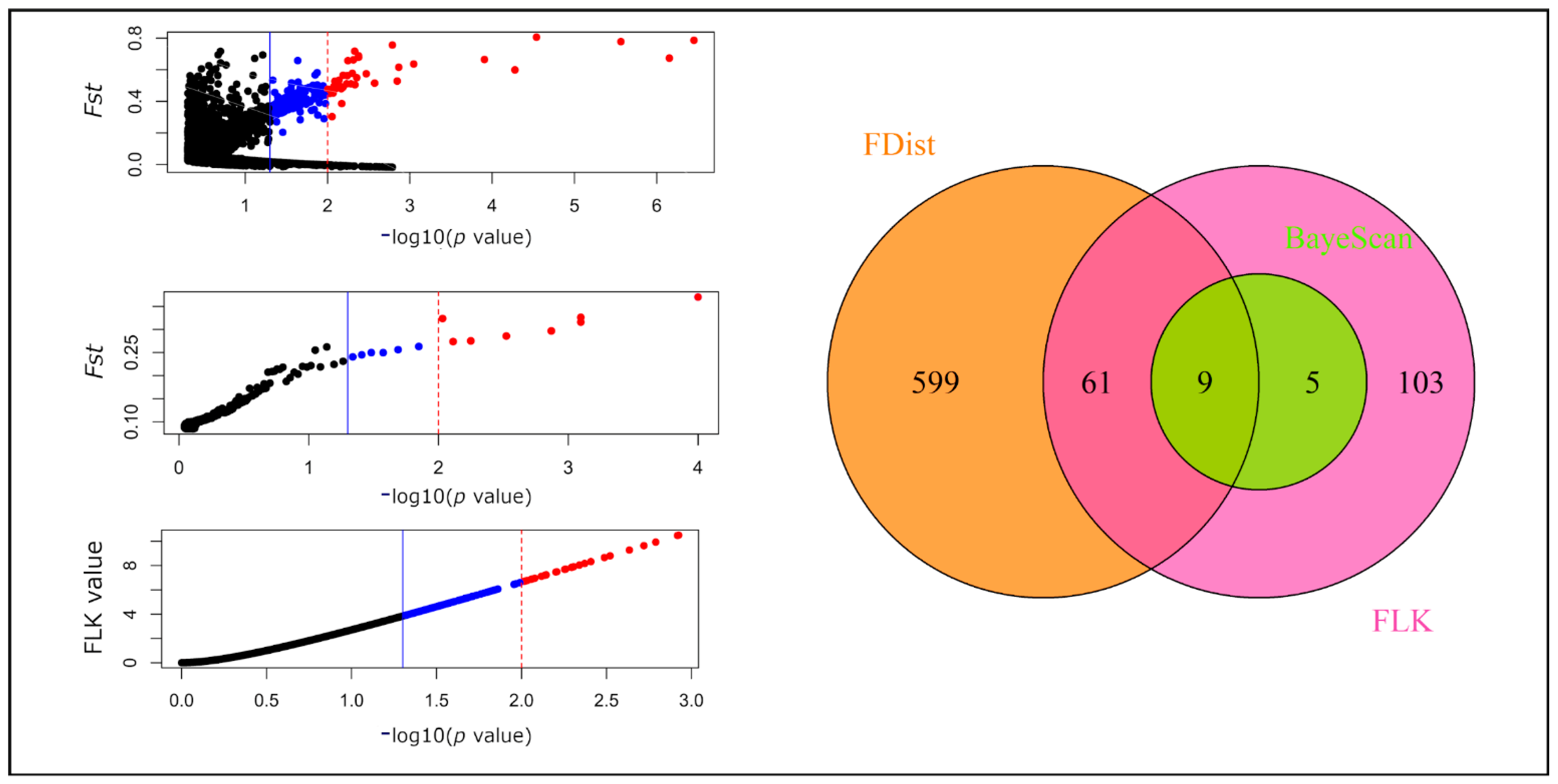
2.3. Outlier SNPs and Their Functional Annotation
3. Discussion
3.1. Discrimination of the Species
3.2. Inner Genetic Variation of Three Pine Species Populations
3.3. Candidates for Drivers of High-Altitude Adaptations in the Studied Subalpine Pines
3.4. Conclusions
4. Materials and Methods
4.1. Materials
4.2. SNP Array Genotyping
4.3. Analysis of Genetic Diversity, Differentiation, and Population Grouping
4.4. Outlier SNP Detection and Functional Annotations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Petit, R.J.; Hampe, A. Some evolutionary consequences of being a tree. Annu. Rev. Ecol. Evol. Syst. 2006, 37, 187–214. [Google Scholar] [CrossRef]
- Billings, W.D. Adaptations and Origins of Alpine Plants. Arct. Alp. Res. 1974, 6, 129–142. [Google Scholar] [CrossRef]
- Wieser, G.; Tausz, M. Trees at Their Upper Limit: Treelife Limitation at the Alpine Timberline; Springer GmbH: Heidelberg, Germany, 2007. [Google Scholar]
- Smith, W.K.; Johnson, D.M.; Reinhardt, K. Alpine Forest. In Encyclopedia of Ecology; Jørgensen, S.E., Fath, B.D., Eds.; Academic Press: Oxford, UK, 2008; pp. 144–153. [Google Scholar]
- Oleksyn, J.; Modrzynski, J.; Tjoelker, M.G.; Zytkowiak, R.; Reich, P.B.; Karolewski, P. Growth and physiology of Picea abies populations from elevational transects: Common garden evidence for altitudinal ecotypes and cold adaptation. Funct. Ecol. 1998, 12, 573–590. [Google Scholar] [CrossRef]
- Keller, F.; Korner, C. The role of photoperiodism in alpine plant development. Arct. Antarct. Alp. Res. 2003, 35, 361–368. [Google Scholar] [CrossRef]
- Coomes, D.A.; Jenkins, K.L.; Cole, L.E.S. Scaling of tree vascular transport systems along gradients of nutrient supply and altitude. Biol. Lett. 2007, 3, 86–89. [Google Scholar] [CrossRef]
- Lutz, C.; Engel, L. Changes in chloroplast ultrastructure in some high-alpine plants: Adaptation to metabolic demands and climate? Protoplasma 2007, 231, 183–192. [Google Scholar] [CrossRef] [PubMed]
- Ohsawa, T.; Ide, Y. Global patterns of genetic variation in plant species along vertical and horizontal gradients on mountains. Glob. Ecol. Biogeogr. 2008, 17, 152–163. [Google Scholar] [CrossRef]
- Gonzalo-Turpin, H.; Hazard, L. Local adaptation occurs along altitudinal gradient despite the existence of gene flow in the alpine plant species Festuca eskia. J. Ecol. 2009, 97, 742–751. [Google Scholar] [CrossRef]
- Stocklin, J.; Kuss, P.; Pluess, A.R. Genetic diversity, phenotypic variation and local adaptation in the alpine landscape: Case studies with alpine plant species. Bot. Helv. 2009, 119, 125–133. [Google Scholar] [CrossRef]
- Farjon, A. The Kew Review: Conifers of the World. Kew Bull. 2018, 73, 8. [Google Scholar] [CrossRef]
- Critchfield, W.B.; Little, E.L. Geographic Distribution of the Pines of the World; Department of Agriculture, Forest Service: Washington, DC, USA, 1966. [Google Scholar]
- Alexandrov, A.H.; von Wühlisch, G.; Vendramin, G.G. EUFORGEN Technical Guidelines for Genetic Conservation and Use of Mountain Pine (Pinus mugo); European Forest Genetic Resources Programme (EUFORGEN), European Forest Institute: Bonn, Germany, 2019. [Google Scholar]
- Monteleone, I.; Ferrazzini, D.; Belletti, P. Effectiveness of neutral RAPD markers to detect genetic divergence between the subspecies uncinata and mugo of Pinus mugo Turra. Silva Fenn. 2006, 40, 391–406. [Google Scholar] [CrossRef][Green Version]
- Heuertz, M.; Teufel, J.; Gonzalez-Martinez, S.C.; Soto, A.; Fady, B.; Alia, R.; Vendramin, G.G. Geography determines genetic relationships between species of mountain pine (Pinus mugo complex) in western Europe. J. Biogeogr. 2010, 37, 541–556. [Google Scholar] [CrossRef]
- Zaborowska, J.; Labiszak, B.; Wachowiak, W. Population history of European mountain pines Pinus mugo and Pinus uncinata revealed by mitochondrial DNA markers. J. Syst. Evol. 2020, 58, 474–486. [Google Scholar] [CrossRef]
- Boratynska, K.; Jasinska, A.K.; Boratynski, A. Taxonomic and geographic differentiation of Pinus mugo complex on the needle characteristics. Syst. Biodivers. 2015, 13, 901–915. [Google Scholar] [CrossRef]
- Jalas, J.; Suominen, J. Atlas Florae Europaeae: Gymnospermae (Pinaceae to Ephedraceae); Committee for Mapping the Flora of Europe & Suomen Biologian Seura Vanamo: Helsinki, Finland, 1972. [Google Scholar]
- Camarero, J.J.; Gutierrez, E.; Fortin, M.J.; Ribbens, E. Spatial patterns of tree recruitment in a relict population of Pinus uncinata: Forest expansion through stratified diffusion. J. Biogeogr. 2005, 32, 1979–1992. [Google Scholar] [CrossRef]
- Boratynska, K.; Marcysiak, K.; Boratynski, A. Pinus mugo (Pinaceae) in the Abruzzi Mountains: High morphological variation in isolated populations. Bot. J. Linn. Soc. 2005, 147, 309–316. [Google Scholar] [CrossRef]
- Wachowiak, W.; Perry, A.; Donnelly, K.; Cavers, S. Early phenology and growth trait variation in closely related European pine species. Ecol. Evol. 2018, 8, 655–666. [Google Scholar] [CrossRef] [PubMed]
- Bogunic, F.; Siljak-Yakovlev, S.; Muratovic, E.; Pustahija, F.; Medjedovic, S. Molecular cytogenetics and flow cytometry reveal conserved genome organization in Pinus mugo and P. uncinata. Ann. For. Sci. 2011, 68, 179–187. [Google Scholar] [CrossRef][Green Version]
- Lewandowski, A.; Boratynski, A.; Mejnartowicz, L. Allozyme investigations on the genetic differentiation between closely related pines—Pinus sylvestris, P. mugo, P. uncinata, and P. uliginosa (Pinaceae). Plant Syst. Evol. 2000, 221, 15–24. [Google Scholar] [CrossRef]
- Wachowiak, W.; Boratynska, K.; Cavers, S. Geographical patterns of nucleotide diversity and population differentiation in three closely related European pine species in the Pinus mugo complex. Bot. J. Linn. Soc. 2013, 172, 225–238. [Google Scholar] [CrossRef][Green Version]
- Wachowiak, W.; Trivedi, U.; Perry, A.; Cavers, S. Comparative transcriptomics of a complex of four European pine species. BMC Genom. 2015, 16, 234. [Google Scholar] [CrossRef]
- Nystedt, B.; Street, N.R.; Wetterbom, A.; Zuccolo, A.; Lin, Y.C.; Scofield, D.G.; Vezzi, F.; Delhomme, N.; Giacomello, S.; Alexeyenko, A.; et al. The Norway spruce genome sequence and conifer genome evolution. Nature 2013, 497, 579–584. [Google Scholar] [CrossRef]
- Neale, D.B.; Wegrzyn, J.L.; Stevens, K.A.; Zimin, A.V.; Puiu, D.; Crepeau, M.W.; Cardeno, C.; Koriabine, M.; Holtz-Morris, A.E.; Liechty, J.D.; et al. Decoding the massive genome of loblolly pine using haploid DNA and novel assembly strategies. Genome Biol. 2014, 15, R59. [Google Scholar] [CrossRef]
- Zimin, A.V.; Stevens, K.A.; Crepeau, M.W.; Puiu, D.; Wegrzyn, J.L.; Yorke, J.A.; Langley, C.H.; Neale, D.B.; Salzberg, S.L. An improved assembly of the loblolly pine mega-genome using long-read single-molecule sequencing. Gigascience 2017, 6, 1–4. [Google Scholar] [CrossRef]
- Dzialuk, A.; Boratynska, K.; Romo, A.; Boratynski, A. Taxonomic and geographic variation of the Pinus mugo complex on chloroplast microsatellite markers. Syst. Biodivers. 2017, 15, 464–479. [Google Scholar] [CrossRef]
- Wachowiak, W.; Zaborowska, J.; Labiszak, B.; Perry, A.; Zucca, G.M.; Gonzalez-Martinez, S.C.; Cavers, S. Molecular signatures of divergence and selection in closely related pine taxa. Tree Genet. Genomes 2018, 14, 83. [Google Scholar] [CrossRef]
- Martinez, I.; Gonzalez-Taboada, F.; Wiegand, T.; Camarero, J.J.; Gutierrez, E. Dispersal limitation and spatial scale affect model based projections of Pinus uncinata response to climate change in the Pyrenees. Glob. Chang. Biol. 2012, 18, 1714–1724. [Google Scholar] [CrossRef]
- Grotkopp, E.; Rejmanek, M.; Sanderson, M.J.; Rost, T.L. Evolution of genome size in pines (Pinus) and its life-history correlates: Supertree analyses. Evolution 2004, 58, 1705–1729. [Google Scholar] [CrossRef] [PubMed]
- Wachowiak, W.; Palme, A.E.; Savolainen, O. Speciation history of three closely related pines Pinus mugo (T.), P. uliginosa (N.) and P. sylvestris (L.). Mol. Ecol. 2011, 20, 1729–1743. [Google Scholar] [CrossRef] [PubMed]
- Perry, A.; Wachowiak, W.; Downing, A.; Talbot, R.; Cavers, S. Development of a SNP array for population genomic studies in four European pine species. Mol. Ecol. Resour. 2020, 20, 1697–1705. [Google Scholar] [CrossRef]
- Wikimedia Commons, the Free Media Repository. Available online: https://commons.wikimedia.org (accessed on 22 September 2009).
- Distribution map of Scots pine (Pinus sylvestris) EUFORGEN 2009, European Forest Genetic Resources Programme. Available online: http://euforgen.org (accessed on 4 October 2018).
- Hamernik, J.; Musil, I. Pinus mugo complex—Its structuring and general overview of the used nomenclature. J. For. Sci. 2007, 53, 253–266. [Google Scholar] [CrossRef]
- Dzialuk, A.; Muchewicz, E.; Boratynski, A.; Montserrat, J.M.; Boratynska, K.; Burczyk, J. Genetic variation of Pinus uncinata (Pinaceae) in the Pyrenees determined with cpSSR markers. Plant Syst. Evol. 2009, 277, 197–205. [Google Scholar] [CrossRef]
- Dainou, K.; Blanc-Jolivet, C.; Degen, B.; Kimani, P.; Ndiade-Bourobou, D.; Donkpegan, A.S.; Tosso, F.; Kaymak, E.; Bourland, N.; Doucet, J.L.; et al. Revealing hidden species diversity in closely related species using nuclear SNPs, SSRs and DNA sequences—A case study in the tree genus Milicia. BMC Evol. Biol. 2016, 16, 259. [Google Scholar] [CrossRef]
- Nicotra, A.B.; Chong, C.; Bragg, J.G.; Ong, C.R.; Aitken, N.C.; Chuah, A.; Lepschi, B.; Borevitz, J.O. Population and phylogenomic decomposition via genotyping-by-sequencing in Australian Pelargonium. Mol. Ecol. 2016, 25, 2000–2014. [Google Scholar] [CrossRef]
- Lee, S.R.; Gaskin, J.F.; Kim, Y.D. Molecular diagnosis for a Tamarix species from two reclaimed lands along the Yellow Sea in Korea inferred from genome wide SNP markers. J. Syst. Evol. 2019, 57, 247–255. [Google Scholar] [CrossRef]
- Zukowska, W.B.; Boratynska, K.; Wachowiak, W. Comparison of range-wide chloroplast microsatellite and needle trait variation patterns in Pinus mugo Turra (dwarf mountain pine). iForest 2017, 10, 250–258. [Google Scholar] [CrossRef]
- Zukowska, W.B.; Wachowiak, W. Nuclear microsatellite markers reveal the low genetic structure of Pinus mugo Turra (dwarf mountain pine) populations in Europe. Plant Syst. Evol. 2017, 303, 641–651. [Google Scholar] [CrossRef]
- Robledo-Arnuncio, J.J. Wind pollination over mesoscale distances: An investigation with Scots pine. New Phytol. 2011, 190, 222–233. [Google Scholar] [CrossRef]
- Soto, A.; Robledo-Arnuncio, J.J.; Gonzalez-Martinez, S.C.; Smouse, P.E.; Alia, R. Climatic niche and neutral genetic diversity of the six Iberian pine species: A retrospective and prospective view. Mol. Ecol. 2010, 19, 1396–1409. [Google Scholar] [CrossRef]
- Polle, A.; Rennenberg, H. Field Studies on Norway Spruce Trees at High-Altitudes. 2. Defense Systems against Oxidative Stress in Needles. New Phytol. 1992, 121, 635–642. [Google Scholar] [CrossRef]
- Streb, P.; Shang, W.; Feierabend, J.; Bligny, R. Divergent strategies of photoprotection in high-mountain plants. Planta 1998, 207, 313–324. [Google Scholar] [CrossRef]
- Polle, A.; Baumbusch, L.O.; Oschinski, C.; Eiblmeier, M.; Kuhlenkamp, V.; Vollrath, B.; Scholz, F.; Rennenberg, H. Growth and protection against oxidative stress in young clones and mature spruce trees (Picea abies L.) at high altitudes. Oecologia 1999, 121, 149–156. [Google Scholar] [CrossRef]
- Oncel, I.; Yurdakulol, E.; Keles, Y.; Kurt, L.; Yildiz, A. Role of antioxidant defense system and biochemical adaptation on stress tolerance of high mountain and steppe plants. Acta Oecol. 2004, 26, 211–218. [Google Scholar] [CrossRef]
- Keles, Y.; Everest, A. Relation to altitude adaptation and antioxidant defence system in five shrubs and trees species from middle Taurus Mountains. Int. J. Nat. Eng. Sci. 2008, 2, 45–49. [Google Scholar]
- Huang, W.; Yang, Y.J.; Hu, H.; Zhang, S.B. Seasonal variations in photosystem I compared with photosystem II of three alpine evergreen broad-leaf tree species. J. Photochem. Photobiol. B 2016, 165, 71–79. [Google Scholar] [CrossRef]
- Wang, H.; Prentice, I.C.; Davis, T.W.; Keenan, T.F.; Wright, I.J.; Peng, C.H. Photosynthetic responses to altitude: An explanation based on optimality principles. New Phytol. 2017, 213, 976–982. [Google Scholar] [CrossRef] [PubMed]
- Gibert, J.M.; Mouchel-Vielh, E.; De Castro, S.; Peronnet, F. Phenotypic Plasticity through Transcriptional Regulation of the Evolutionary Hotspot Gene tan in Drosophila melanogaster. PLoS Genet. 2016, 12, e1006218. [Google Scholar] [CrossRef] [PubMed]
- Ecker, S.; Pancaldi, V.; Valencia, A.; Beck, S.; Paul, D.S. Epigenetic and Transcriptional Variability Shape Phenotypic Plasticity. Bioessays 2018, 40, 1700148. [Google Scholar] [CrossRef]
- Hirst, J.; Bright, N.A.; Rous, B.; Robinson, M.S. Characterization of a fourth adaptor-related protein complex. Mol. Biol. Cell 1999, 10, 2787–2802. [Google Scholar] [CrossRef] [PubMed]
- Nakatsu, F.; Ohno, H. Adaptor protein complexes as the key regulators of protein sorting in the post-Golgi network. Cell Struct. Funct. 2003, 28, 419–429. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Sun, X.D.; Kong, X.X.; Galvan, J.V.; Li, X.; Yang, S.H.; Yang, Y.Q.; Yang, Y.P.; Hu, X.Y. Physiological, biochemical and proteomics analysis reveals the adaptation strategies of the alpine plant Potentilla saundersiana at altitude gradient of the Northwestern Tibetan Plateau. J. Proteom. 2015, 112, 63–82. [Google Scholar] [CrossRef]
- Eveno, E.; Collada, C.; Guevara, M.A.; Leger, V.; Soto, A.; Diaz, L.; Leger, P.; Gonzalez-Martinez, S.C.; Cervera, M.T.; Plomion, C.; et al. Contrasting patterns of selection at Pinus pinaster Ait. drought stress candidate genes as revealed by genetic differentiation analyses. Mol. Biol. Evol. 2008, 25, 417–437. [Google Scholar] [CrossRef]
- Behringer, D.; Zimmermann, H.; Ziegenhagen, B.; Liepelt, S. Differential Gene Expression Reveals Candidate Genes for Drought Stress Response in Abies alba (Pinaceae). PLoS ONE 2015, 10, e0124564. [Google Scholar] [CrossRef]
- Heer, K.; Behringer, D.; Piermattei, A.; Bassler, C.; Brandl, R.; Fady, B.; Jehl, H.; Liepelt, S.; Lorch, S.; Piotti, A.; et al. Linking dendroecology and association genetics in natural populations: Stress responses archived in tree rings associate with SNP genotypes in silver fir (Abies alba Mill.). Mol. Ecol. 2018, 27, 1428–1438. [Google Scholar] [CrossRef]
- Zhou, Y.F.; Zhang, L.R.; Liu, J.Q.; Wu, G.L.; Savolainen, O. Climatic adaptation and ecological divergence between two closely related pine species in Southeast China. Mol. Ecol. 2014, 23, 3504–3522. [Google Scholar] [CrossRef]
- De Abreu-Neto, J.B.; Turchetto-Zolet, A.C.; de Oliveira, L.F.V.; Zanettini, M.H.B.; Margis-Pinheiro, M. Heavy metal-associated isoprenylated plant protein (HIPP): Characterization of a family of proteins exclusive to plants. FEBS J. 2013, 280, 1604–1616. [Google Scholar] [CrossRef] [PubMed]
- Hicks, M.A.; Barber, A.E.; Giddings, L.A.; Caldwell, J.; O’Connor, S.E.; Babbitt, P.C. The evolution of function in strictosidine synthase-like proteins. Proteins 2011, 79, 3082–3098. [Google Scholar] [CrossRef] [PubMed]
- Hicks, M.A.; Barber, A.E.; Babbitt, P.C. The Nucleophilic Attack Six-Bladed β-Propeller (N6P) Superfamily. In Protein Families: Relating Protein Sequence, Structure, and Function; Orengo, C., Bateman, A., Eds.; John Wiley & Sons, Inc.: Chichester, UK, 2013; pp. 125–158. [Google Scholar]
- Nakamura, Y.; Koizumi, R.; Shui, G.H.; Shimojima, M.; Wenk, M.R.; Ito, T.; Ohta, H. Arabidopsis lipins mediate eukaryotic pathway of lipid metabolism and cope critically with phosphate starvation. Proc. Natl. Acad. Sci. USA 2009, 106, 20978–20983. [Google Scholar] [CrossRef]
- Nakamura, Y. Phosphate starvation and membrane lipid remodeling in seed plants. Prog. Lipid Res. 2013, 52, 43–50. [Google Scholar] [CrossRef]
- Cumming, J.R. Phosphate-limitation physiology in ectomycorrhizal pitch pine (Pinus rigida) seedlings. Tree Physiol. 1996, 16, 977–983. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gerlitz, T.G.M.; Gerlitz, A. Phosphate uptake and polyphosphate metabolism of mycorrhizal and nonmycorrhizal roots of pine and of Suillus bovinus at varying external pH measured by in vivo P-31-NMR. Mycorrhiza 1997, 7, 101–106. [Google Scholar] [CrossRef]
- Anacker, B.L.; Strauss, S.Y. The geography and ecology of plant speciation: Range overlap and niche divergence in sister species. Proc. R. Soc. B Biol. Sci. 2014, 281, 20132980. [Google Scholar] [CrossRef] [PubMed]
- Cudlin, P.; Klopcic, M.; Tognetti, R.; Malis, F.; Alados, C.L.; Bebi, P.; Grunewald, K.; Zhiyanski, M.; Andonowski, V.; La Porta, N.; et al. Drivers of treeline shift in different European mountains. Clim. Res. 2017, 73, 135–150. [Google Scholar] [CrossRef]
- Basset, G.J.C.; Ravanel, S.; Quinlivan, E.P.; White, R.; Giovannoni, J.J.; Rebeille, F.; Nichols, B.P.; Shinozaki, K.; Seki, M.; Gregory, J.F.; et al. Folate synthesis in plants: The last step of the p-aminobenzoate branch is catalyzed by a plastidial aminodeoxychorismate lyase. Plant J. 2004, 40, 453–461. [Google Scholar] [CrossRef]
- Hanson, A.D.; Gregory, J.F. Synthesis and turnover of folates in plants. Curr. Opin. Plant Biol. 2002, 5, 244–249. [Google Scholar] [CrossRef]
- Gorelova, V.; Ambach, L.; Rebeille, F.; Stove, C.; Van Der Straeten, D. Folates in Plants: Research Advances and Progress in Crop Biofortification. Front. Chem. 2017, 5, 21. [Google Scholar] [CrossRef]
- Xu, N.; Gao, X.Q.; Zhao, X.Y.; Zhu, D.Z.; Zhou, L.Z.; Zhang, X.S. Arabidopsis AtVPS15 is essential for pollen development and germination through modulating phosphatidylinositol 3-phosphate formation. Plant Mol. Biol. 2011, 77, 251–260. [Google Scholar] [CrossRef]
- Zhou, X.M.; Zhao, P.; Wang, W.; Zou, J.; Cheng, T.H.; Peng, X.B.; Sun, M.X. A comprehensive, genome-wide analysis of autophagy-related genes identified in tobacco suggests a central role of autophagy in plant response to various environmental cues. DNA Res. 2015, 22, 245–257. [Google Scholar] [CrossRef]
- Schomburg, F.M.; Bizzell, C.M.; Lee, D.J.; Zeevaart, J.A.D.; Amasino, R.M. Overexpression of a novel class of gibberellin 2-oxidases decreases gibberellin levels and creates dwarf plants. Plant Cell 2003, 15, 151–163. [Google Scholar] [CrossRef]
- Luo, Y.H.; Dong, X.W.; Yu, T.Y.; Shi, X.; Li, Z.Y.; Yang, W.C.; Widmer, A.; Karrenberg, S. A Single Nucleotide Deletion in Gibberellin20-oxidase1 Causes Alpine Dwarfism in Arabidopsis. Plant Physiol. 2015, 168, 930–937. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.L.; Ogawa, M.; Fleet, C.M.; Zentella, R.; Hu, J.H.; Heo, J.O.; Lim, J.; Kamiya, Y.; Yamaguchi, S.; Sun, T.P. SCARECROW-LIKE 3 promotes gibberellin signaling by antagonizing master growth repressor DELLA in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 2160–2165. [Google Scholar] [CrossRef]
- Takahashi, K.; Yoshida, S. How the scrub height of dwarf pine Pinus pumila decreases at the treeline. Ecol. Res. 2009, 24, 847–854. [Google Scholar] [CrossRef]
- Phinney, B.O. Gibberellin A1 dwarfism and shoot elongation in higher plants. Biol. Plant. 1985, 27, 172–179. [Google Scholar] [CrossRef]
- Junttila, O. Gibberellins and the Regulation of Shoot Elongation in Woody Plants. In Gibberellins; Springer: New York, NY, USA, 1991; pp. 199–210. [Google Scholar]
- Little, C.H.A.; MacDonald, J.E. Effects of exogenous gibberellin and auxin on shoot elongation and vegetative bud development in seedlings of Pinus sylvestris and Picea glauca. Tree Physiol. 2003, 23, 73–83. [Google Scholar] [CrossRef]
- Wachowiak, W.; Balk, P.A.; Savolainen, O. Search for nucleotide diversity patterns of local adaptation in dehydrins and other cold-related candidate genes in Scots pine (Pinus sylvestris L.). Tree Genet. Genomes 2009, 5, 117–132. [Google Scholar] [CrossRef]
- Kujala, S.T.; Savolainen, O. Sequence variation patterns along a latitudinal cline in Scots pine (Pinus sylvestris): Signs of clinal adaptation? Tree Genet. Genomes 2012, 8, 1451–1467. [Google Scholar] [CrossRef]
- Mosca, E.; Eckert, A.J.; Di Pierro, E.A.; Rocchini, D.; La Porta, N.; Belletti, P.; Neale, D.B. The geographical and environmental determinants of genetic diversity for four alpine conifers of the European Alps. Mol. Ecol. 2012, 21, 5530–5545. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. Available online: http://pngu.mgh.harvard.edu/purcell/plink/ (accessed on 29 August 2019). [CrossRef]
- Lischer, H.E.L.; Excoffier, L. PGDSpider: An automated data conversion tool for connecting population genetics and genomics programs. Bioinformatics 2012, 28, 298–299. Available online: http://www.cmpg.unibe.ch/software/PGDSpider/ (accessed on 30 June 2017). [CrossRef]
- RStudio Team. RStudio: Integrated Development for R. 2018. Available online: http://www.rstudio.com/ (accessed on 22 July 2019).
- R Core Team. R: A Language and Environment for Statistical Computing. 2019. Available online: http://www.R-project.org/ (accessed on 22 July 2019).
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 2012, 28, 2537–2539. Available online: https://biology-assets.anu.edu.au/GenAlEx/Welcome.html (accessed on 4 December 2016). [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. Available online: https://www.megasoftware.net/ (accessed on 15 March 2017). [CrossRef]
- Excoffier, L.; Lischer, H.E.L. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. Available online: http://cmpg.unibe.ch/software/arlequin35/ (accessed on 15 March 2017). [CrossRef] [PubMed]
- Jombart, T.; Pontier, D.; Dufour, A.B. Genetic markers in the playground of multivariate analysis. Heredity 2009, 102, 330–341. [Google Scholar] [CrossRef] [PubMed]
- Jombart, T. adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics 2008, 24, 1403–1405. [Google Scholar] [CrossRef] [PubMed]
- Jombart, T.; Ahmed, I. adegenet 1.3-1: New tools for the analysis of genome-wide SNP data. Bioinformatics 2011, 27, 3070–3071. Available online: http://adegenet.r-forge.r-project.org/ (accessed on 13 October 2020). [CrossRef] [PubMed]
- Mantel, N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967, 27, 209–220. [Google Scholar]
- Hijimans, R.; Williams, E.; Vennes, C. Geosphere. 2019. Available online: https://cran.r-project.org/package=geosphere (accessed on 12 October 2020).
- Ripley, B.; Venables, B.; Bates, D.M.; Hornik, K.; Gebhardt, A.; Firth, D. MASS. 2020. Available online: https://CRAN.R-project.org/package=MASS (accessed on 12 October 2020).
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [PubMed]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of population structure using multilocus genotype data: Dominant markers and null alleles. Mol. Ecol. Notes 2007, 7, 574–578. [Google Scholar] [CrossRef] [PubMed]
- Hubisz, M.J.; Falush, D.; Stephens, M.; Pritchard, J.K. Inferring weak population structure with the assistance of sample group information. Mol. Ecol. Resour. 2009, 9, 1322–1332. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Earl, D.A.; Vonholdt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. Available online: http://taylor0.biology.ucla.edu/structureHarvester/ (accessed on 26 June 2020). [CrossRef]
- Ramasamy, R.K.; Ramasamy, S.; Bindroo, B.B.; Naik, V.G. STRUCTURE PLOT: A program for drawing elegant STRUCTURE bar plots in user friendly interface. Springerplus 2014, 3, 431. Available online: http://omicsspeaks.com/strplot2/ (accessed on 11 July 2020). [CrossRef] [PubMed]
- Weigand, H.; Leese, F. Detecting signatures of positive selection in non-model species using genomic data. Zool. J. Linn. Soc. Lond. 2018, 184, 528–583. [Google Scholar] [CrossRef]
- Foll, M.; Gaggiotti, O. A Genome-Scan Method to Identify Selected Loci Appropriate for Both Dominant and Codominant Markers: A Bayesian Perspective. Genetics 2008, 180, 977–993. [Google Scholar] [CrossRef]
- Fariello, M.I.; Boitard, S.; Naya, H.; SanCristobal, M.; Servin, B. Detecting Signatures of Selection Through Haplotype Differentiation Among Hierarchically Structured Populations. Genetics 2013, 193, 929–941. [Google Scholar] [CrossRef] [PubMed]
- BioBam Bioinformatics SL. OmicsBox—Bioinformatics Made Easy. Available online: https://www.biobam.com/omicsbox (accessed on 11 June 2020).
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Carbon, S.; Douglass, E.; Dunn, N.; Good, B.; Harris, N.L.; Lewis, S.E.; Mungall, C.J.; Basu, S.; Chisholm, R.L.; Dodson, R.J.; et al. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 2019, 47, D330–D338. [Google Scholar] [CrossRef]
| Group | N | %P | Sp | d | Ho | uHe | F |
|---|---|---|---|---|---|---|---|
| M8 | 7 | 75.4 | 1 | 1262.7 | 0.338 | 0.278 | −0.272 |
| M12 | 22 | 90.2 | 2 | 1325.5 | 0.313 | 0.274 | −0.123 |
| M14 | 20 | 82.5 | 1 | 1193.4 | 0.292 | 0.253 | −0.141 |
| M16 | 30 | 91.3 | 4 | 1282.4 | 0.285 | 0.261 | −0.060 |
| U17 | 23 | 85.2 | 0 | 1368.4 | 0.287 | 0.259 | −0.105 |
| U18 | 42 | 87.5 | 0 | 1437.1 | 0.277 | 0.256 | −0.075 |
| U23 | 47 | 84.9 | 0 | 1393.2 | 0.245 | 0.247 | −0.006 |
| U24 | 33 | 85.9 | 1 | 1264.1 | 0.298 | 0.257 | −0.137 |
| U28 | 37 | 88.8 | 1 | 1236.1 | 0.316 | 0.264 | −0.158 |
| S30 | 22 | 61.1 | 0 | 906.8 | 0.182 | 0.168 | −0.085 |
| S31 | 37 | 70.6 | 2 | 946.0 | 0.192 | 0.177 | −0.072 |
| S37 | 44 | 62.5 | 1 | 867.4 | 0.160 | 0.156 | −0.035 |
| S39 | 32 | 80.3 | 1 | 994.8 | 0.182 | 0.180 | 0.037 |
| S43 | 42 | 79.6 | 0 | 994.5 | 0.176 | 0.177 | 0.058 |
| P. mugo | 79 | 97.5 | 75 | 1308.7 | 0.299 | 0.272 | −0.065 |
| P. uncinata | 182 | 96.8 | 12 | 1426.8 | 0.280 | 0.270 | −0.030 |
| P. sylvestris | 177 | 92.8 | 25 | 1003.5 | 0.177 | 0.182 | 0.067 |
| Total | 438 | 100 | na | 1459.6 | 0.239 | 0.274 | 0.081 |
| Comparison | dxy | Fst (a/b) | Outlier SNPs (2**/2*/3*) |
|---|---|---|---|
| P. mugo vs. P. uncinata | 1523.5 | 0.1061/0.1510 | 12/75/9 |
| P. mugo vs. P. sylvestris | 1761.9 | 0.3612/0.3937 | 0/1/0 |
| P. uncinata vs. P. sylvestris | 1552.3 | 0.2162/0.2629 | 12/29/6 |
| mountain pines vs. P. sylvestris | 1615.7 | 0.2324/0.2969 | 7/35/2 |
| Sequence Name | Sequence Description and Definitions of Related GO Terms (Order: BP/MF/CC) |
|---|---|
| comp20176_c0_seq1 | AP-4 complex subunit mu: protein targeting; Golgi to lysosome transport/nd/cytosol; cytoplasmic vesicle; trans-Golgi network; AP-4 adaptor complex; clathrin adaptor complex |
| comp39941_c0_seq1 | glucan endo-1,3-beta-glucosidase 8-like: carbohydrate metabolic process/hydrolase activity, hydrolyzing O-glycosyl compounds/anchored component of plasma membrane |
| comp41821_c0_seq1 | heavy metal-associated isoprenylated plant protein 36-like: metal ion transport/metal ion binding/nd |
| comp44835_c0_seq1 | protein strictosidine synthase-like 3 biosynthetic process/strictosidine synthase activity/nd |
| comp50552_c0_seq1 | phosphatidate phosphatase PAH2-like: hydrolase activity/cellular metabolic process/nd |
| comp52994_c0_seq1 | unknown: transcription coregulator activity/regulation of transcription, DNA-templated/nd |
| comp53610_c0_seq1 | aminodeoxychorismate synthase, chloroplastic isoform X1: biosynthetic process; carboxylic acid metabolic process/nd/nd |
| comp54487_c0_seq1 | serine/threonine-protein kinase VPS15-like isoform X1: protein kinase activity; protein binding; endosome; autophagy/vacuolar transport; phosphorylation/phosphatidylinositol 3-kinase complex, class III; intracellular transport |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zaborowska, J.; Łabiszak, B.; Perry, A.; Cavers, S.; Wachowiak, W. Candidate Genes for the High-Altitude Adaptations of Two Mountain Pine Taxa. Int. J. Mol. Sci. 2021, 22, 3477. https://doi.org/10.3390/ijms22073477
Zaborowska J, Łabiszak B, Perry A, Cavers S, Wachowiak W. Candidate Genes for the High-Altitude Adaptations of Two Mountain Pine Taxa. International Journal of Molecular Sciences. 2021; 22(7):3477. https://doi.org/10.3390/ijms22073477
Chicago/Turabian StyleZaborowska, Julia, Bartosz Łabiszak, Annika Perry, Stephen Cavers, and Witold Wachowiak. 2021. "Candidate Genes for the High-Altitude Adaptations of Two Mountain Pine Taxa" International Journal of Molecular Sciences 22, no. 7: 3477. https://doi.org/10.3390/ijms22073477
APA StyleZaborowska, J., Łabiszak, B., Perry, A., Cavers, S., & Wachowiak, W. (2021). Candidate Genes for the High-Altitude Adaptations of Two Mountain Pine Taxa. International Journal of Molecular Sciences, 22(7), 3477. https://doi.org/10.3390/ijms22073477






