Untargeted In Silico Compound Classification—A Novel Metabolomics Method to Assess the Chemodiversity in Bryophytes
Abstract
1. Introduction
2. Results
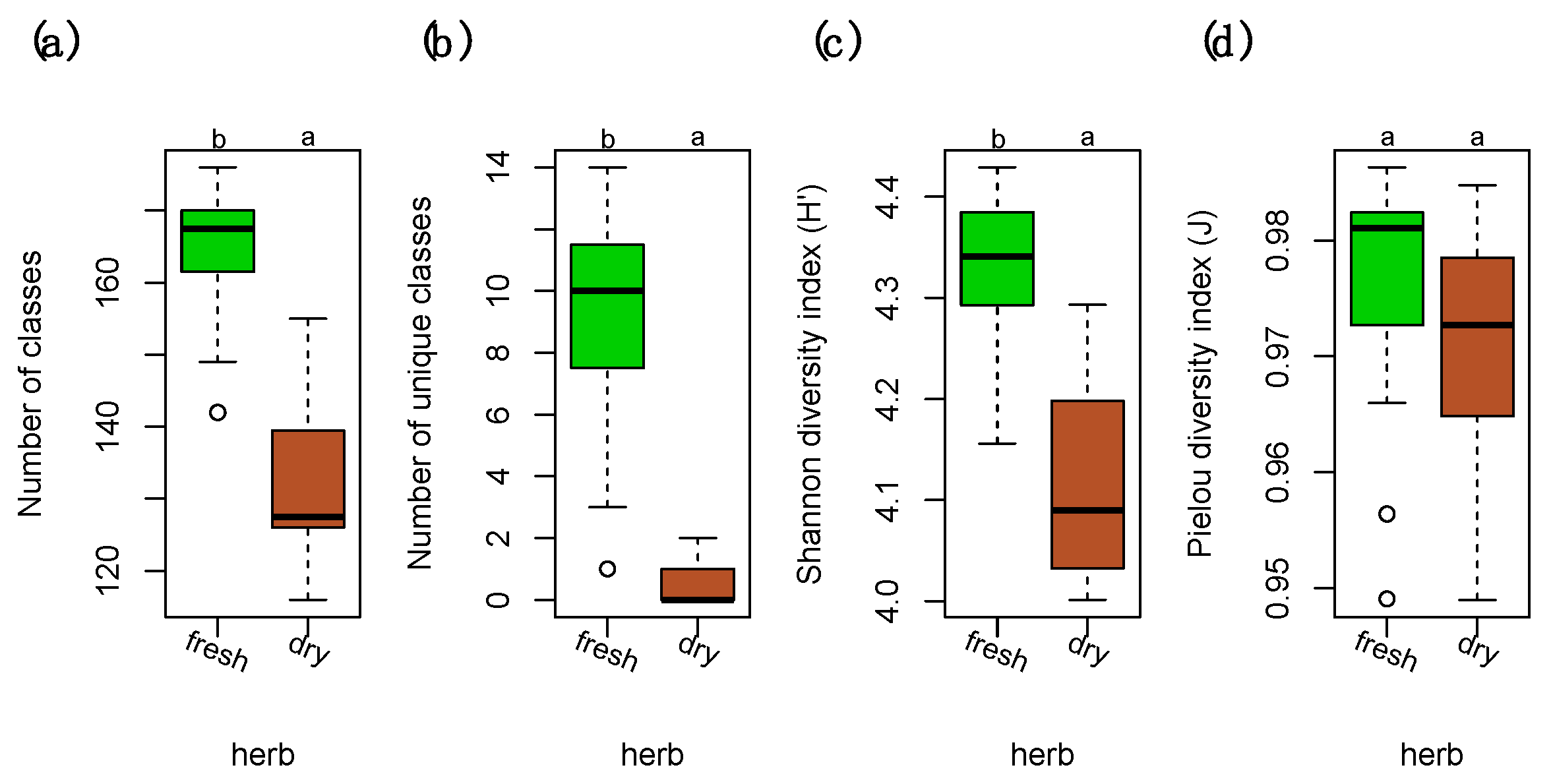
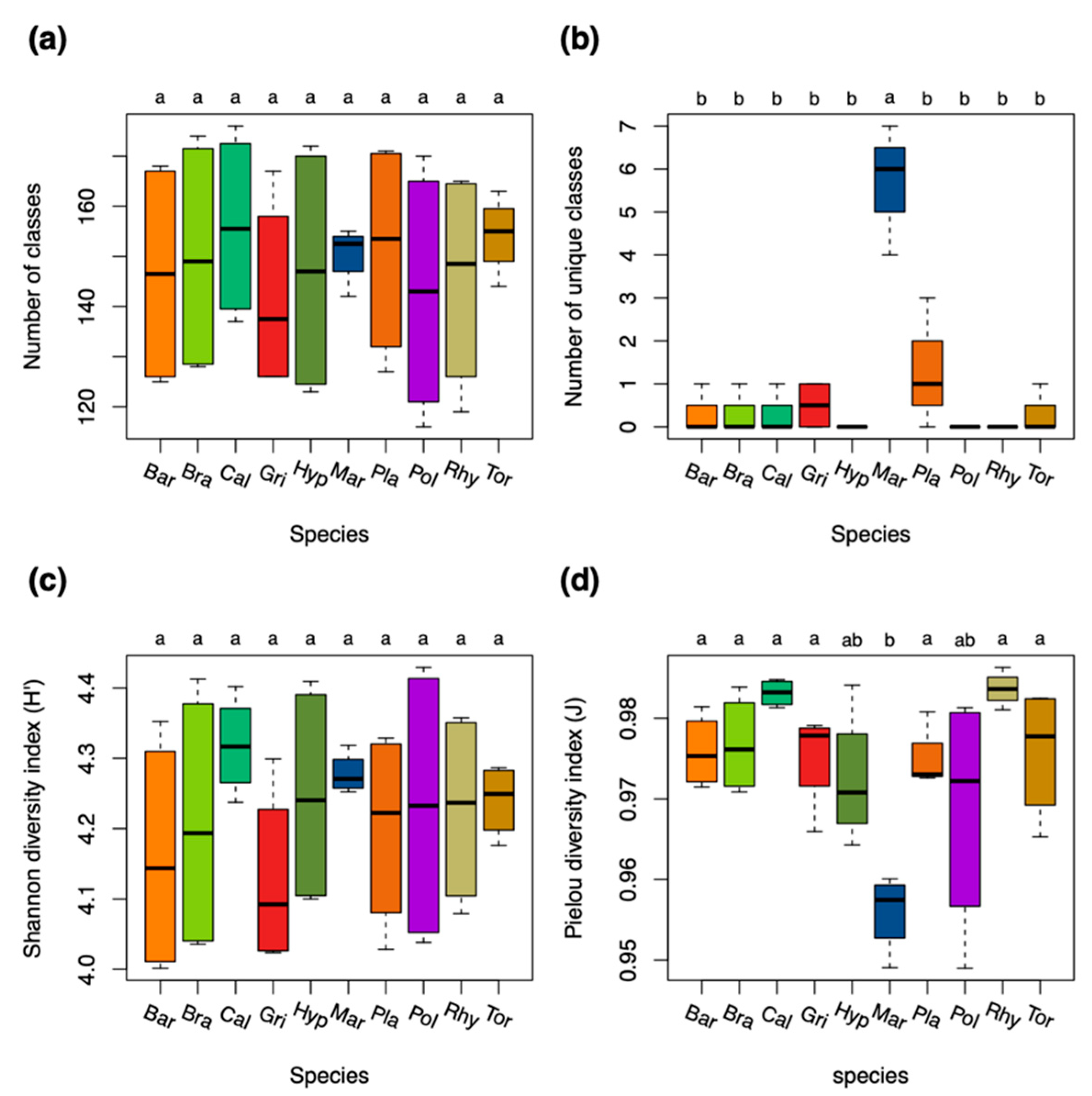
2.1. Chemical Diversity Analyses
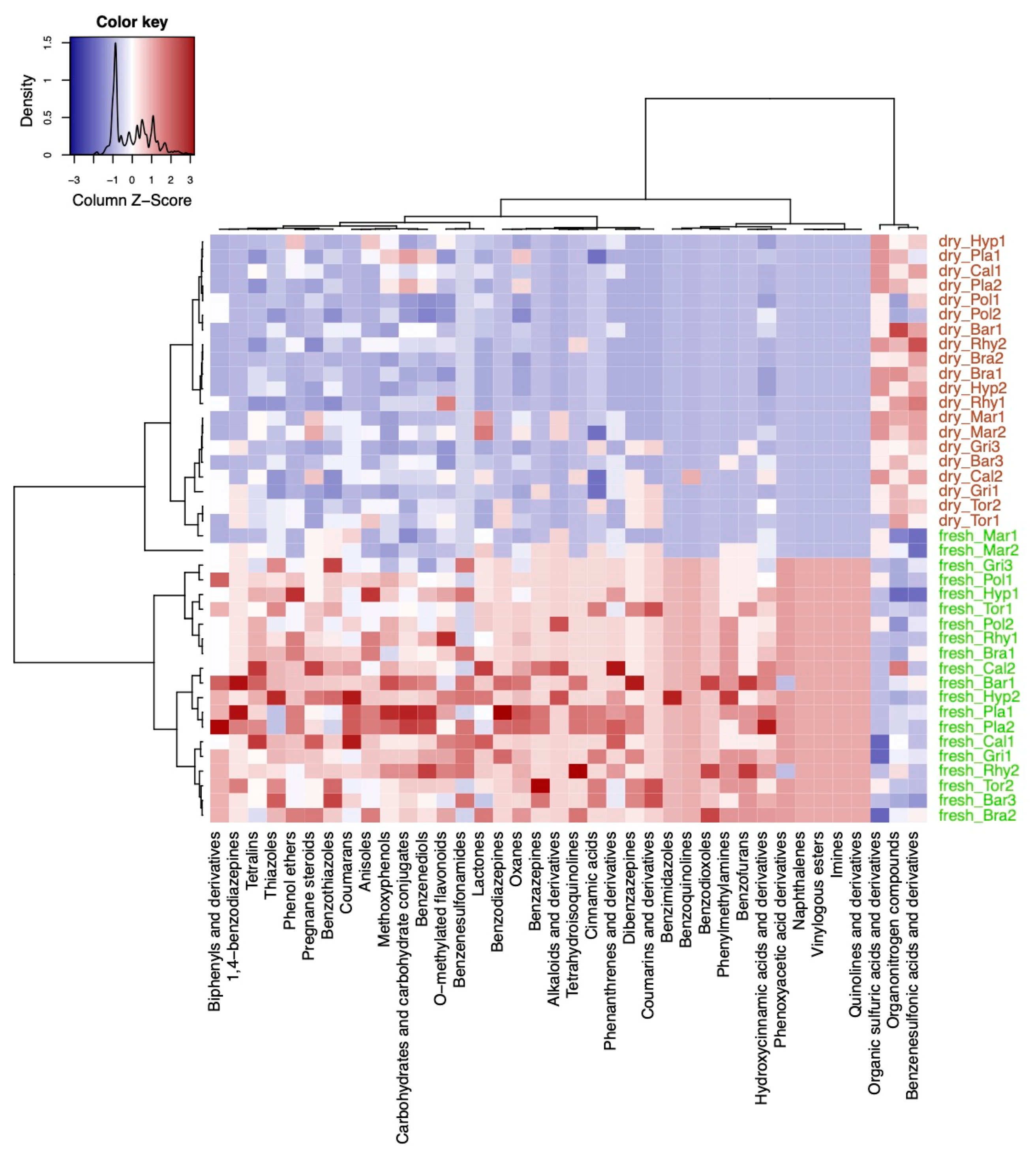
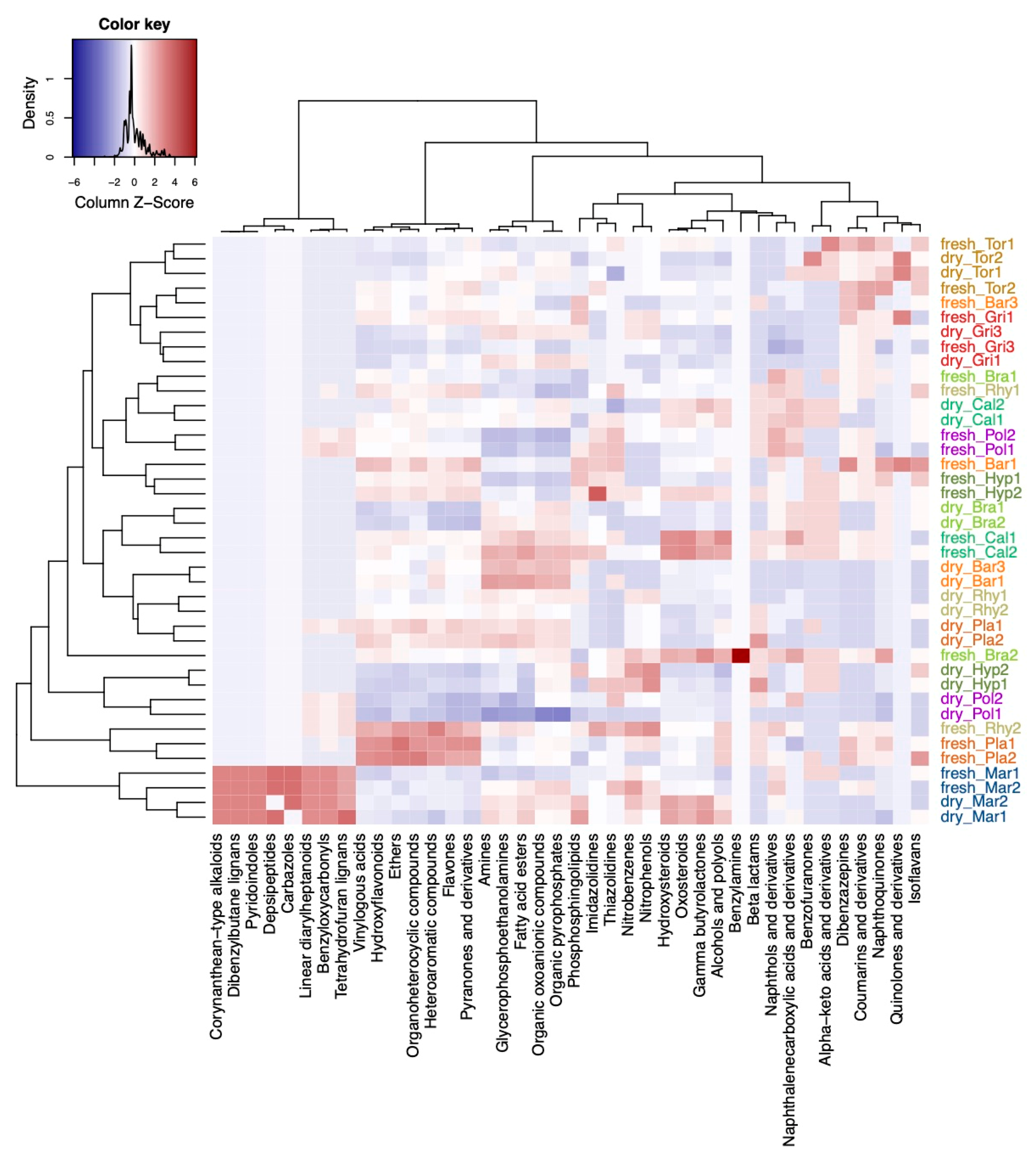
2.2. Exploring Differences in the Composition of Metabolite Families
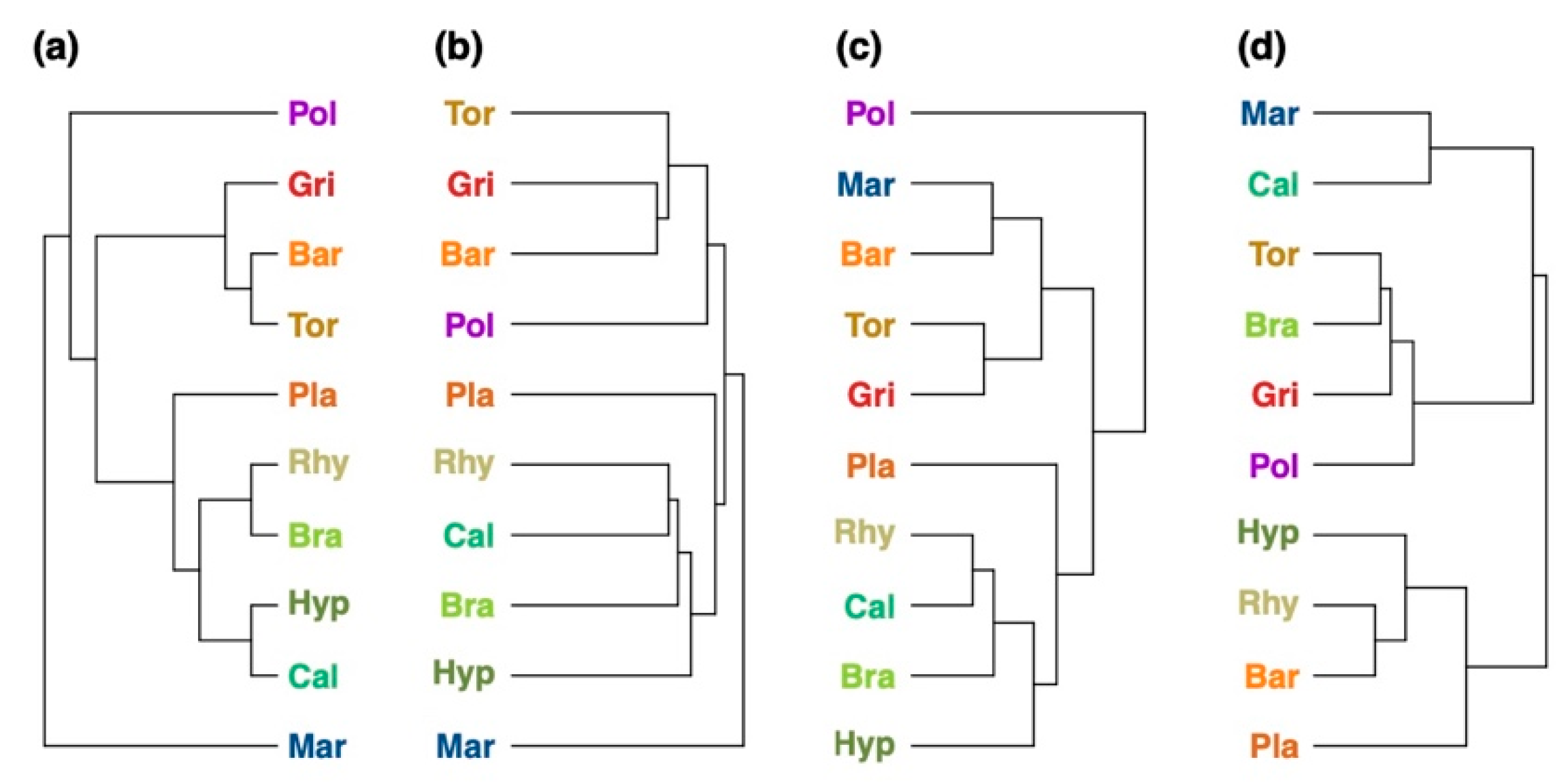
2.3. Comparison of Chemotaxonomic Analysis with Phylogeny
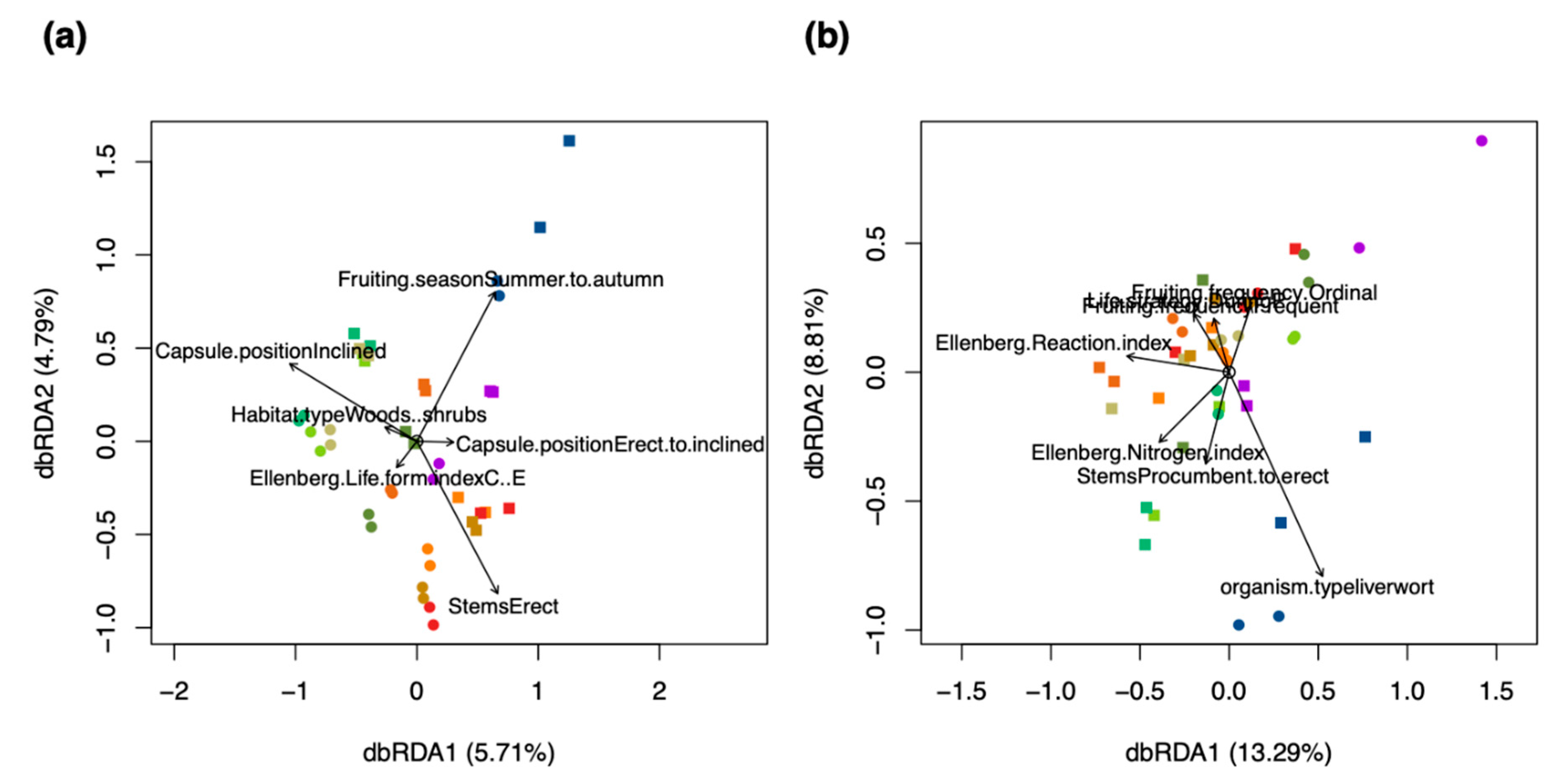
2.4. Trait Analysis
3. Discussion
4. Materials and Methods
4.1. Sampling
4.2. Metabolite Extraction
4.3. Metabolite Separation
4.4. Untargeted Mass Spectrometry
4.5. Raw Data Acquisition
4.6. Peak Detection
4.7. In Silico Classification
4.8. Statistical and Diversity Analyses
4.9. Data Records
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| acrocarpous | Phylogenetic clade of bryophytes having mostly an erect stem and sporophytes originate at the top of stems |
| ANOVA | Analysis of Variance |
| AUC | Area Under receiver operating characteristic Curve |
| AUC-PR | Area Under Precision Recall Curve |
| Bar | Species code for Barbula unguiculata Hedw., ott1027526 |
| Bra | Species code for Brachythecium rutabulum (Hedw.) Schimp., ott734784 |
| Bryophytina | Subdivision of plants on which the phylogenetic tree was constructed, ott471195 |
| Cal | Species code for Calliergonella cuspidata (Hedw.) Loeske, ott405418 |
| dbRDA | Distance-based Redundancy Analysis |
| DIA | Data Independent Acquisition |
| dry | Factor level: Samples which were taken under herbarium conditions |
| fresh | Factor level: Samples which were taken under fresh conditions |
| Gri | Species code for Grimmia pulvinata (Hedw.) Sm., ott604969 |
| Hyp | Species code for Hypnum cupressiforme Hedw., ott160023 |
| LC/MS | Liquid chromatography coupled to a Mass Spectrometer |
| Mar | Species code for Marchantia polymorpha L., ott56596 |
| MS1 | Mass-spectral analysis |
| MS/MS | Tandem mass-spectral analysis where two MS are performed resulting in fragment spectra |
| ott | Phylogenetic identifier for species and phylogenetic groups |
| PCA | Principal Component Analysis |
| Pla | Species code for Plagiomnium undulatum (Hedw.) T.J. Kop., ott744339 |
| pleurocarpous | Phylogenetic clade of bryophytes having mostly a branched prostrate stem and sporophytes are produced in branches |
| Pol | Species code for Polytrichum strictum Menzies ex Brid., ott101288 |
| PR | Precision and Recall |
| R | R statistical programming language |
| R2 | R-squared |
| Rhy | Species code for Rhytidiadelphus squarrosus (Hedw.) Warnst., ott734791 |
| ROC | Receiver Operating Characteristic |
| sPLS-DA | Sparse Partial Least Squares Discriminant Analysis |
| SWATH | Sequential Windowed Acquisition of All Theoretical Fragment Ion Mass Spectra |
| Tor | Species code for Tortula muralis Hedw., ott318814 |
| Tukey HSD | Honestly significant difference test |
| XCMS | Framework to perform detection of chromatographic peaks |
References
- Sardans, J.; Peñuelas, J.; Rivas-Ubach, A. Ecological metabolomics: Overview of current developments and future challenges. Chemoecology 2011, 21, 191–225. [Google Scholar] [CrossRef]
- Van Dam, N.M.; van der Meijden, E. A Role for Metabolomics in Plant Ecology. In Annual Plant Reviews Volume 43; Hall, R.D., Ed.; Wiley-Blackwell: Oxford, UK, 2011; pp. 87–107. ISBN 978-1-4443-3995-6. [Google Scholar]
- Metabolomics Tools for Natural Product Discovery; Roessner, U., Dias, D.A., Eds.; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2013; Volume 1055, ISBN 978-1-62703-576-7. [Google Scholar]
- Peters, K.; Worrich, A.; Weinhold, A.; Alka, O.; Balcke, G.; Birkemeyer, C.; Bruelheide, H.; Calf, O.; Dietz, S.; Dührkop, K.; et al. Current Challenges in Plant Eco-Metabolomics. Int. J. Mol. Sci. 2018, 19, 1385. [Google Scholar] [CrossRef]
- Fiehn, O. Metabolomics—The link between genotypes and phenotypes. In Functional Genomics; Town, C., Ed.; Springer: Berlin/Heidelberg, Germany, 2002; pp. 155–171. ISBN 978-94-010-3903-1. [Google Scholar]
- Uthe, H.; van Dam, N.M.; Hervé, M.R.; Sorokina, M.; Peters, K.; Weinhold, A. A practical guide to implementing metabolomics in plant ecology and biodiversity research. In Advances in Botanical Research; Elsevier: Amsterdam, The Netherlands, 2020; p. S0065229620300732. [Google Scholar]
- Goffinet, B.; Shaw, A.J. Bryophyte Biology; Cambridge University Press: Cambridge, UK, 2009; ISBN 978-0-511-45773-9. [Google Scholar]
- Shi, X.-M.; Song, L.; Liu, W.-Y.; Lu, H.-Z.; Qi, J.-H.; Li, S.; Chen, X.; Wu, J.-F.; Liu, S.; Wu, C.-S. Epiphytic bryophytes as bio-indicators of atmospheric nitrogen deposition in a subtropical montane cloud forest: Response patterns, mechanism, and critical load. Environ. Pollut. 2017, 229, 932–941. [Google Scholar] [CrossRef] [PubMed]
- Bennett, J. Bioindicators & Biomonitors, Principles, Concepts and Applications. Sci. Total Environ. 2004, 328, 295. [Google Scholar] [CrossRef]
- Roos, R.E.; Zuijlen, K.; Birkemoe, T.; Klanderud, K.; Lang, S.I.; Bokhorst, S.; Wardle, D.A.; Asplund, J. Contrasting drivers of community-level trait variation for vascular plants, lichens and bryophytes across an elevational gradient. Funct. Ecol. 2019, 33, 2430–2446. [Google Scholar] [CrossRef]
- Cornelissen, J.H.C.; Lang, S.I.; Soudzilovskaia, N.A.; During, H.J. Comparative Cryptogam Ecology: A Review of Bryophyte and Lichen Traits that Drive Biogeochemistry. Ann. Bot. 2007, 99, 987–1001. [Google Scholar] [CrossRef] [PubMed]
- Streitberger, M.; Schmidt, C.; Fartmann, T. Contrasting response of vascular plant and bryophyte species assemblages to a soil-disturbing ecosystem engineer in calcareous grasslands. Ecol. Eng. 2017, 99, 391–399. [Google Scholar] [CrossRef]
- Wulf, P.; Pearson, R.G. Mossy stones gather more bugs: Moss as habitat, nurseries and refugia for tropical stream invertebrates. Hydrobiologia 2017, 790, 167–182. [Google Scholar] [CrossRef]
- Vanderpoorten, A.; Goffinet, B. Introduction to Bryophytes; Cambridge University Press: Cambridge, UK, 2009; ISBN 978-0-511-65080-2. [Google Scholar]
- Müller, C.; Caspers, B.A.; Gadau, J.; Kaiser, S. The Power of Infochemicals in Mediating Individualized Niches. Trends Ecol. Evol. 2020, 35, 981–989. [Google Scholar] [CrossRef] [PubMed]
- Cronberg, N. Microarthropods Mediate Sperm Transfer in Mosses. Science 2006, 313, 1255. [Google Scholar] [CrossRef] [PubMed]
- Rosenstiel, T.N.; Shortlidge, E.E.; Melnychenko, A.N.; Pankow, J.F.; Eppley, S.M. Sex-specific volatile compounds influence microarthropod-mediated fertilization of moss. Nature 2012, 489, 431–433. [Google Scholar] [CrossRef] [PubMed]
- Asakawa, Y.; Ludwiczuk, A.; Nagashima, F. Phytochemical and biological studies of bryophytes. Phytochemistry 2013, 91, 52–80. [Google Scholar] [CrossRef] [PubMed]
- Chemical Constituents of Bryophytes: Bio- and Chemical Diversity, Biological Activity, and Chemosystematics; Asakawa, Y., Ludwiczuk, A., Nagashima, F., Eds.; Fortschritte der Chemie organischer Naturstoffe = Progress in the chemistry of organic natural products; Springer: Berlin/Heidelberg, Germany, 2013; ISBN 978-3-7091-1083-6. [Google Scholar]
- Bristol, B.M. On the remarkable retention of vitality of moss protonema. New Phytol. 1916, 15, 137–143. [Google Scholar] [CrossRef]
- Maheu, M.J. Régénération du Barbula muralis, après quatorze ans de sécheresse, par protonémas foliaires primaires propagulifères et protonémas secondaires bulbigènes. Bull. Société Bot. Fr. 1922, 69, 330–334. [Google Scholar] [CrossRef]
- Breuil-Sée, A. Recorded desiccation-survival times in bryophytes. J. Bryol. 1993, 17, 679–680. [Google Scholar] [CrossRef]
- Oliver, M.J.; Tuba, Z.; Mishler, B.D. The evolution of vegetative desiccation tolerance in land plants. Plant Ecol. 2000, 151, 85–100. [Google Scholar] [CrossRef]
- Oliver, M.J. Desiccation Tolerance in Bryophytes: A Reflection of the Primitive Strategy for Plant Survival in Dehydrating Habitats? Integr. Comp. Biol. 2005, 45, 788–799. [Google Scholar] [CrossRef]
- Garcia, E.L.; Rosenstiel, T.N.; Graves, C.; Shortlidge, E.E.; Eppley, S.M. Distribution drivers and physiological responses in geothermal bryophyte communities. Am. J. Bot. 2016, 103, 625–634. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.; Ohtani, M.; Yamaguchi, M.; Toyooka, K.; Wakazaki, M.; Sato, M.; Kubo, M.; Nakano, Y.; Sano, R.; Hiwatashi, Y.; et al. Contribution of NAC Transcription Factors to Plant Adaptation to Land. Science 2014, 343, 1505–1508. [Google Scholar] [CrossRef]
- Niklas, K.J.; Cobb, E.D.; Matas, A.J. The evolution of hydrophobic cell wall biopolymers: From algae to angiosperms. J. Exp. Bot. 2017, 68, 5261–5269. [Google Scholar] [CrossRef] [PubMed]
- Proctor, M.C.F.; Smirnoff, N. Ecophysiology of photosynthesis in bryophytes: Major roles for oxygen photoreduction and non-photochemical quenching? Physiol. Plant. 2011, 141, 130–140. [Google Scholar] [CrossRef] [PubMed]
- Asakawa, Y.; Ludwiczuk, A. Chemical Constituents of Bryophytes: Structures and Biological Activity. J. Nat. Prod. 2018, 81, 641–660. [Google Scholar] [CrossRef] [PubMed]
- Charron, A.J.; Quatrano, R.S. Between a Rock and a Dry Place: The Water-Stressed Moss. Mol. Plant 2009, 2, 478–486. [Google Scholar] [CrossRef] [PubMed]
- Proctor, M.C.F.; Oliver, M.J.; Wood, A.J.; Alpert, P.; Stark, L.R.; Cleavitt, N.L.; Mishler, B.D. Desiccation-tolerance in bryophytes: A review. Bryologist 2007, 110, 595–621. [Google Scholar] [CrossRef]
- Wang, R.; Yin, Y.; Zhu, Z.-J. Advancing untargeted metabolomics using data-independent acquisition mass spectrometry technology. Anal. Bioanal. Chem. 2019, 411, 4349–4357. [Google Scholar] [CrossRef] [PubMed]
- Hopfgartner, G.; Tonoli, D.; Varesio, E. High-resolution mass spectrometry for integrated qualitative and quantitative analysis of pharmaceuticals in biological matrices. Anal. Bioanal. Chem. 2012, 402, 2587–2596. [Google Scholar] [CrossRef] [PubMed]
- Tsugawa, H.; Cajka, T.; Kind, T.; Ma, Y.; Higgins, B.; Ikeda, K.; Kanazawa, M.; VanderGheynst, J.; Fiehn, O.; Arita, M. MS-DIAL: Data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat. Methods 2015, 12, 523–526. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.A.; Want, E.J.; O’Maille, G.; Abagyan, R.; Siuzdak, G. XCMS: Processing Mass Spectrometry Data for Metabolite Profiling Using Nonlinear Peak Alignment, Matching, and Identification. Anal. Chem. 2006, 78, 779–787. [Google Scholar] [CrossRef] [PubMed]
- Tsugawa, H.; Nakabayashi, R.; Mori, T.; Yamada, Y.; Takahashi, M.; Rai, A.; Sugiyama, R.; Yamamoto, H.; Nakaya, T.; Yamazaki, M.; et al. A cheminformatics approach to characterize metabolomes in stable-isotope-labeled organisms. Nat. Methods 2019, 16, 295–298. [Google Scholar] [CrossRef] [PubMed]
- Bonner, R.; Hopfgartner, G. SWATH data independent acquisition mass spectrometry for metabolomics. TrAC Trends Anal. Chem. 2019, 120, 115278. [Google Scholar] [CrossRef]
- Zhu, X.; Chen, Y.; Subramanian, R. Comparison of Information-Dependent Acquisition, SWATH, and MS All Techniques in Metabolite Identification Study Employing Ultrahigh-Performance Liquid Chromatography–Quadrupole Time-of-Flight Mass Spectrometry. Anal. Chem. 2014, 86, 1202–1209. [Google Scholar] [CrossRef]
- Balcke, G.U.; Bennewitz, S.; Bergau, N.; Athmer, B.; Henning, A.; Majovsky, P.; Jiménez-Gómez, J.M.; Hoehenwarter, W.; Tissier, A. Multi-Omics of Tomato Glandular Trichomes Reveals Distinct Features of Central Carbon Metabolism Supporting High Productivity of Specialized Metabolites. Plant Cell 2017, 29, 960–983. [Google Scholar] [CrossRef]
- Van Dam, N.M.; Bouwmeester, H.J. Metabolomics in the Rhizosphere: Tapping into Belowground Chemical Communication. Trends Plant Sci. 2016, 21, 256–265. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S. Computational strategies for metabolite identification in metabolomics. Bioanalysis 2009, 1, 1579–1596. [Google Scholar] [CrossRef] [PubMed]
- Peters, K.; Treutler, H.; Döll, S.; Kindt, A.S.; Hankemeier, T.; Neumann, S. Neumann Chemical Diversity and Classification of Secondary Metabolites in Nine Bryophyte Species. Metabolites 2019, 9, 222. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, R.R.; Dorrestein, P.C.; Quinn, R.A. Illuminating the dark matter in metabolomics. Proc. Natl. Acad. Sci. USA 2015, 112, 12549–12550. [Google Scholar] [CrossRef] [PubMed]
- Ruttkies, C.; Schymanski, E.L.; Wolf, S.; Hollender, J.; Neumann, S. MetFrag relaunched: Incorporating strategies beyond in silico fragmentation. J. Cheminform. 2016, 8. [Google Scholar] [CrossRef] [PubMed]
- Dührkop, K.; Shen, H.; Meusel, M.; Rousu, J.; Böcker, S. Searching molecular structure databases with tandem mass spectra using CSI:FingerID. Proc. Natl. Acad. Sci. USA 2015, 112, 12580–12585. [Google Scholar] [CrossRef] [PubMed]
- Scholz, M.; Gatzek, S.; Sterling, A.; Fiehn, O.; Selbig, J. Metabolite fingerprinting: Detecting biological features by independent component analysis. Bioinformatics 2004, 20, 2447–2454. [Google Scholar] [CrossRef] [PubMed]
- Treutler, H.; Tsugawa, H.; Porzel, A.; Gorzolka, K.; Tissier, A.; Neumann, S.; Balcke, G.U. Discovering Regulated Metabolite Families in Untargeted Metabolomics Studies. Anal. Chem. 2016, 88, 8082–8090. [Google Scholar] [CrossRef]
- Rogers, S.; Ong, C.W.; Wandy, J.; Ernst, M.; Ridder, L.; van der Hooft, J.J.J. Deciphering complex metabolite mixtures by unsupervised and supervised substructure discovery and semi-automated annotation from MS/MS spectra. Faraday Discuss. 2019, 218, 284–302. [Google Scholar] [CrossRef] [PubMed]
- Dührkop, K.; Nothias, L.-F.; Fleischauer, M.; Reher, R.; Ludwig, M.; Hoffmann, M.A.; Petras, D.; Gerwick, W.H.; Rousu, J.; Dorrestein, P.C.; et al. Systematic classification of unknown metabolites using high-resolution fragmentation mass spectra. Nat. Biotechnol. 2020. [Google Scholar] [CrossRef]
- Van der Hooft, J.J.J.; Wandy, J.; Barrett, M.P.; Burgess, K.E.V.; Rogers, S. Topic modeling for untargeted substructure exploration in metabolomics. Proc. Natl. Acad. Sci. USA 2016, 113, 13738–13743. [Google Scholar] [CrossRef]
- Barbas, C.; Moraes, E.P.; Villaseñor, A. Capillary electrophoresis as a metabolomics tool for non-targeted fingerprinting of biological samples. J. Pharm. Biomed. Anal. 2011, 55, 823–831. [Google Scholar] [CrossRef] [PubMed]
- Hastings, J.; Chepelev, L.; Willighagen, E.; Adams, N.; Steinbeck, C.; Dumontier, M. The Chemical Information Ontology: Provenance and Disambiguation for Chemical Data on the Biological Semantic Web. PLoS ONE 2011, 6, e25513. [Google Scholar] [CrossRef]
- Djoumbou Feunang, Y.; Eisner, R.; Knox, C.; Chepelev, L.; Hastings, J.; Owen, G.; Fahy, E.; Steinbeck, C.; Subramanian, S.; Bolton, E.; et al. ClassyFire: Automated chemical classification with a comprehensive, computable taxonomy. J. Cheminform. 2016, 8. [Google Scholar] [CrossRef] [PubMed]
- Van der Hooft, J.J.J.; Wandy, J.; Young, F.; Padmanabhan, S.; Gerasimidis, K.; Burgess, K.E.V.; Barrett, M.P.; Rogers, S. Unsupervised Discovery and Comparison of Structural Families Across Multiple Samples in Untargeted Metabolomics. Anal. Chem. 2017, 89, 7569–7577. [Google Scholar] [CrossRef]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [PubMed]
- Edmands, W.M.B.; Petrick, L.; Barupal, D.K.; Scalbert, A.; Wilson, M.J.; Wickliffe, J.K.; Rappaport, S.M. compMS2Miner: An Automatable Metabolite Identification, Visualization, and Data-Sharing R Package for High-Resolution LC–MS Data Sets. Anal. Chem. 2017, 89, 3919–3928. [Google Scholar] [CrossRef] [PubMed]
- Blaženović, I.; Kind, T.; Sa, M.R.; Ji, J.; Vaniya, A.; Wancewicz, B.; Roberts, B.S.; Torbašinović, H.; Lee, T.; Mehta, S.S.; et al. Structure Annotation of All Mass Spectra in Untargeted Metabolomics. Anal. Chem. 2019, 91, 2155–2162. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, A.; Vázquez-Baeza, Y.; Gauglitz, J.M.; Wang, M.; Dührkop, K.; Nothias-Esposito, M.; Acharya, D.D.; Ernst, M.; van der Hooft, J.J.J.; Zhu, Q.; et al. Chemically informed analyses of metabolomics mass spectrometry data with Qemistree. Nat. Chem. Biol. 2021, 17, 146–151. [Google Scholar] [CrossRef] [PubMed]
- Haug, K.; Salek, R.M.; Conesa, P.; Hastings, J.; de Matos, P.; Rijnbeek, M.; Mahendraker, T.; Williams, M.; Neumann, S.; Rocca-Serra, P.; et al. MetaboLights—An open-access general-purpose repository for metabolomics studies and associated meta-data. Nucleic Acids Res. 2013, 41, D781–D786. [Google Scholar] [CrossRef]
- Peters, K.; Gorzolka, K.; Bruelheide, H.; Neumann, S. Seasonal variation of secondary metabolites in nine different bryophytes. Ecol. Evol. 2018, 8, 9105–9117. [Google Scholar] [CrossRef] [PubMed]
- Peters, K.; Gorzolka, K.; Bruelheide, H.; Neumann, S. Computational workflow to study the seasonal variation of secondary metabolites in nine different bryophytes. Sci. Data 2018, 5, 180179. [Google Scholar] [CrossRef]
- Schymanski, E.L.; Ruttkies, C.; Krauss, M.; Brouard, C.; Kind, T.; Dührkop, K.; Allen, F.; Vaniya, A.; Verdegem, D.; Böcker, S.; et al. Critical Assessment of Small Molecule Identification 2016: Automated methods. J. Cheminform. 2017, 9, 22. [Google Scholar] [CrossRef]
- Horai, H.; Arita, M.; Kanaya, S.; Nihei, Y.; Ikeda, T.; Suwa, K.; Ojima, Y.; Tanaka, K.; Tanaka, S.; Aoshima, K.; et al. MassBank: A public repository for sharing mass spectral data for life sciences. J. Mass Spectrom. 2010, 45, 703–714. [Google Scholar] [CrossRef] [PubMed]
- Shahaf, N.; Rogachev, I.; Heinig, U.; Meir, S.; Malitsky, S.; Battat, M.; Wyner, H.; Zheng, S.; Wehrens, R.; Aharoni, A. The WEIZMASS spectral library for high-confidence metabolite identification. Nat. Commun. 2016, 7, 12423. [Google Scholar] [CrossRef] [PubMed]
- Vinaixa, M.; Schymanski, E.L.; Neumann, S.; Navarro, M.; Salek, R.M.; Yanes, O. Mass spectral databases for LC/MS- and GC/MS-based metabolomics: State of the field and future prospects. TrAC Trends Anal. Chem. 2016, 78, 23–35. [Google Scholar] [CrossRef]
- Dührkop, K.; Fleischauer, M.; Ludwig, M.; Aksenov, A.A.; Melnik, A.V.; Meusel, M.; Dorrestein, P.C.; Rousu, J.; Böcker, S. SIRIUS 4: A rapid tool for turning tandem mass spectra into metabolite structure information. Nat. Methods 2019, 16, 299–302. [Google Scholar] [CrossRef]
- Djoumbou-Feunang, Y.; Pon, A.; Karu, N.; Zheng, J.; Li, C.; Arndt, D.; Gautam, M.; Allen, F.; Wishart, D.S. CFM-ID 3.0: Significantly Improved ESI-MS/MS Prediction and Compound Identification. Metabolites 2019, 9, 72. [Google Scholar] [CrossRef]
- Maksimova, V.; Klavina, L.; Bikovens, O.; Zicmanis, A.; Purmalis, O. Structural Characterization and Chemical Classification of Some Bryophytes Found in Latvia. Chem. Biodivers. 2013, 10, 1284–1294. [Google Scholar] [CrossRef] [PubMed]
- Klavina, L. A study on bryophyte chemical composition–search for new applications. Agron. Res. 2015, 13, 969–978. [Google Scholar]
- Sardans, J.; Gargallo-Garriga, A.; Urban, O.; Klem, K.; Walker, T.W.N.; Holub, P.; Janssens, I.A.; Peñuelas, J. Ecometabolomics for a Better Understanding of Plant Responses and Acclimation to Abiotic Factors Linked to Global Change. Metabolites 2020, 10, 239. [Google Scholar] [CrossRef] [PubMed]
- Araújo, W.L.; Tohge, T.; Ishizaki, K.; Leaver, C.J.; Fernie, A.R. Protein degradation—An alternative respiratory substrate for stressed plants. Trends Plant Sci. 2011, S1360138511001063. [Google Scholar] [CrossRef]
- Rico-Reséndiz, F.; Cervantes-Pérez, S.A.; Espinal-Centeno, A.; Dipp-Álvarez, M.; Oropeza-Aburto, A.; Hurtado-Bautista, E.; Cruz-Hernández, A.; Bowman, J.L.; Ishizaki, K.; Arteaga-Vázquez, M.A.; et al. Transcriptional and morpho-physiological responses of Marchantia polymorpha upon phosphate starvation. Int. J. Mol. Sci. 2020, 21, 8354. [Google Scholar] [CrossRef] [PubMed]
- Kranner, I.; Beckett, R.; Hochman, A. Desiccation-tolerance in lichens: A review. Bryologist 2008, 111, 576–593. [Google Scholar] [CrossRef]
- Erxleben, A.; Gessler, A.; Vervliet-Scheebaum, M.; Reski, R. Metabolite profiling of the moss Physcomitrella patens reveals evolutionary conservation of osmoprotective substances. Plant Cell Rep. 2012, 31, 427–436. [Google Scholar] [CrossRef] [PubMed]
- Brunetti, C.; George, R.M.; Tattini, M.; Field, K.; Davey, M.P. Metabolomics in plant environmental physiology. J. Exp. Bot. 2013, 64, 4011–4020. [Google Scholar] [CrossRef]
- Yang, L.; Wen, K.S.; Ruan, X.; Zhao, Y.X.; Wei, F.; Wang, Q. Response of Plant Secondary Metabolites to Environmental Factors. Molecules 2018, 23, 762. [Google Scholar] [CrossRef] [PubMed]
- Davies, K.M.; Jibran, R.; Zhou, Y.; Albert, N.W.; Brummell, D.A.; Jordan, B.R.; Bowman, J.L.; Schwinn, K.E. The Evolution of Flavonoid Biosynthesis: A Bryophyte Perspective. Front. Plant Sci. 2020, 11, 7. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Pandey, A.K. Chemistry and Biological Activities of Flavonoids: An Overview. Sci. World J. 2013, 2013, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Lambers, H.; Chapin, F.S.; Pons, T.L. Plant Physiological Ecology, 2nd ed.; Springer: Berlin/Heidelberg, Germany, 2008; ISBN 978-0-387-78340-6. [Google Scholar]
- Chopra, R.N.; Bhatla, S.C. Bryophyte Development: Physiology and Biochemistry, 1st ed.; Chopra, R.N., Bhatla, S.C., Eds.; CRC Press: Boca Raton, FL, USA, 2019; ISBN 978-0-429-26056-8. [Google Scholar]
- Mithöfer, A.; Boland, W. Plant Defense against Herbivores: Chemical Aspects. Annu. Rev. Plant Biol. 2012, 63, 431–450. [Google Scholar] [CrossRef] [PubMed]
- Nandy, S.; Dey, A. Bibenzyls and bisbybenzyls of bryophytic origin as promising source of novel therapeutics: Pharmacology, synthesis and structure-activity. DARU J. Pharm. Sci. 2020. [Google Scholar] [CrossRef]
- Tanaka, M.; Esaki, T.; Kenmoku, H.; Koeduka, T.; Kiyoyama, Y.; Masujima, T.; Asakawa, Y.; Matsui, K. Direct evidence of specific localization of sesquiterpenes and marchantin A in oil body cells of Marchantia polymorpha L. Phytochemistry 2016, 130, 77–84. [Google Scholar] [CrossRef]
- Xie, C.-F.; Lou, H.-X. Secondary Metabolites in Bryophytes: An Ecological Aspect. Chem. Biodivers. 2009, 6, 303–312. [Google Scholar] [CrossRef]
- Commisso, M.; Guarino, F.; Marchi, L.; Muto, A.; Piro, A.; Degola, F. Bryo-Activities: A Review on How Bryophytes Are Contributing to the Arsenal of Natural Bioactive Compounds against Fungi. Plants 2021, 10, 203. [Google Scholar] [CrossRef] [PubMed]
- Lang, S.I.; Cornelissen, J.H.C.; Klahn, T.; van Logtestijn, R.S.P.; Broekman, R.; Schweikert, W.; Aerts, R. An experimental comparison of chemical traits and litter decomposition rates in a diverse range of subarctic bryophyte, lichen and vascular plant species. J. Ecol. 2009, 97, 886–900. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, X.; Bao, W. Higher photosynthetic capacity and different functional trait scaling relationships in erect bryophytes compared with prostrate species. Oecologia 2016, 180, 359–369. [Google Scholar] [CrossRef]
- Züst, T.; Agrawal, A.A. Trade-Offs Between Plant Growth and Defense against Insect Herbivory: An Emerging Mechanistic Synthesis. Annu. Rev. Plant Biol. 2017, 68, 513–534. [Google Scholar] [CrossRef] [PubMed]
- Vieira, C.; Séneca, A.; Sérgio, C.; Ferreira, M.T. Bryophyte taxonomic and functional groups as indicators of fine scale ecological gradients in mountain streams. Ecol. Indic. 2012, 18, 98–107. [Google Scholar] [CrossRef]
- Díaz, S.; Kattge, J.; Cornelissen, J.H.C.; Wright, I.J.; Lavorel, S.; Dray, S.; Reu, B.; Kleyer, M.; Wirth, C.; Colin Prentice, I.; et al. The global spectrum of plant form and function. Nature 2016, 529, 167–171. [Google Scholar] [CrossRef]
- Asner, G.P.; Anderson, C.B.; Martin, R.E.; Tupayachi, R.; Knapp, D.E.; Sinca, F. Landscape biogeochemistry reflected in shifting distributions of chemical traits in the Amazon forest canopy. Nat. Geosci. 2015, 8, 567–573. [Google Scholar] [CrossRef]
- Tsunoda, T.; van Dam, N.M. Root chemical traits and their roles in belowground biotic interactions. Pedobiologia 2017, 65, 58–67. [Google Scholar] [CrossRef]
- Descombes, P.; Kergunteuil, A.; Glauser, G.; Rasmann, S.; Pellissier, L. Plant physical and chemical traits associated with herbivory in situ and under a warming treatment. J. Ecol. 2020, 108, 733–749. [Google Scholar] [CrossRef]
- Wilkinson, M.D.; Dumontier, M.; Aalbersberg, I.J.; Appleton, G.; Axton, M.; Baak, A.; Blomberg, N.; Boiten, J.-W.; da Silva Santos, L.B.; Bourne, P.E.; et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data 2016, 3, 160018. [Google Scholar] [CrossRef] [PubMed]
- Ludwiczuk, A.; Odrzykoski, I.J.; Asakawa, Y. Identification of cryptic species within liverwort Conocephalum conicum based on the volatile components. Phytochemistry 2013, 95, 234–241. [Google Scholar] [CrossRef] [PubMed]
- Kohn, G.; Demmerle, S.; Vandekerkhove, O.; Hartmann, E.; Beutelmann, P. Distribution and chemotaxonomic significance of acetylenic fatty acids in mosses of the dicranales. Phytochemistry 1987, 26, 2271–2275. [Google Scholar] [CrossRef]
- Brodo, I.M. Interpreting Chemical Variation in Lichens for Systematic Purposes. Bryologist 1986, 89, 132. [Google Scholar] [CrossRef]
- Culberson, W.L. The use of chemistry in the systematics of the lichens. TAXON 1969, 18, 152–166. [Google Scholar] [CrossRef]
- Eisenreich, W.; Knispel, N.; Beck, A. Advanced methods for the study of the chemistry and the metabolism of lichens. Phytochem. Rev. 2011, 10, 445–456. [Google Scholar] [CrossRef]
- Lumbsch, H.T. Analysis of Phenolic Products in Lichens for Identification and Taxonomy. In Protocols in Lichenology; Kranner, I.C., Beckett, R.P., Varma, A.K., Eds.; Springer: Berlin/Heidelberg, Germany, 2002; pp. 281–295. ISBN 978-3-540-41139-0. [Google Scholar]
- Liu, X.; Li, N.; Liu, S.; Wang, J.; Zhang, N.; Zheng, X.; Leung, K.-S.; Cheng, L. Normalization Methods for the Analysis of Unbalanced Transcriptome Data: A Review. Front. Bioeng. Biotechnol. 2019, 7, 358. [Google Scholar] [CrossRef] [PubMed]
- Risso, D.; Massa, M.S.; Chiogna, M.; Romualdi, C. A modified LOESS normalization applied to microRNA arrays: A comparative evaluation. Bioinformatics 2009, 25, 2685–2691. [Google Scholar] [CrossRef]
- Smola, A.J.; Schölkopf, B. A tutorial on support vector regression. Stat. Comput. 2004, 14, 199–222. [Google Scholar] [CrossRef]
- Balcke, G.U.; Bennewitz, S.; Zabel, S.; Tissier, A. Isoprenoid and Metabolite Profiling of Plant Trichomes. In Plant Isoprenoids; Rodríguez-Concepción, M., Ed.; Methods in Molecular Biology; Springer: Berlin/Heidelberg, Germany, 2014; Volume 1153, pp. 189–202. ISBN 978-1-4939-0605-5. [Google Scholar]
- Adusumilli, R.; Mallick, P. Data Conversion with ProteoWizard msConvert. In Proteomics; Comai, L., Katz, J.E., Mallick, P., Eds.; Methods in Molecular Biology; Springer: Berlin/Heidelberg, Germany, 2017; Volume 1550, pp. 339–368. ISBN 978-1-4939-6745-2. [Google Scholar]
- Sansone, S.-A.; Rocca-Serra, P.; Field, D.; Maguire, E.; Taylor, C.; Hofmann, O.; Fang, H.; Neumann, S.; Tong, W.; Amaral-Zettler, L.; et al. Toward interoperable bioscience data. Nat. Genet. 2012, 44, 121–126. [Google Scholar] [CrossRef]
- Spicer, R.A.; Salek, R.; Steinbeck, C. Compliance with minimum information guidelines in public metabolomics repositories. Sci. Data 2017, 4, 170137. [Google Scholar] [CrossRef] [PubMed]
- Borcard, D.; Legendre, P.; Drapeau, P. Partialling out the Spatial Component of Ecological Variation. Ecology 1992, 73, 1045–1055. [Google Scholar] [CrossRef]
- Rohart, F.; Gautier, B.; Singh, A.; Lê Cao, K.-A. mixOmics: An R package for omics feature selection and multiple data integration. PLOS Comput. Biol. 2017, 13, e1005752. [Google Scholar] [CrossRef] [PubMed]
- Robin, X.; Turck, N.; Hainard, A.; Tiberti, N.; Lisacek, F.; Sanchez, J.-C.; Müller, M. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 2011, 12, 77. [Google Scholar] [CrossRef]
- Grau, J.; Grosse, I.; Keilwagen, J. PRROC: Computing and visualizing precision-recall and receiver operating characteristic curves in R. Bioinformatics 2015, 31, 2595–2597. [Google Scholar] [CrossRef]
- Fawcett, T. An introduction to ROC analysis. Pattern Recognit. Lett. 2006, 27, 861–874. [Google Scholar] [CrossRef]
- Tharwat, A. Classification assessment methods. Appl. Comput. Inform. 2020. [Google Scholar] [CrossRef]
- Hurlbert, S.H. The Nonconcept of Species Diversity: A Critique and Alternative Parameters. Ecology 1971, 52, 577–586. [Google Scholar] [CrossRef] [PubMed]
- Heip, C.H.R.; Herman, P.M.J.; Soetaert, K. Indices of diversity and evenness. Océanis 1998, 24, 61–87. [Google Scholar]
- McCune, B.; Grace, J.B.; Urban, D.L. Analysis of Ecological Communities, 2nd ed.; MjM Software Design: Gleneden Beach, OR, USA, 2002; ISBN 978-0-9721290-0-8. [Google Scholar]
- Gu, Z.; Gu, L.; Eils, R.; Schlesner, M.; Brors, B. circlize implements and enhances circular visualization in R. Bioinformatics 2014, 30, 2811–2812. [Google Scholar] [CrossRef] [PubMed]
- Fisher, R.A. On the Interpretation of χ2 from Contingency Tables, and the Calculation of P. J. R. Stat. Soc. 1922, 85, 87. [Google Scholar] [CrossRef]
- Agresti, A. A Survey of Exact Inference for Contingency Tables. Stat. Sci. 1992, 7, 131–153. [Google Scholar] [CrossRef]
- Redelings, B.D.; Holder, M.T. A supertree pipeline for summarizing phylogenetic and taxonomic information for millions of species. PeerJ 2017, 5, e3058. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peters, K.; Balcke, G.; Kleinenkuhnen, N.; Treutler, H.; Neumann, S. Untargeted In Silico Compound Classification—A Novel Metabolomics Method to Assess the Chemodiversity in Bryophytes. Int. J. Mol. Sci. 2021, 22, 3251. https://doi.org/10.3390/ijms22063251
Peters K, Balcke G, Kleinenkuhnen N, Treutler H, Neumann S. Untargeted In Silico Compound Classification—A Novel Metabolomics Method to Assess the Chemodiversity in Bryophytes. International Journal of Molecular Sciences. 2021; 22(6):3251. https://doi.org/10.3390/ijms22063251
Chicago/Turabian StylePeters, Kristian, Gerd Balcke, Niklas Kleinenkuhnen, Hendrik Treutler, and Steffen Neumann. 2021. "Untargeted In Silico Compound Classification—A Novel Metabolomics Method to Assess the Chemodiversity in Bryophytes" International Journal of Molecular Sciences 22, no. 6: 3251. https://doi.org/10.3390/ijms22063251
APA StylePeters, K., Balcke, G., Kleinenkuhnen, N., Treutler, H., & Neumann, S. (2021). Untargeted In Silico Compound Classification—A Novel Metabolomics Method to Assess the Chemodiversity in Bryophytes. International Journal of Molecular Sciences, 22(6), 3251. https://doi.org/10.3390/ijms22063251







