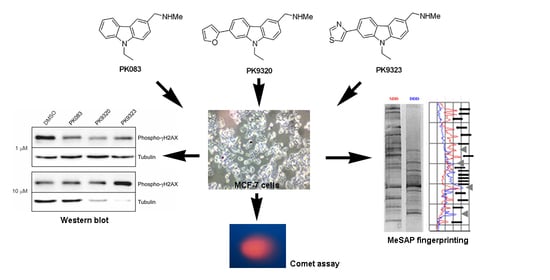
Genotoxicity and Epigenotoxicity of Carbazole-Derived Molecules on MCF-7 Breast Cancer Cells
Abstract
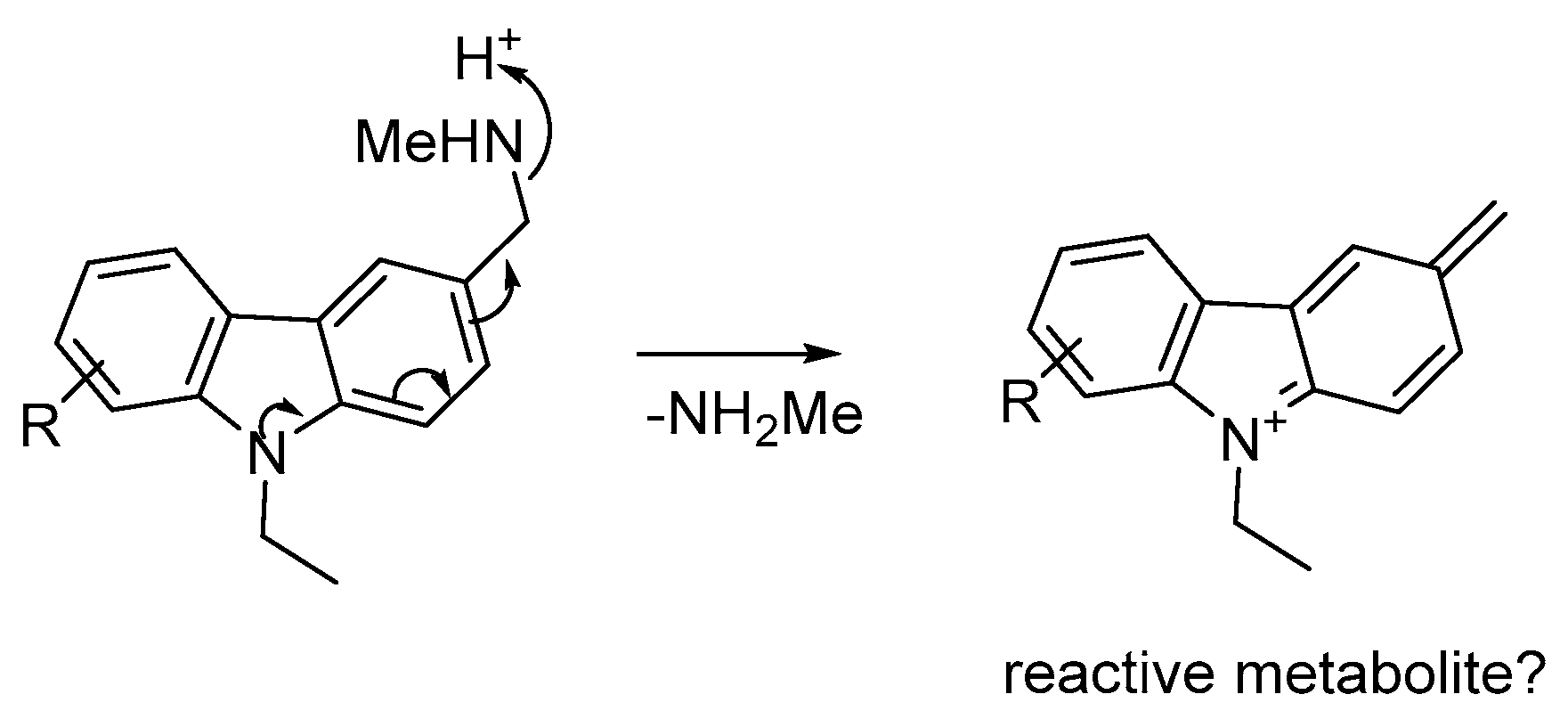
:1. Introduction
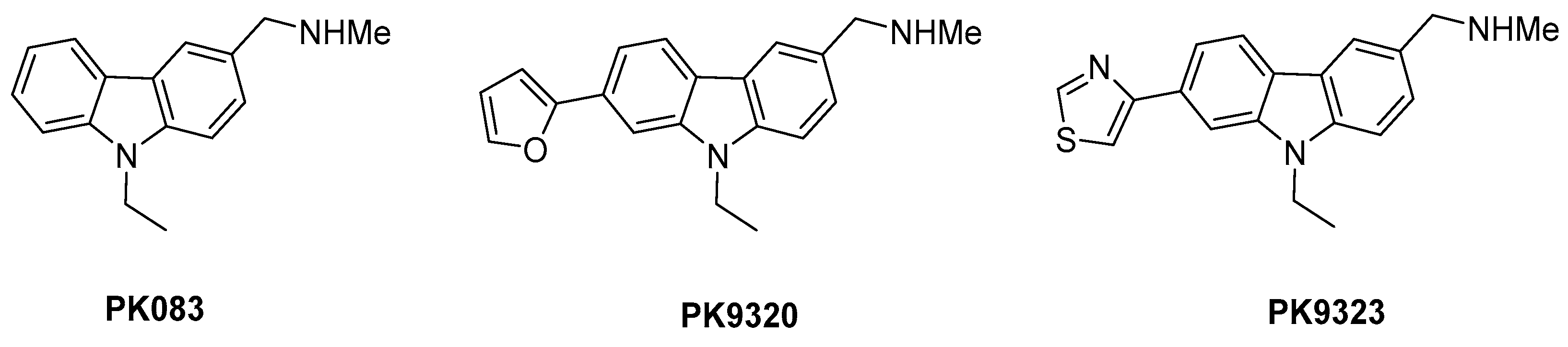
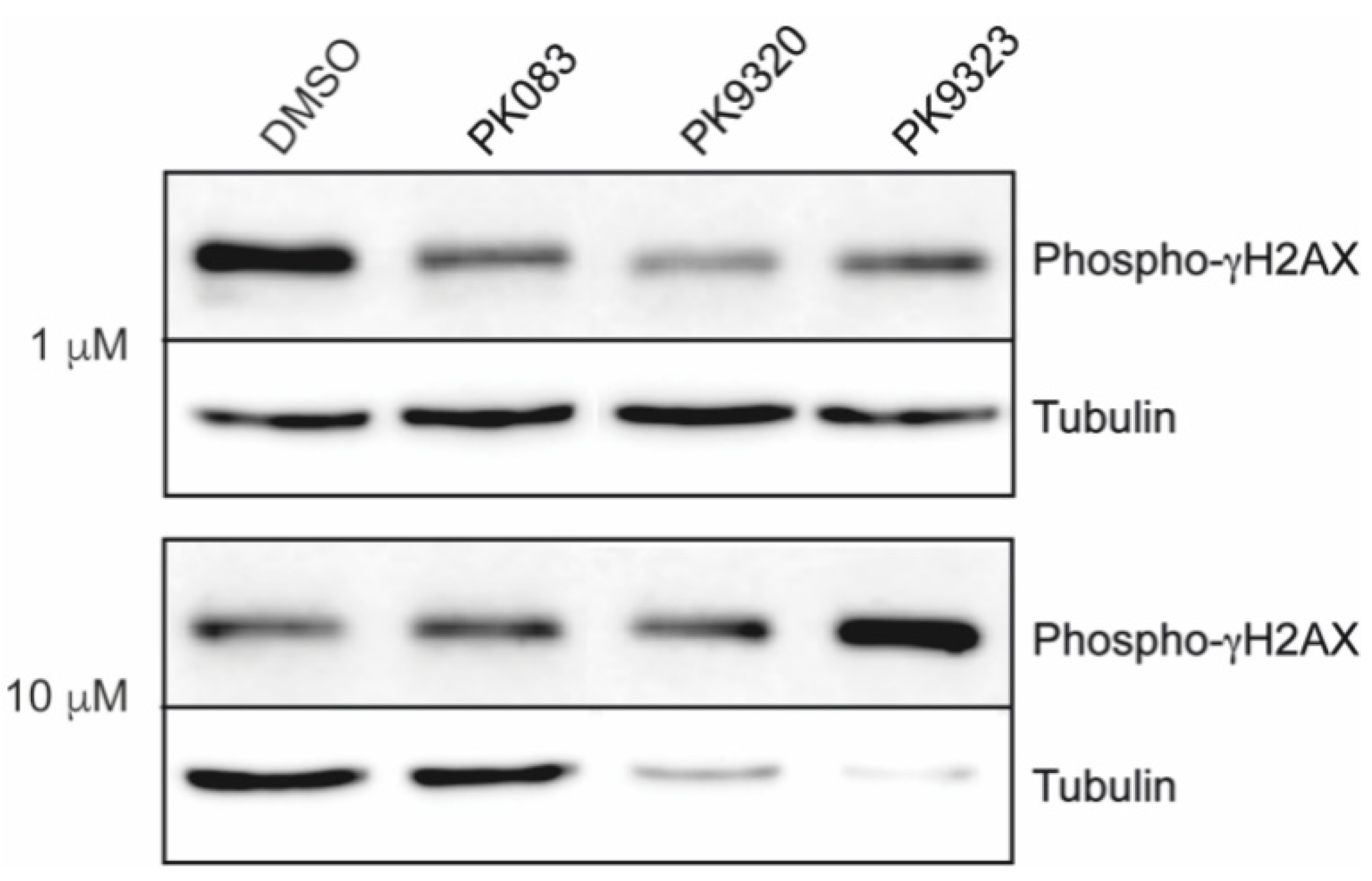
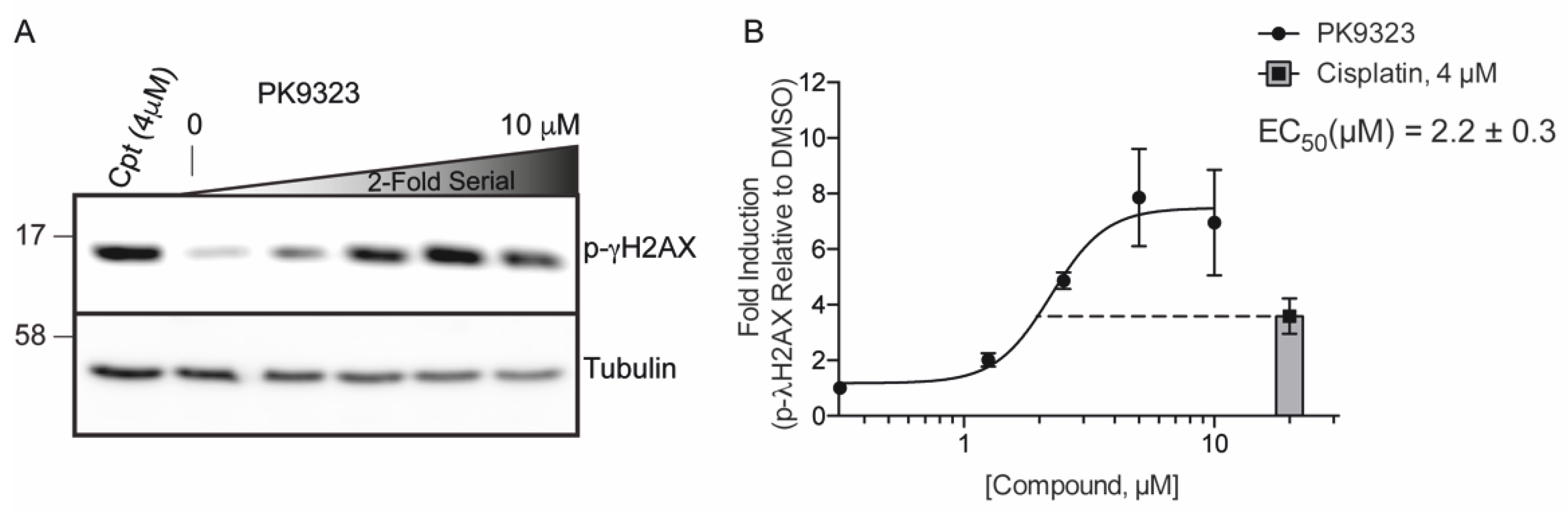
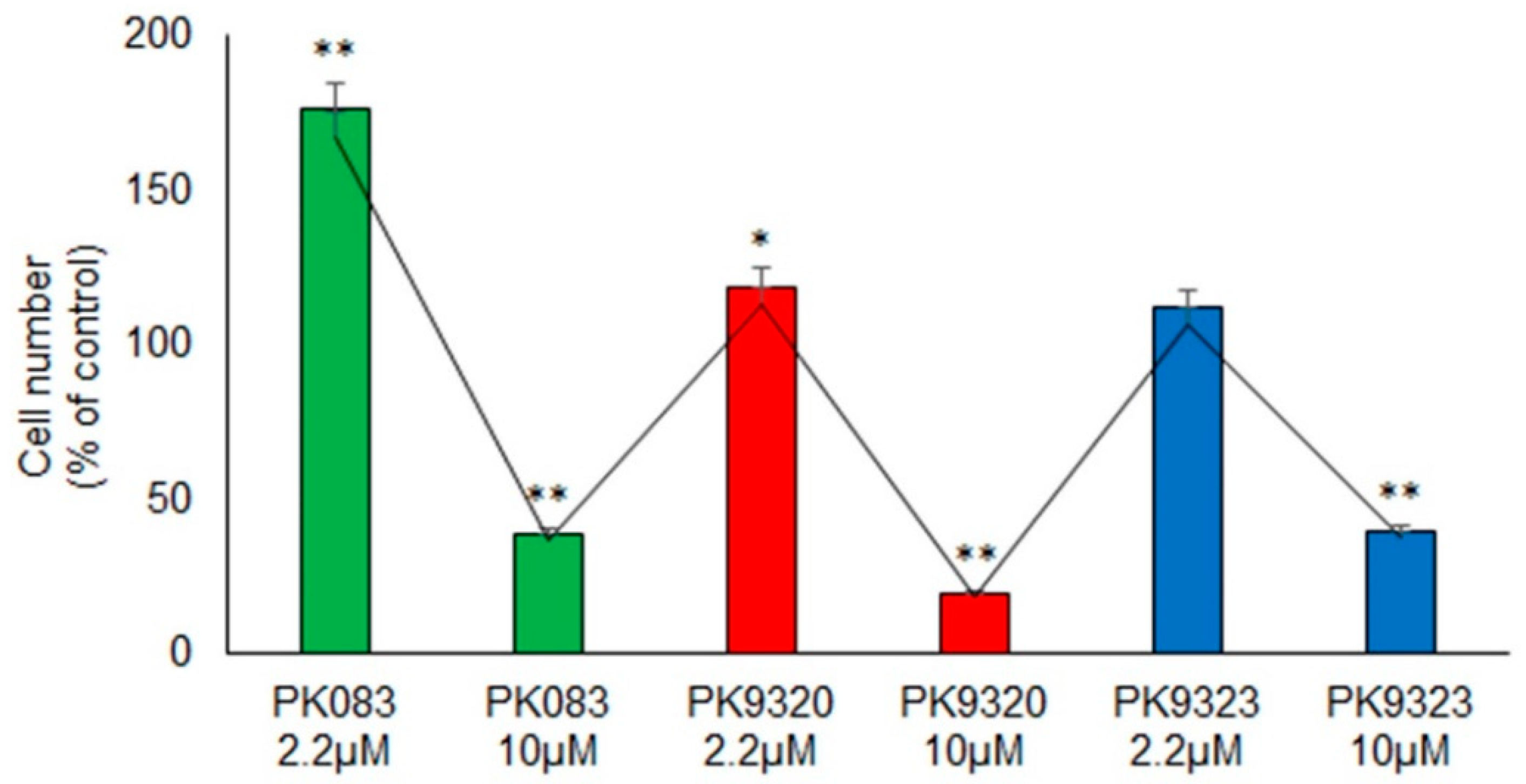
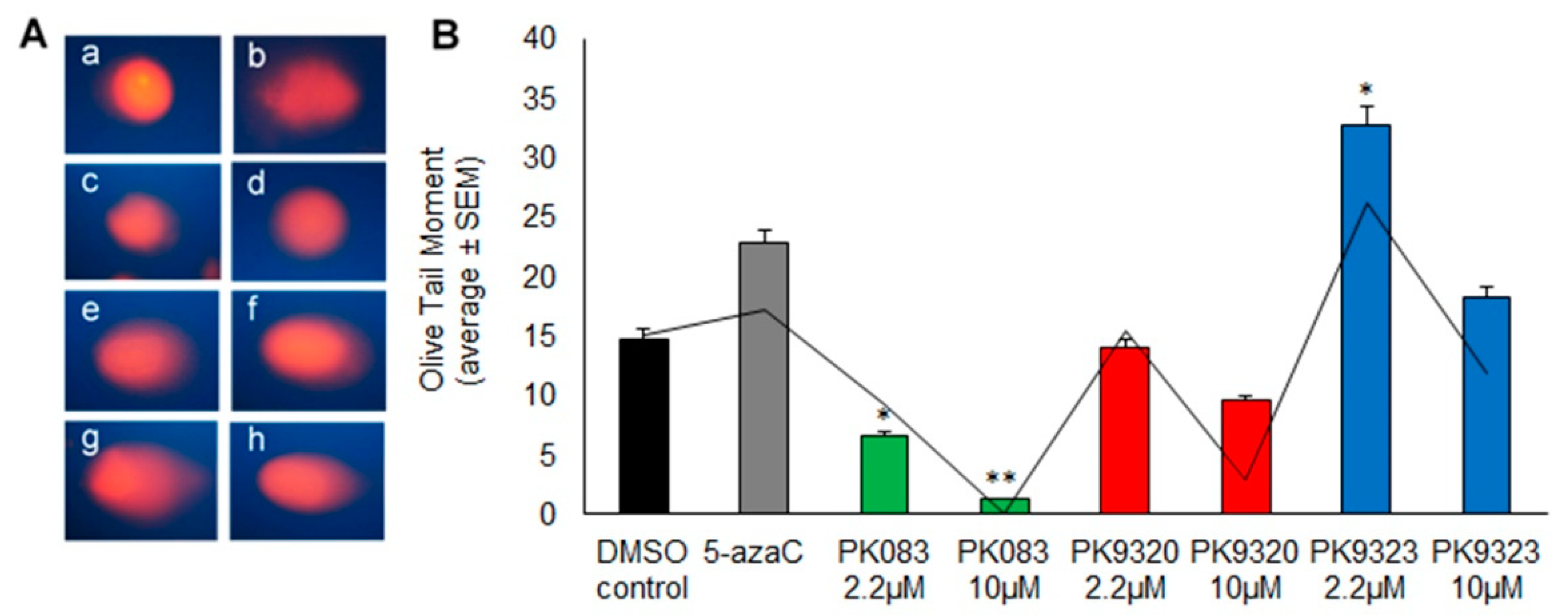
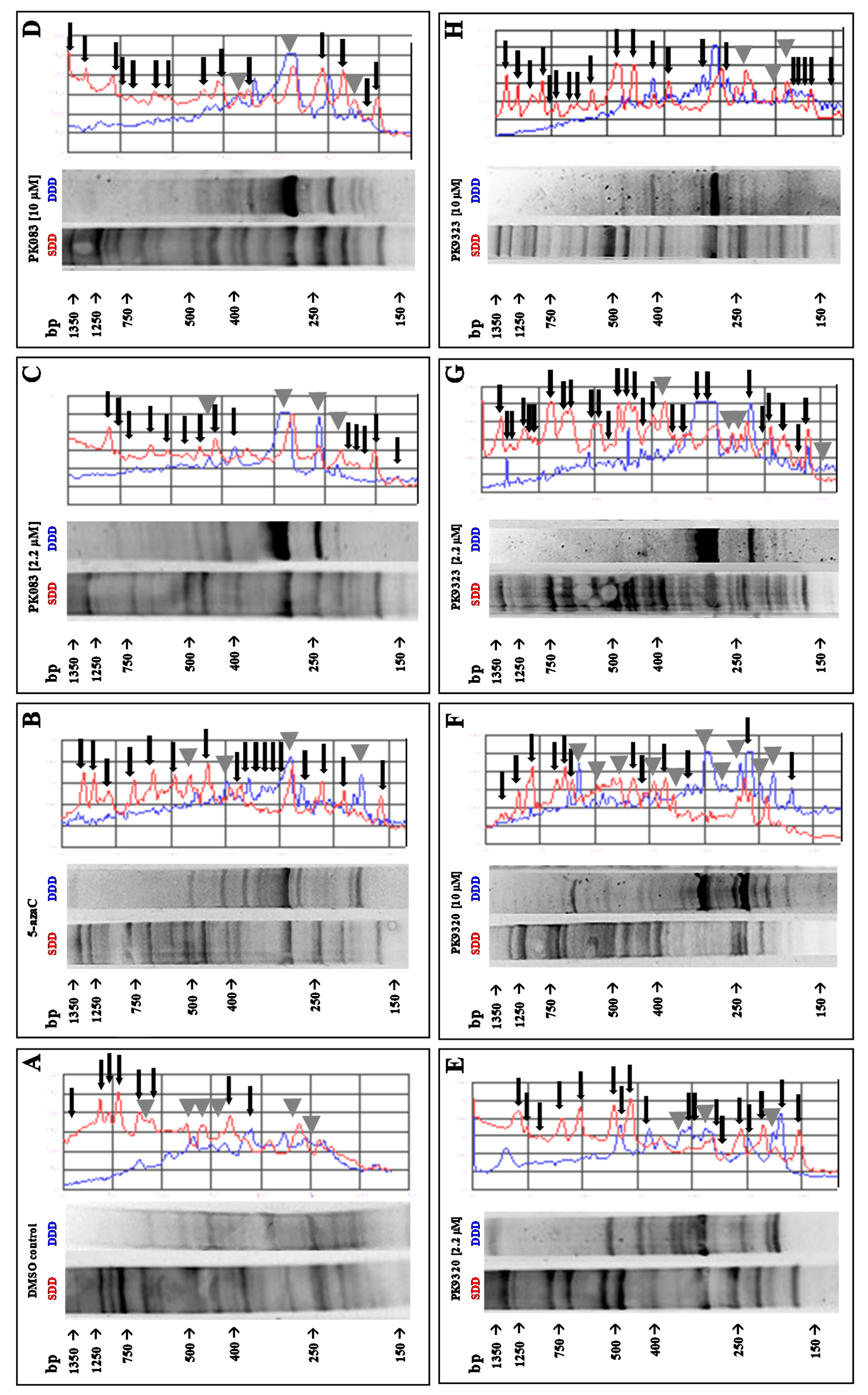
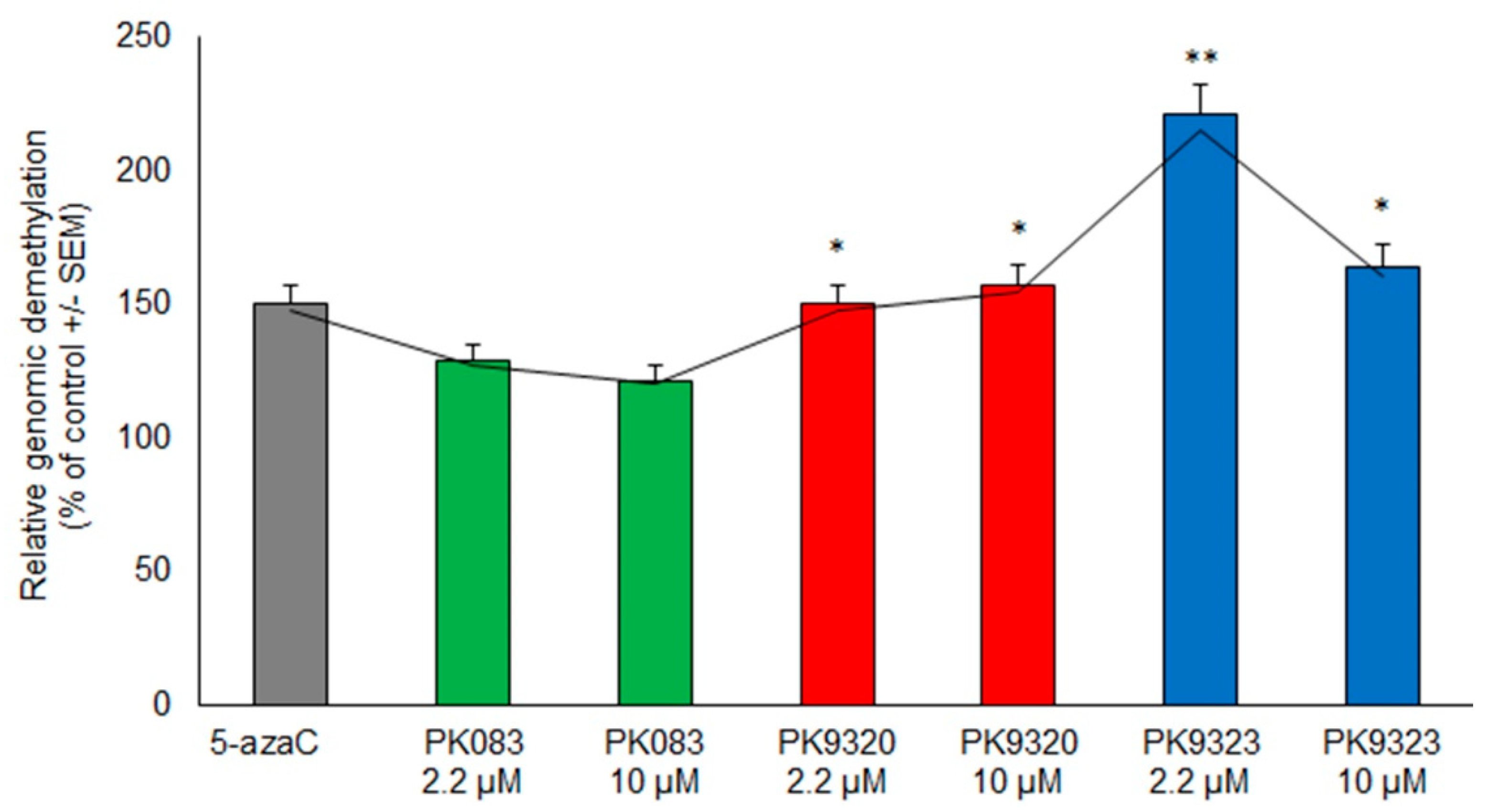
2. Results
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Treatments
4.2. Morphological Observations and Cell Number Evaluation
4.3. DNA Damage Assays
4.3.1. Western Blot for Phospho-γH2AX Histone
4.3.2. Alkaline Comet Assay
4.4. DNA Extraction and Methylation-Sensitive Arbitrarily Primed Polymerase Chain Reaction (MeSAP PCR)
4.5. Pharmacokinetic (PK) Studies
4.6. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jha, A.N.; Cheung, V.V.; Foulkes, M.E.; Hill, S.J.; Depledge, M.H. Detection of genotoxins in the marine environment: Adoption and evaluation of an integrated approach using the embryo-larval stages of the marine mussel, Mytilus edulis. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2000, 464, 213–228. [Google Scholar] [CrossRef]
- Goodwin, S.; Smith, A.F.; Horning, E.C. Alkaloids of Ochrosia elliptica Labill. J. Am. Chem. Soc. 1959, 81, 1903–1908. [Google Scholar] [CrossRef]
- Das, K.C.; Chakraborty, D.P.; Bose, P.K. Antifungal activity of some constituents of Murraya koenigii spreng. Experientia 1965, 21, 340. [Google Scholar] [CrossRef]
- Bashir, M.; Bano, A.; Ijaz, A.S.; Chaudhary, B.A. Recent developments and biological activities of n-substituted carbazole derivatives: A review. Molecules 2015, 20, 13496–13517. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.-Z.; Gan, L.-L.; Wang, H.; Zhou, C.-H. New progress in azole compounds as antimicrobial agents. Mini Rev. Med. Chem. 2017, 17, 122–166. [Google Scholar] [CrossRef] [PubMed]
- Issa, S.; Prandina, A.; Bedel, N.; Rongved, P.; Yous, S.; Le Borgne, M.; Bouaziz, Z. Carbazole scaffolds in cancer therapy: A review from 2012 to 2018. J. Enzyme Inhib. Med. Chem. 2019, 34, 1321–1346. [Google Scholar] [CrossRef] [Green Version]
- Agarwal, S.; Amin, K.S.; Jagadeesh, S.; Baishay, G.; Rao, P.G.; Barua, N.C.; Bhattacharya, S.; Banerjee, P.P. Mahanine restores RASSF1A expression by down-regulating DNMT1 and DNMT3B in prostate cancer cells. Mol. Cancer 2013, 12, 99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, H.J.; Yi, Y.W.; Hou, S.J.; Kim, H.J.; Kong, Y.; Bae, I.; Brown, M.L. Disruption of STAT3-DNMT1 interaction by SH-I-14 induces re-expression of tumor suppressor genes and inhibits growth of triple-negative breast tumor. Oncotarget 2017, 8, 83457. [Google Scholar] [CrossRef] [Green Version]
- Ghaedi, A.; Keshavarzi, M.; Ghafarian Bahraman, A.; Mohammadi-Bardbori, A. Selective cytochrome P450 1A1 but not 1B1 promoterCpG island DNA methylation by 6-formylindolo[3,2-b]carbazole (FICZ). J. Biochem. Mol. Toxicol. 2020, 34, e22414. [Google Scholar] [CrossRef]
- Ji, C.; Yue, S.; Gu, J.; Kong, Y.; Chen, H.; Yu, C.; Sun, Z.; Zhao, M. 2,7-Dibromocarbazole interferes with tube formation in HUVECs by altering Ang2 promoter DNA methylation status. Sci. Total Environ. 2019, 697, 134156. [Google Scholar] [CrossRef]
- Valovičová, Z.; Mesárošová, M.; Trilecová, L.; Hrubá, E.; Marvanová, S.; Krčmář, P.; Milcová, A.; Schmuczerová, J.; Vondráček, J.; Machala, M.; et al. Genotoxicity of 7H-dibenzo[c,g]carbazole and its methyl derivatives in human keratinocytes. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2012, 743, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Zhanataev, A.K.; Eremina, N.V.; Chayka, Z.V.; Kazey, V.I.; Andrianova, E.L.; Purmal, A.A.; Rydkina, E.B.; Durnev, A.D. Genotoxicity of two new carbazole derivatives with antifungal activity. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2017, 816–817, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Joerger, A.C.; Fersht, A.R. The p53 pathway: Origins, inactivation in cancer, and emerging therapeutic approaches. Annu. Rev. Biochem. 2016, 85, 375–404. [Google Scholar] [CrossRef]
- Boeckler, F.M.; Joerger, A.C.; Jaggi, G.; Rutherford, T.J.; Veprintsev, D.B.; Fersht, A.R. Targeted rescue of a destabilized mutant of p53 by an in silico screened drug. Proc. Natl. Acad. Sci. USA 2008, 105, 10360–10365. [Google Scholar] [CrossRef] [Green Version]
- Bauer, M.R.; Jones, R.N.; Tareque, R.K.; Springett, B.; Dingler, F.A.; Verduci, L.; Patel, K.J.; Fersht, A.R.; Joerger, A.C.; Spencer, J. A structure-guided molecular chaperone approach for restoring the transcriptional activity of the p53 cancer mutant Y220C. Future Med. Chem. 2019, 11, 2491–2504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bauer, M.R.; Krämer, A.; Settanni, G.; Jones, R.N.; Ni, X.; Khan Tareque, R.; Fersht, A.R.; Spencer, J.; Joerger, A.C. Targeting cavity-creating p53 cancer mutations with small-molecule stabilizers: The Y220X paradigm. ACS Chem. Biol. 2020, 15, 657–668. [Google Scholar] [CrossRef]
- Sharma, A.; Singh, K.; Almasan, A. Histone H2AX phosphorylation: A marker for DNA damage. Methods Mol. Biol. 2012, 920, 613–626. [Google Scholar] [CrossRef]
- Plappert-Helbig, U.; Libertini, S.; Frieauff, W.; Theil, D.; Martus, H.J. Gamma-H2AX immunofluorescence for the detection of tissue-specific genotoxicity in vivo. Environ. Mol. Mutagen. 2019, 60, 4–16. [Google Scholar] [CrossRef] [Green Version]
- Ceramella, J.; Iacopetta, D.; Barbarossa, A.; Caruso, A.; Grande, F.; Bonomo, M.G.; Mariconda, A.; Longo, P.; Carmela, S.; Sinicropi, M.S. Carbazole derivatives as kinase-targeting inhibitors for cancer treatment. Mini-Rev. Med. Chem. 2020, 20, 444–465. [Google Scholar] [CrossRef] [PubMed]
- Sciandrello, G.; Mauro, M.; Caradonna, F.; Catanzaro, I.; Saverini, M.; Barbata, G. Acrylamide catalytically inhibits topoisomerase II in V79 cells. Toxicol. Vitro 2010, 24, 830–834. [Google Scholar] [CrossRef]
- Caradonna, F.; Mauro, M. Role of the antioxidant defence system and telomerase in arsenic-induced genomic instability. Mutagenesis 2016, 31, 661–667. [Google Scholar] [CrossRef]
- Griffiths, E.A.; Carraway, H.E.; Chandhok, N.S.; Prebet, T. Advances in non-intensive chemotherapy treatment options for adults diagnosed with acute myeloid leukemia. Leuk. Res. 2020, 91, 106339. [Google Scholar] [CrossRef] [PubMed]
- Gabelova, A. 7H-Dibenzo[c,g]carbazole: Metabolic pathways and toxicity. Chem. Biol. Interact. 2020, 323, 109077. [Google Scholar] [CrossRef] [PubMed]
- Catanzaro, I.; Naselli, F.; Saverini, M.; Giacalone, A.; Montalto, G.; Caradonna, F. Cytochrome P450 2E1 variable number tandem repeat polymorphisms and health risks: A genotype-phenotype study in cancers associated with drinking and/or smoking. Mol. Med. Rep. 2012, 6, 416–420. [Google Scholar] [CrossRef] [Green Version]
- Naselli, F.; Catanzaro, I.; Bellavia, D.; Perez, A.; Sposito, L.; Caradonna, F. Role and importance of polymorphisms with respect to DNA methylation for the expression of CYP2E1 enzyme. Gene 2014, 536, 29–39. [Google Scholar] [CrossRef]
- Petrović, N.; Nakashidze, I.; Nedeljković, M. Breast cancer response to therapy: Can microRNAs lead the way? J. Mammary Gland Biol. Neoplasia 2021, 25, 1–22. [Google Scholar]
- Kinner, A.; Wu, W.; Staudt, C.; Iliakis, G. Gamma-H2AX in recognition and signaling of DNA double-strand breaks in the context of chromatin. Nucleic Acids Res. 2008, 36, 5678–5694. [Google Scholar] [CrossRef]
- Sciandrello, G.; Mauro, M.; Catanzaro, I.; Saverini, M.; Caradonna, F.; Barbata, G. Long-lasting genomic instability following arsenite exposure in mammalian cells: The role of reactive oxygen species. Environ. Mol. Mutagen. 2011, 52, 562–568. [Google Scholar] [CrossRef] [PubMed]
- Kaneti, J.; Georgieva, M.; Rangelov, M.; Philipova, I.; Vasileva, B.; Angelov, I.; Staneva, D.; Miloshev, G.; Bakalova, S. Biological activity of quinazoline analogues and molecular modeling of their interactions with G-quadruplexes. Biochim. Biophys. Acta Gen. Subj. 2021, 1865, 129773. [Google Scholar] [CrossRef]
- Choi, H.; Joe, S.; Nam, H. Development of tissue-specific age predictors using DNA methylation data. Genes 2019, 10, 888. [Google Scholar] [CrossRef] [Green Version]
- Ghasemi, S. Cancer’s epigenetic drugs: Where are they in the cancer medicines? Pharmacogenom. J. 2020, 20, 367–379. [Google Scholar] [CrossRef]
- Lu, C.; Wang, W.; El-Deiry, W.S. Non-genotoxic anti-neoplastic effects of ellipticine derivative NSC176327 in p53-deficient human colon carcinoma cells involve stimulation of p73. Cancer Biol. Ther. 2008, 7, 2039–2046. [Google Scholar] [CrossRef] [Green Version]
- Luparello, C.; Asaro, D.M.L.; Cruciata, I.; Hassell-Hart, S.; Sansook, S.; Spencer, J.; Caradonna, F. Cytotoxic activity of the histone deacetylase 3-selective inhibitor Pojamide on MDA-MB-231 triple-negative breast cancer cells. Int. J. Mol. Sci. 2019, 20, 804. [Google Scholar] [CrossRef] [Green Version]
- Librizzi, M.; Caradonna, F.; Cruciata, I.; Debski, J.; Sansook, S.; Dadlez, M.; Spencer, J.; Luparello, C. Molecular signatures associated with treatment of triple-negative MDA-MB231 breast cancer cells with histone deacetylase inhibitors JAHA and SAHA. Chem. Res. Toxicol. 2017, 30, 2187–2196. [Google Scholar] [CrossRef] [PubMed]
- Mannino, G.; Caradonna, F.; Cruciata, I.; Lauria, A.; Perrone, A.; Gentile, C. Melatonin reduces inflammatory response in human intestinal epithelial cells stimulated by interleukin-1β. J. Pineal Res. 2019, 67, e12598. [Google Scholar] [CrossRef]
- Caradonna, F.; Cruciata, I.; Schifano, I.; La Rosa, C.; Naselli, F.; Chiarelli, R.; Perrone, A.; Gentile, C. Methylation of cytokines gene promoters in IL-1β-treated human intestinal epithelial cells. Inflamm. Res. 2018, 67, 327–337. [Google Scholar] [CrossRef] [PubMed]
- Librizzi, M.; Chiarelli, R.; Bosco, L.; Sansook, S.; Gascon, J.M.; Spencer, J.; Caradonna, F.; Luparello, C. The histone deacetylase inhibitor JAHA down-regulates pERK and global DNA methylation in MDA-MB231 breast cancer cells. Materials 2015, 8, 7041–7047. [Google Scholar] [CrossRef] [Green Version]
- Bordoni, L.; Nasuti, C.; Mirto, M.; Caradonna, F.; Gabbianelli, R. Intergenerational effect of early life exposure to permethrin: Changes in global DNA methylation and in Nurr1 gene expression. Toxics 2015, 3, 451–461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hassell-Hart, S.; Runcie, A.; Krojer, T.; Doyle, J.; Lineham, E.; Ocasio, C.A.; Neto, B.A.D.; Fedorov, O.; Marsh, G.; Maple, H.; et al. Synthesis and biological investigation of (+)-JD1, an Organometallic BET Bromodomain Inhibitor. Organometallics 2020, 39, 408–416. [Google Scholar] [CrossRef]
- Miret, S.; De Groene, E.M.; Klaffke, W. Comparison of in vitro assays of cellular toxicity in the human hepatic cell line HepG2. J. Biomol. Screen. 2006, 11, 184–193. [Google Scholar] [CrossRef]
- Bauer, M.R.; Jones, R.N.; Baud, M.G.; Wilcken, R.; Boeckler, F.M.; Fersht, A.R.; Joerger, A.C.; Spencer, J. Harnessing fluorine-sulfur contacts and multipolar interactions for the design of p53 mutant Y220C rescue drugs. ACS Chem. Biol. 2016, 19, 2265–2274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luparello, C.; Cruciata, I.; Joerger, A.C.; Ocasio, C.A.; Jones, R.; Tareque, R.K.; Bagley, M.C.; Spencer, J.; Walker, M.; Austin, C.; et al. Genotoxicity and Epigenotoxicity of Carbazole-Derived Molecules on MCF-7 Breast Cancer Cells. Int. J. Mol. Sci. 2021, 22, 3410. https://doi.org/10.3390/ijms22073410
Luparello C, Cruciata I, Joerger AC, Ocasio CA, Jones R, Tareque RK, Bagley MC, Spencer J, Walker M, Austin C, et al. Genotoxicity and Epigenotoxicity of Carbazole-Derived Molecules on MCF-7 Breast Cancer Cells. International Journal of Molecular Sciences. 2021; 22(7):3410. https://doi.org/10.3390/ijms22073410
Chicago/Turabian StyleLuparello, Claudio, Ilenia Cruciata, Andreas C. Joerger, Cory A. Ocasio, Rhiannon Jones, Raysa Khan Tareque, Mark C. Bagley, John Spencer, Martin Walker, Carol Austin, and et al. 2021. "Genotoxicity and Epigenotoxicity of Carbazole-Derived Molecules on MCF-7 Breast Cancer Cells" International Journal of Molecular Sciences 22, no. 7: 3410. https://doi.org/10.3390/ijms22073410
APA StyleLuparello, C., Cruciata, I., Joerger, A. C., Ocasio, C. A., Jones, R., Tareque, R. K., Bagley, M. C., Spencer, J., Walker, M., Austin, C., Ferrara, T., D′Oca, P., Bellina, R., Branni, R., & Caradonna, F. (2021). Genotoxicity and Epigenotoxicity of Carbazole-Derived Molecules on MCF-7 Breast Cancer Cells. International Journal of Molecular Sciences, 22(7), 3410. https://doi.org/10.3390/ijms22073410