Garcinol—A Natural Histone Acetyltransferase Inhibitor and New Anti-Cancer Epigenetic Drug
Abstract
1. Introduction
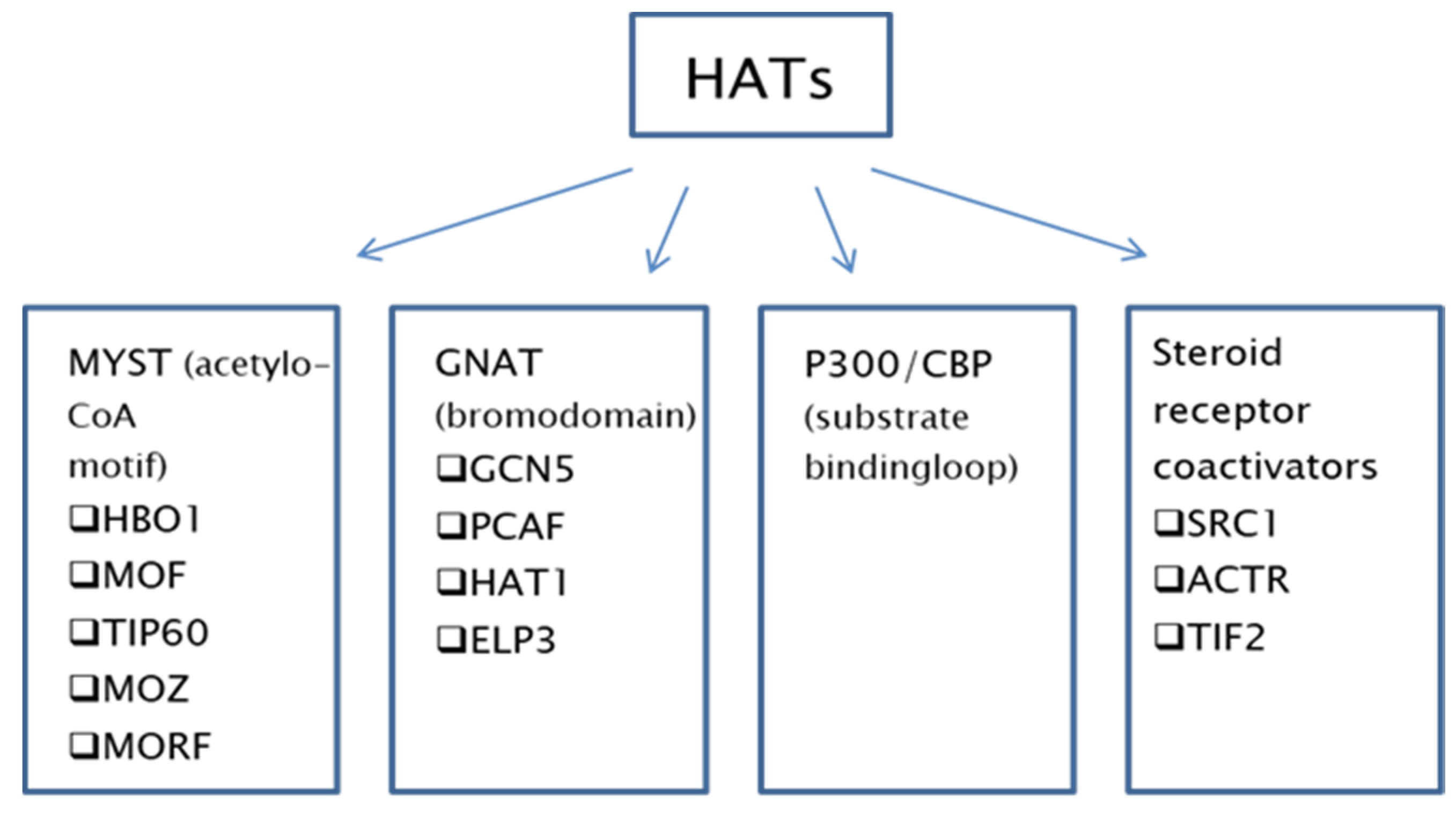
2. Histone Acetyltransferases
| Pathway/ Element | Pathway Role | Cell Line | Inhibitor/Silencing of p300 or CBP | Reference |
|---|---|---|---|---|
| ↓Cyclin A2 ↑CDKN1A | Cell cycle | Human melanoma | Silencing | [27] |
| ↑NKG2D-L | Innate immune response NK cells | HUVEC,911, WI-38, SKOV3, HepG2, MCF7, MDA-231 | Inhibition (C646) | [28] |
| ↓PD-L1 | Innate immune response CD8+ T cells | Prostate cancer cell lines: DU145, Pc3, 22Rv1, LNCaP | Inhibition (A485) | [29] |
| ↓PIK3R1/P50 | Cell proliferation, differentiation and survival | Primary breast cancer patient samples, cell lines MCf7, MDA231, ZR75-1, T47D, nude balb/c mice | Not applicable | [30] |
| E2F1 | Double strand break repair | MEFs from FVB mice (E2f1Knock in) | Not applicable | [31] |
3. Garcinol
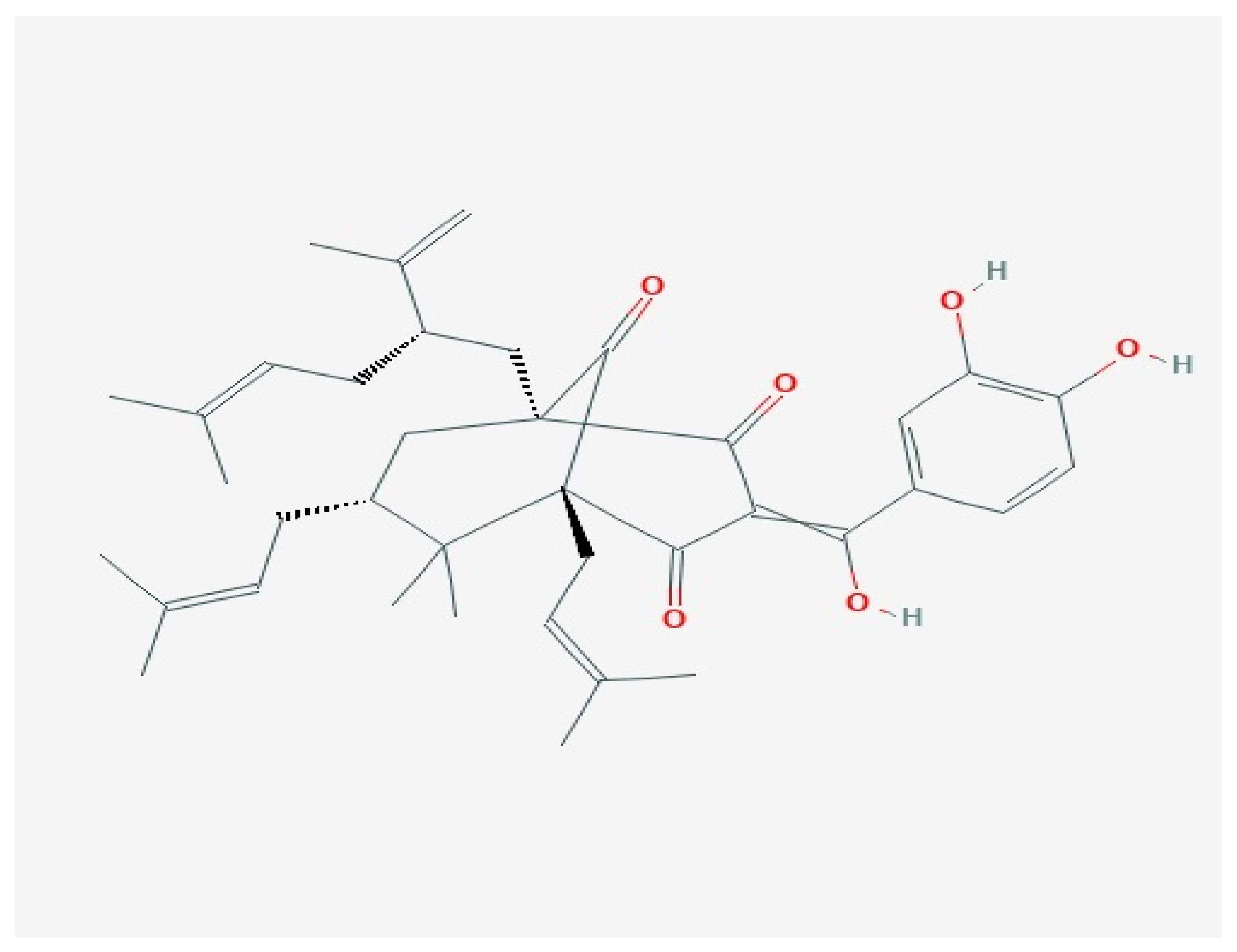
3.1. Anti-Cancer Properties of Garcinol
3.2. Garcinol as an Effective Inhibitor of HATs and Putative Epigenetic Drug
3.3. Garcinol Affects miRNA Expression
| miRNA | Cell Type | Effect of Garcinol | Reference |
|---|---|---|---|
| miR-200b, miR-200c,let-7 | MDA-MB-231 BT-549 breast cancer lines | Upregulated- reversed EMT to MET | [56] |
| PANC-1-SP pancreatic cancer cells | Upregulated- caused ↓Notch1 and ↓Oct4 | [58] | |
| A549 chemo-resistant lung cancer line | Upregulated- reversed EMT | [65] | |
| miR-218, miR-101, miR-205 | A549 chemo-resistant lung cancer line | Upregulated-reversed EMT at a lesser degree than miR-200 and let-7 | [65] |
| miR-181 | glioblastoma | Upregulated-caused ↓STAT, ↓migration | [66] |
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Egger, G.; Liang, G.; Aparicio, A.; Jones, P.A. Epigenetics in human disease and prospects for epigenetic therapy. Nat. Cell Biol. 2004, 429, 457–463. [Google Scholar] [CrossRef]
- Berger, S.L.; Kouzarides, T.; Shiekhattar, R.; Shilatifard, A. An operational definition of epigenetics. Genes Dev. 2009, 23, 781–783. [Google Scholar] [CrossRef]
- Reik, W.; Dean, W.; Walter, J. Epigenetic Reprogramming in Mammalian Development. Science 2001, 293, 1089–1093. [Google Scholar] [CrossRef] [PubMed]
- Morgan, H.D.; Sutherland, H.G.; Martin, D.I.; Whitelaw, E. Epigenetic inheritance at the agouti locus in the mouse. Nat. Genet. 1999, 23, 314–318. [Google Scholar] [CrossRef] [PubMed]
- Perri, F.; Longo, F.; Giuliano, M.; Sabbatino, F.; Favia, G.; Ionna, F.; Addeo, R.; Scarpati, G.D.V.; Di Lorenzo, G.; Pisconti, S. Epigenetic control of gene expression: Potential implications for cancer treatment. Crit. Rev. Oncol. 2017, 111, 166–172. [Google Scholar] [CrossRef] [PubMed]
- Barnes, C.E.; English, D.M.; Cowley, S.M. Acetylation & Co: An expanding repertoire of histone acylations regulates chromatin and transcription. Essays Biochem. 2019, 63, 97–107. [Google Scholar] [CrossRef] [PubMed]
- Eckschlager, T.; Plch, J.; Stiborova, M.; Hrabeta, J. Histone Deacetylase Inhibitors as Anticancer Drugs. Int. J. Mol. Sci. 2017, 18, 1414. [Google Scholar] [CrossRef]
- Farria, A.; Li, W.; Dent, S.Y.R. KATs in cancer: Functions and therapies. Oncogene 2015, 34, 4901–4913. [Google Scholar] [CrossRef]
- Kleff, S.; Andrulis, E.D.; Anderson, C.W.; Sternglanz, R.J. Identification of a gene encoding a yeast histone H4 acetyl-transferase. Biol. Chem. 1995, 270, 24674–24677. [Google Scholar] [CrossRef]
- Sun, X.J.; Man, N.; Tan, Y.; Nimer, S.D.; Wang, L. The Role of Histone Acetyltransferases in Normal and Malignant Hem-atopoiesis. Front. Oncol. 2015, 5, 108–118. [Google Scholar] [CrossRef]
- Yang, X.-J. The diverse superfamily of lysine acetyltransferases and their roles in leukemia and other diseases. Nucleic Acids Res. 2004, 32, 959–976. [Google Scholar] [CrossRef]
- Roth, S.Y.; Denu, J.M.; Allis, C.D. Histone Acetyltransferases. Annu. Rev. Biochem. 2001, 70, 81–120. [Google Scholar] [CrossRef]
- Bowers, E.M.; Yan, G.; Mukherjee, C.; Orry, A.; Wang, L.; Holbert, M.A.; Crump, N.T.; Hazzalin, C.A.; Liszczak, G.; Yuan, H.; et al. Virtual Ligand Screening of the p300/CBP Histone Acetyltransferase: Identification of a Selective Small Molecule Inhibitor. Chem. Biol. 2010, 17, 471–482. [Google Scholar] [CrossRef]
- Voss, A.K.; Thomas, T. MYST family histone acetyltransferases take center stage in stem cells and development. BioEssays 2009, 31, 1050–1061. [Google Scholar] [CrossRef]
- Varga, J.; Korbai, S.; Neller, A.; Zsindely, N.; Bodai, L. Hat1 acetylates histone H4 and modulates the transcriptional program in Drosophila embryogenesis. Sci. Rep. 2019, 9, 17973. [Google Scholar] [CrossRef]
- Huang, M.; Huang, J.; Zheng, Y.; Sun, Q. Histone acetyltransferase inhibitors: An overview in synthesis, structure-activity relationship and molecular mechanism. Eur. J. Med. Chem. 2019, 178, 259–286. [Google Scholar] [CrossRef] [PubMed]
- Avvakumov, N.; Cote, J. The MYST family of histone acetyltransferases and their intimate links to cancer. Oncogene 2007, 26, 5395–5407. [Google Scholar] [CrossRef] [PubMed]
- Utley, R.T.; Côté, J. The MYST Family of Histone Acetyltransferases. Curr. Top. Microbiol. Immunol. 2003, 274, 203–236. [Google Scholar] [CrossRef] [PubMed]
- Voss, A.K.; Thomas, T. Histone Lysine and Genomic Targets of Histone Acetyltransferases in Mammals. BioEssays 2018, 40, e1800078. [Google Scholar] [CrossRef] [PubMed]
- Chan, H.M.; La Thangue, N.B. p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J. Cell Sci. 2001, 114, 2363–2373. [Google Scholar]
- Spencer, T.E.; Jenster, G.; Burcin, M.M.; Allis, C.D.; Zhou, J.; Mizzen, C.A.; McKenna, N.J.; Onate, S.A.; Tsai, S.Y.; Tsai, M.-J.; et al. Steroid receptor coactivator-1 is a histone acetyltransferase. Nat. Cell Biol. 1997, 389, 194–198. [Google Scholar] [CrossRef]
- Wapenaar, H.; Dekker, F.J. Histone acetyltransferases: Challenges in targeting bi-substrate enzymes. Clin. Epigenetics 2016, 8, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.; Zucconi, B.E.; Wu, M.; Nocco, S.E.; Meyers, D.J.; McGee, J.S.; Venkatesh, S.; Cohen, D.L.; Gonzalez, E.C.; Ryu, B.; et al. MITF Expression Predicts Therapeutic Vulnerability to p300 Inhibition in Human Melano-ma. Cancer Res. 2019, 79, 649–2661. [Google Scholar] [CrossRef]
- Zhang, X.; Zegar, T.; Lucas, A.; Morrison-Smith, C.; Knox, T.; French, C.A.; Knapp, S.; Müller, S.; Siveke, J.T. Therapeutic targeting of p300/CBP HAT domain for the treatment of NUT midline carcinoma. Oncogene 2020, 39, 4770–4779. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; He, Y.; Robinson, V.; Yang, Z.; Hessler, P.; Lasko, L.M.; Lu, X.; Bhathena, A.; Lai, A.; Uziel, T.; et al. Targeting Lineage-specific MITF Pathway in Human Melanoma Cell Lines by A-485, the Selective Small-molecule Inhibitor of p300/CBP. Mol. Cancer Ther. 2018, 17, 2543–2550. [Google Scholar] [CrossRef]
- Chi, Y.; Xue, J.; Huang, S.; Xiu, B.; Su, Y.; Wang, W.; Guo, R.; Wang, L.; Li, L.; Shao, Z.; et al. CapG promotes resistance to paclitaxel in breast cancer through transactivation of PIK3R1/P50. Theranostics 2019, 9, 6840–6855. [Google Scholar] [CrossRef] [PubMed]
- Brugge, J.; Hung, M.C.; Mills, G.B. a new mutational activation in the PI3K pathway. Cancer Cell 2007, 12, 104–107. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, H.R.; Lindén, S.K. The aspirin metabolite salicylate inhibits lysine acetyltransferases and MUC1 induced epithelial to mesenchymal transition. Sci. Rep. 2017, 7, 5626. [Google Scholar] [CrossRef] [PubMed]
- Lasko, L.M.; Jakob, C.G.; Edalji, R.P.; Qiu, W.; Montgomery, D.; Digiammarino, E.L.; Hansen, T.M.; Risi, R.M.; Frey, R.; Manaves, V.; et al. Discovery of a selective catalytic p300/CBP inhibitor that targets lineage-specific tumours. Nat. Cell Biol. 2017, 550, 128–132. [Google Scholar] [CrossRef] [PubMed]
- Kong, D.; Ying, B.; Zhang, J.; Ying, H. PCAF regulates H3 phosphorylation and promotes autophagy in osteosarcoma cells. Biomed. Pharmacother. 2019, 118, 109395. [Google Scholar] [CrossRef]
- Falahi, F.; Ssro, A.; Blancafort, P. Epigenome engeneering in cancer: Fairytale or a realistic path to the clinic? Front. Oncol. 2015, 6, 22. [Google Scholar]
- Sauer, M.; Schuldner, M.; Hoffmann, N.; Cetintas, A.; Reiners, K.S.; Shatnyeva, O.; Hallek, M.; Hansen, H.P.; Gasser, S.; Von Strandmann, E.P. CBP/p300 acetyltransferases regulate the expression of NKG2D ligands on tumor cells. Oncogene 2017, 36, 933–941. [Google Scholar] [CrossRef]
- Liu, J.; He, D.; Cheng, L.; Huang, C.; Zhang, Y.; Rao, X.; Kong, Y.; Li, C.; Zhang, Z.; Liu, J.; et al. p300/CBP inhibition enhances the efficacy of programmed death-ligand 1 blockade treatment in prostate cancer. Oncogene 2020, 39, 3939–3951. [Google Scholar] [CrossRef]
- Manickavinayaham, S.; Vélez-Cruz, R.; Biswas, A.K.; Bedford, E.; Klein, B.J.; Kutateladze, T.G.; Liu, B.; Bedford, M.T.; Johnson, D.G. E2F1 acetylation directs p300/CBP-mediated histone acetylation at DNA double-strand breaks to facilitate repair. Nat. Commun. 2019, 10, 1–14. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, E.J.; Shi, C.; Mou, P.K.; Zhang, B.; Wu, C.; Lyu, J.; Shim, J.S. Histone Acetyltransferase (HAT) P300/CBP Inhibitors Induce Synthetic Lethality in PTEN-Deficient Colorectal Cancer Cells through Destabilizing AKT. Int. J. Biol. Sci. 2020, 16, 1774–1784. [Google Scholar] [CrossRef] [PubMed]
- Fiorentino, F.; Mai, A.; Rotili, D. Lysine acetyltransferase inhibitors: Structure-activity relationships and potential therapeutic implications. Future Med. Chem. 2018, 10, 1067–1091. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Guo, C.; Zhang, X.; Wang, X.; Ma, A. Garcinol acts as an antineoplastic agent in human gastric cancer by inhibiting the PI3K/AKT signaling pathway. Oncol. Lett. 2020, 20, 667–676. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, V.; Tuli, H.S.; Kaur, J.; Aggarwal, D.; Parashar, G.; Parashar, N.C.; Kulkarni, S.; Kaur, G.; Sak, K.; Kumar, M.; et al. Garcinol Exhibits Anti-Neoplastic Effects by Targeting Diverse Oncogenic Factors in Tumor Cells. Biomedicines 2020, 8, 103. [Google Scholar] [CrossRef]
- Garcinol structure. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/Garcinol (accessed on 1 February 2021).
- Schobert, R.; Biersack, B. Chemical and Biological Aspects of Garcinol and Isogarcinol: Recent Developments. Chem. Biodivers. 2019, 16, e1900366. [Google Scholar] [CrossRef] [PubMed]
- Han, C.-M.; Zhou, X.-Y.; Cao, J.; Zhang, X.-Y.; Chen, X. 13,14-Dihydroxy groups are critical for the anti-cancer effects of garcinol. Bioorganic Chem. 2015, 60, 123–129. [Google Scholar] [CrossRef]
- Zhou, X.-Y.; Cao, J.; Han, C.-M.; Li, S.-W.; Zhang, C.; Du, Y.-D.; Zhou, Q.-Q.; Zhang, X.-Y.; Chen, X. The C8 side chain is one of the key functional group of Garcinol for its anti-cancer effects. Bioorganic Chem. 2017, 71, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Balasubramanyam, K.; Altaf, M.; Varier, R.A.; Swaminathan, V.; Ravindran, A.; Sadhale, P.P.; Kundu, T.K. Polyisoprenylated Benzophenone, Garcinol, a Natural Histone Acetyltransferase Inhibitor, Represses Chromatin Transcription and Alters Global Gene Expression. J. Biol. Chem. 2004, 279, 33716–33726. [Google Scholar] [CrossRef]
- Yu, S.-Y.; Liao, C.-H.; Chien, M.-H.; Tsai, T.-Y.; Lin, J.-K.; Weng, M.-S. Induction of p21Waf1/Cip1 by Garcinol via Downregulation of p38-MAPK Signaling in p53-Independent H1299 Lung Cancer. J. Agric. Food Chem. 2014, 62, 2085–2095. [Google Scholar] [CrossRef]
- Zhang, G.; Fu, J.; Su, Y.; Zhang, X. Opposite Effects of Garcinol on Tumor Energy Metabolism in Oral Squamous Cell Carcinoma Cells. Nutr. Cancer 2019, 71, 1403–1411. [Google Scholar] [CrossRef]
- Zhang, M.; Lu, Q.; Hou, H.; Sun, D.; Chen, M.; Ning, F.; Wu, P.; Wei, D.; Duan, Y.; Pan, Y.; et al. Garcinol inhibits the proliferation of endometrial cancer cells by inducing cell cycle arrest. Oncol. Rep. 2020, 45, 630–640. [Google Scholar] [CrossRef]
- Parasamka, M.A.; Gupta, S.V. Garcinol inhibits cell proliferation and promotes apoptosis in pancreatic adenocarcinoma cells. Nutr. Cancer 2011, 63, 456–465. [Google Scholar] [CrossRef] [PubMed]
- Paul, B.; Gaonkar, R.H.; Mukhopadhyay, R.; Ganguly, S.; Debnath, M.C.; Mukherjee, B. Garcinol-loaded novel cationic nanoliposomes: In vitro and in vivo study against B16F10 melanoma tumor model. Nanomedicine 2019, 14, 2045–2065. [Google Scholar] [CrossRef]
- Kim, S.; Seo, S.U.; Min, K.J.; Woo, S.M.; Nam, J.O.; Kubatka, P.; Kim, S.; Park, J.W.; Kwon, T.K. Garcinol EnhancesTRAIL-Induced Apoptotic Cell Death through Up-Regulation of DR5 and Down-Regulation of c-FLIP Expression. Molecules 2018, 23, 1614. [Google Scholar] [CrossRef]
- Bai, J.; Kwok, W.C.; Thiery, J.-P. Traditional Chinese Medicine and regulatory roles on epithelial–mesenchymal transitions. Chin. Med. 2019, 14, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Parasramka, M.A.; Ali, S.; Banerjee, S.; Deryavoush, T.; Sarkar, F.H.; Gupta, S. Garcinol sensitizes human pancreatic adenocarcinoma cells to gemcitabine in association with microRNA signatures. Mol. Nutr. Food Res. 2013, 57, 235–248. [Google Scholar] [CrossRef] [PubMed]
- Ranjbarnejad, T.; Saidijam, M.; Tafakh, M.S.; Pourjafar, M.; Talebzadeh, F.; Najafi, R. Garcinol exhibits anti-proliferative activities by targeting microsomal prostaglandin E synthase-1 in human colon cancer cells. Hum. Exp. Toxicol. 2016, 36, 692–700. [Google Scholar] [CrossRef]
- Duan, Y.-T.; Yang, X.-A.; Fang, L.-Y.; Wang, J.-H.; Liu, Q. Anti-proliferative and anti-invasive effects of garcinol from Garcinia indica on gallbladder carcinoma cells. Die Pharm. 2018, 73, 413–417. [Google Scholar]
- Saadat, N.; Akhtar, S.; Goja, A.; Razalli, N.H.; Geamanu, A.; David, D.; Shen, Y.; Gupta, S.V. Dietary Garcinol Arrests Pancreatic Cancer in p53 and K-ras Conditional Mutant Mouse Model. Nutr. Cancer 2018, 70, 1075–1087. [Google Scholar] [CrossRef] [PubMed]
- Tu, S.H.; Chiou, Y.S.; Kalyanam, N.; Ho, C.T.; Chen, L.C.; Pan, M.H. Garcinol sensitizes breast cancer cells to Taxol through the suppression of caspase-3/iPLA2 and NF-κB/Twist1 signaling pathways in a mouse 4T1breast tumor model. Food Funct. 2017, 8, 1067. [Google Scholar] [CrossRef]
- Ahmad, A.; Sarkar, S.H.; Bitar, B.; Ali, S.; Aboukameel, A.; Sethi, S.; Li, Y.; Bao, B.; Kong, D.; Banerjee, S.; et al. Garcinol Regulates EMT and Wnt Signaling Pathways In Vitro and In Vivo, Leading to Anticancer Activity against Breast Cancer Cells. Mol. Cancer Ther. 2012, 11, 2193–2201. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, L.; Ho, C.-T.; Zhang, K.; Liu, Q.; Zhao, H. Garcinol from Garcinia indica Downregulates Cancer Stem-like Cell Biomarker ALDH1A1 in Nonsmall Cell Lung Cancer A549 Cells through DDIT3 Activation. J. Agric. Food Chem. 2017, 65, 3675–3683. [Google Scholar] [CrossRef]
- Huang, C.C.; Lin, C.M.; Huang, Y.J.; Wei, L.; Ting, L.L.; Kuo, C.C.; Hsu, C.; Chiou, J.F.; Wu, A.T.; Lee, W.H. Gar-cinoldownregulates Notch1 signaling via modulating miR-200c and suppresses oncogenic properties of PANC-1 cancer stem-like cells. Biotechnol. Appl. Biochem. 2017, 64, 165–173. [Google Scholar] [CrossRef] [PubMed]
- Collins, H.M.; Abdelghany, M.K.; Messmer, M.; Yue, B.; Deeves, S.E.; Kindle, K.B.; Mantelingu, K.; Aslam, A.; Winkler, G.S.; Kundu, T.K.; et al. Differential effects of garcinol and curcumin on histone and p53 modifications in tumour cells. BMC Cancer 2013, 13, 37. [Google Scholar] [CrossRef]
- Du, T.; Nagai, Y.; Xiao, Y.; Greene, M.I.; Zhang, H. Lysosome-dependent p300/FOXP3 degradation and limits Treg cell functions and enhances targeted therapy against cancers. Exp. Mol. Pathol. 2013, 95, 38–45. [Google Scholar] [CrossRef]
- Wang, J.; Wu, M.; Zheng, D.; Zhang, H.; Lv, Y.; Zhang, L.; Tan, H.-S.; Zhou, H.; Lao, Y.-Z.; Xu, H.-X. Garcinol inhibits esophageal cancer metastasis by suppressing the p300 and TGF-β1 signaling pathways. Acta Pharmacol. Sin. 2019, 41, 82–92. [Google Scholar] [CrossRef]
- Srivastava, S.; Mohibi, S.; Mirza, S.; Band, H.; Band, V. Epidermal Growth Factor Receptor activation promotes ADA3 acetylation through the AKT-p300 pathway. Cell Cycle 2017, 16, 1515–1525. [Google Scholar] [CrossRef] [PubMed]
- Bertoli, G.; Cava, C.; Castiglioni, I. MicroRNAs: New Biomarkers for Diagnosis, Prognosis, Therapy Prediction and Therapeutic Tools for Breast Cancer. Theranostics 2015, 5, 1122–1143. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.; Yadav, T.; Rani, V. Exploring miRNA based approaches in cancer diagnostics and therapeutics. Crit. Rev. Oncol. 2016, 98, 12–23. [Google Scholar] [CrossRef]
- Farhan, M.; Malik, A.; Ullah, M.F.; Afaq, S.; Faisal, M.; Farooqi, A.A.; Biersack, B.; Schobert, R.; Ahmad, A. Garcinol Sensitizes NSCLC Cells to Standard Therapies by Regulating EMT-Modulating miRNAs. Int. J. Mol. Sci. 2019, 20, 800. [Google Scholar] [CrossRef]
- Liu, H.-W.; Lee, P.M.; Bamodu, O.A.; Su, Y.-K.; Fong, I.-H.; Yeh, C.-T.; Chien, M.-H.; Kan, I.-H.; Lin, C.M. Enhanced Hsa-miR-181d/p-STAT3and Hsa-miR-181d/p-STAT5A Ratios Mediate the Anticancer effect of Garcinol in STAT3/5A-Addicted Glioblastoma. Cancers 2019, 11, 1888. [Google Scholar] [CrossRef] [PubMed]
| Inhibitor | Inhibited HAT |
|---|---|
| Bi-substrate inhibitors | Non-selective |
| Garcinol | P300 |
| Curcumin | P300 |
| Anacardic acid | Non-selective |
| TH1834 | TIP60 |
| Benzylidene barbituric acid | P300 |
| Isothiazolones | Various |
| Thiazinesulfonamide | P300 |
| C646 | P300 |
| ICG-001 | CBP/β-catenin |
| Ischemin (bromodomain inhibitor) | Gcn5, PCAF, p300/CBP |
| Cyclicpeptide bromodomain inhibitors | Targets p53 |
| N1-aryl-propane-1,3-diamine derivatives (bromodomain inhibitors) | Targets HIF-1 |
| A-485 | P300/CBP (high selectivity) |
| Effect | Type of Cancer (Cell Line) | Garcinol Concentration |
|---|---|---|
| Increased apoptosis | Melanoma, glioblastoma, cervical cancer, breast cancer, leukaemia, lung cancer, hepatocellular carcinoma, pancreatic cancer, colon cancer, prostate cancer | 2.5–50 μM |
| ↑caspase-3,↑caspase-9 | Melanoma, leukaemia, hepatocellular carcinoma, pancreatic cancer, colon cancer | 0–50 μM |
| Cell cycle arrest, ↓cyclins B,D1, D3, and E | Breast cancer, lung cancer, hepatocellular carcinoma | 0–50 μM, 500ppm |
| ↑Bax, ↑Bad, ↓Bcl-2, ↓Bcl-xl | Melanoma, glioblastoma, breast cancer, leukaemia, hepatocellular carcinoma, colon cancer | 0–50 μM |
| ↓NF-κBsignalling pathway | Oral squamous cell carcinoma, breast cancer, pancreatic cancer, prostate cancer | 0–50 μM |
| ↓MMP2,↓MMP9 | Breast cancer, pancreatic cancer, gallbladder cancer, colon cancer, prostate cancer | 0–30 μM |
| ↓p-STAT3 and ↓STAT 3 signalling pathway | Breast cancer, hepatocellular carcinoma, pancreatic cancer, prostate cancer | 0–50 μM |
| ↓VEGF | Oral squamous cell carcinoma, breast cancer, hepatocellular carcinoma, pancreatic cancer, colon cancer, prostate cancer | 0–25 μM |
| ↓IL-6 | Hepatocellular carcinoma, pancreatic cancer, prostate cancer | 0–25 μM |
| ↑mi RNA | Glioblastoma, breast cancer, lung cancer, pancreatic cancer | 0–40 μM |
| HAT inhibition | Oesophageal cancer, breast cancer | 0–50 μM |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kopytko, P.; Piotrowska, K.; Janisiak, J.; Tarnowski, M. Garcinol—A Natural Histone Acetyltransferase Inhibitor and New Anti-Cancer Epigenetic Drug. Int. J. Mol. Sci. 2021, 22, 2828. https://doi.org/10.3390/ijms22062828
Kopytko P, Piotrowska K, Janisiak J, Tarnowski M. Garcinol—A Natural Histone Acetyltransferase Inhibitor and New Anti-Cancer Epigenetic Drug. International Journal of Molecular Sciences. 2021; 22(6):2828. https://doi.org/10.3390/ijms22062828
Chicago/Turabian StyleKopytko, Patrycja, Katarzyna Piotrowska, Joanna Janisiak, and Maciej Tarnowski. 2021. "Garcinol—A Natural Histone Acetyltransferase Inhibitor and New Anti-Cancer Epigenetic Drug" International Journal of Molecular Sciences 22, no. 6: 2828. https://doi.org/10.3390/ijms22062828
APA StyleKopytko, P., Piotrowska, K., Janisiak, J., & Tarnowski, M. (2021). Garcinol—A Natural Histone Acetyltransferase Inhibitor and New Anti-Cancer Epigenetic Drug. International Journal of Molecular Sciences, 22(6), 2828. https://doi.org/10.3390/ijms22062828






