Characterization of Biological Pathways Regulating Acute Cold Resistance of Zebrafish
Abstract
1. Introduction
2. Results
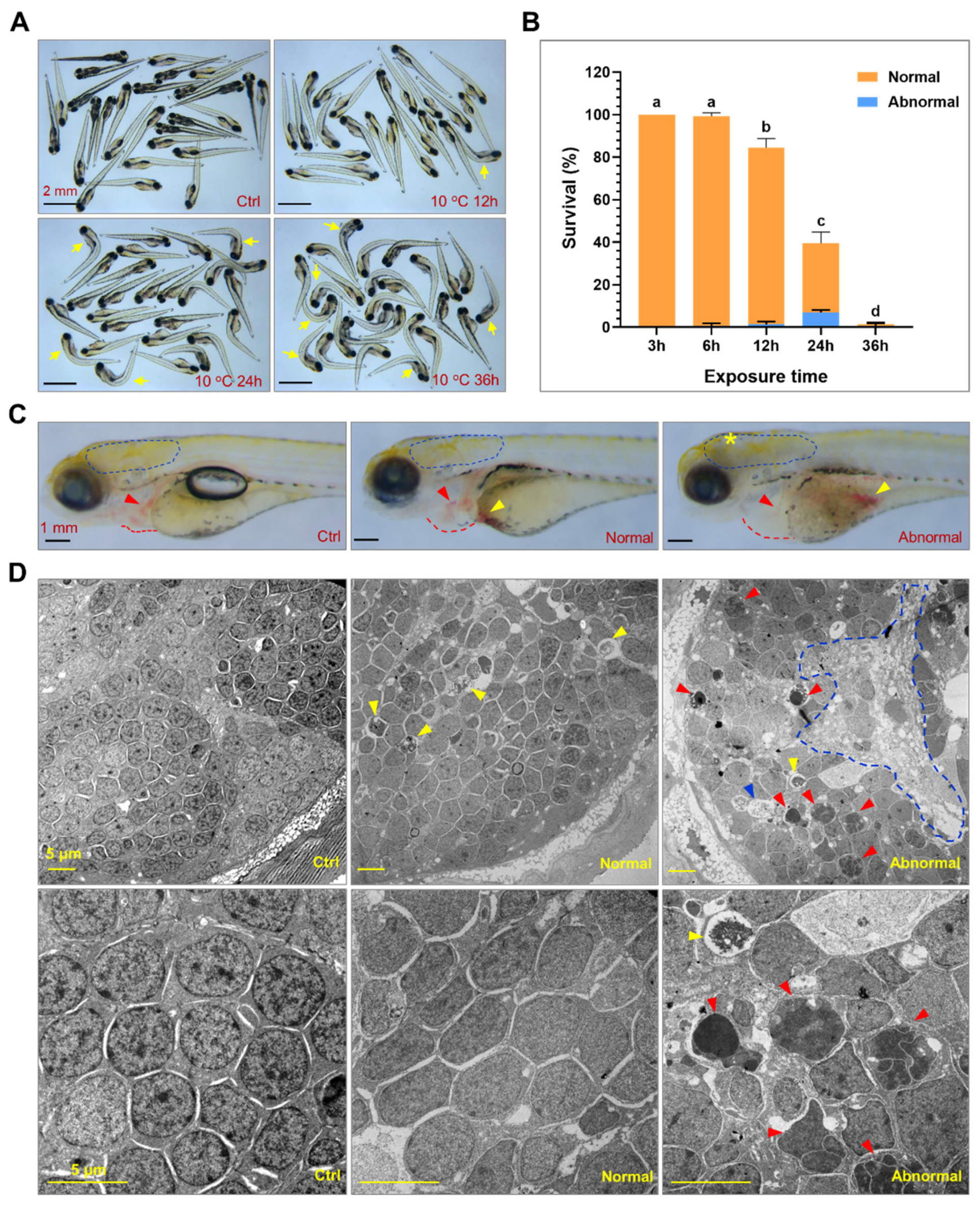
2.1. Exposure to Lethal Cold Stress Caused Irreversible Tissue Damage
2.2. Transcriptome Landscape of Zebrafish Larvae upon Cold Stress and Rewarming
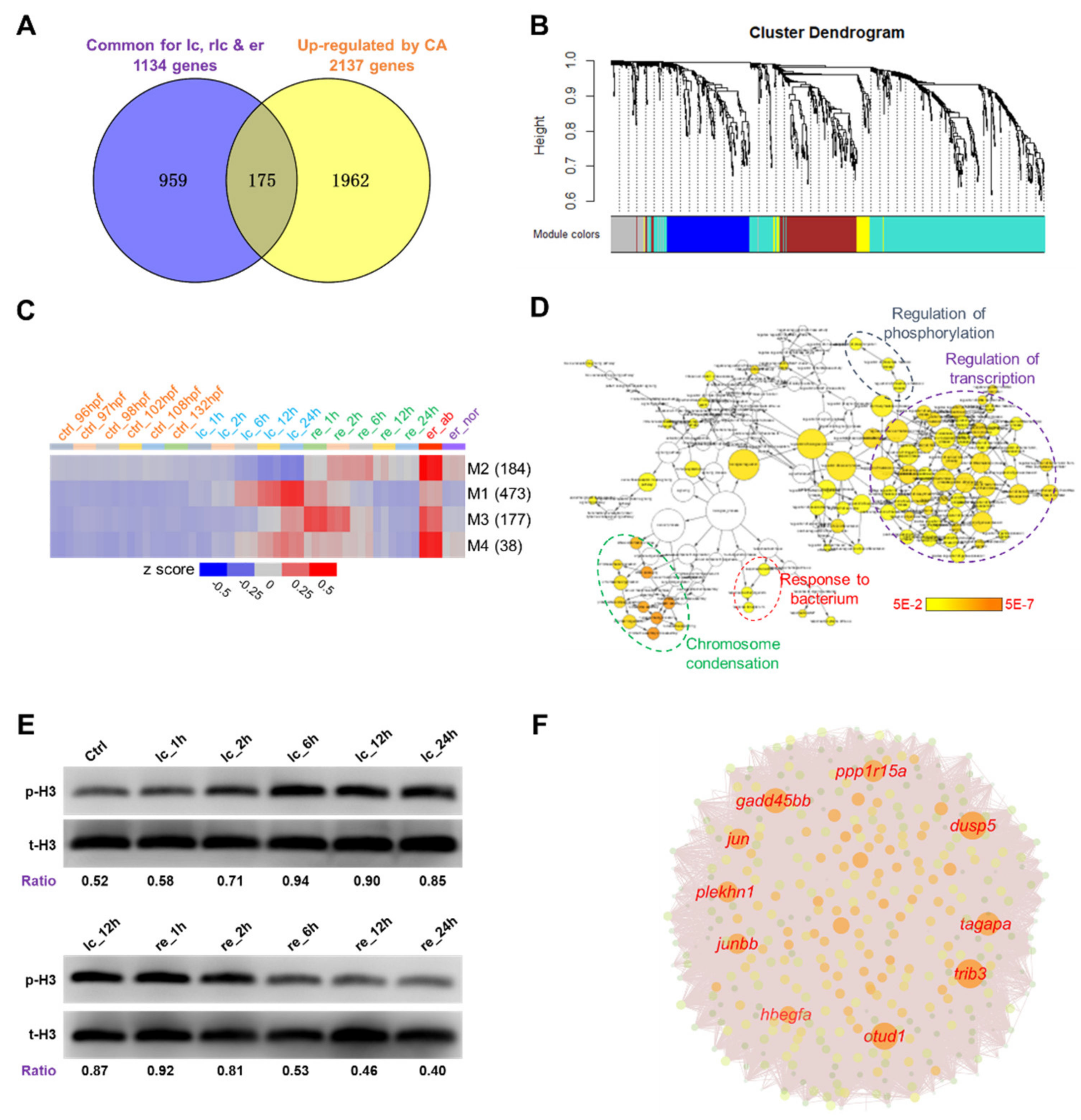
2.3. DEG Clusters and the Hub Genes
2.4. Identification of Potential Molecular Markers for Cold-Induced Damage
2.5. Biological Pathways Affected by Cold Stress and Rewarming
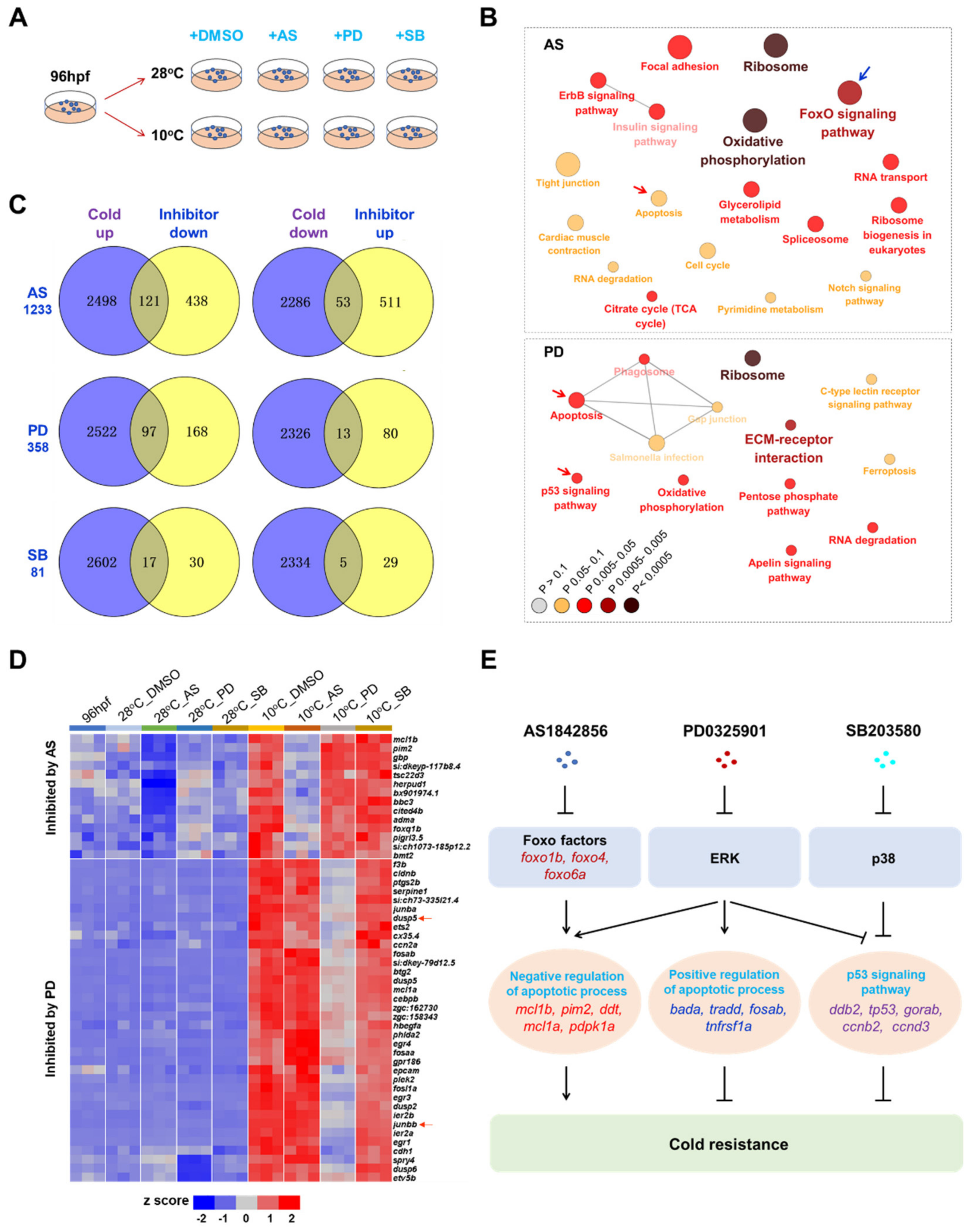
2.6. Identification of Survival Pathways for Cold Resistance and Death Pathways for Cold-Induced Mortality
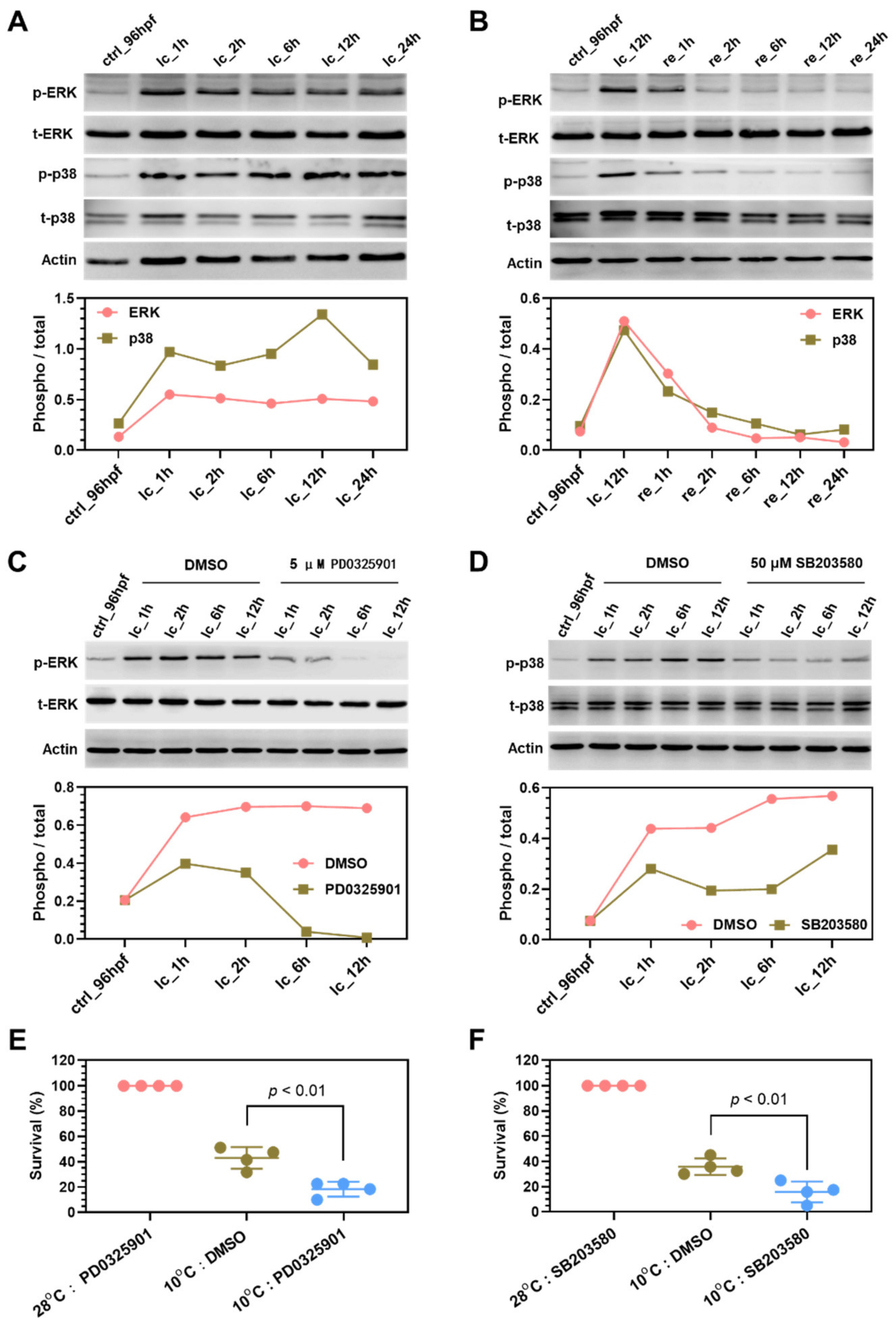
2.7. Mechanisms Underlying Functions of the Survival Pathways
3. Discussion
4. Materials and Methods
4.1. Experimental Fish
4.2. Chemicals and Small Molecule Inhibitors
4.3. Cold Sensitivity Assays and Measurements
4.4. Transmission Electron Microscopy
4.5. Preparation of Samples for RNA-Seq
4.6. Total RNA Extraction and RNA Sequencing
4.7. Differential Gene Expression Analyses
4.8. Bioinformatic Analyses
4.9. Quantitative Real-Time PCR Assays
4.10. Western Blotting
4.11. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| RNA-seq | RNA sequencing |
| ERK | Extracellular signal-regulated kinase |
| MAPK | Mitogen-activated protein kinase |
| ROS | Reactive oxygen species |
| CRG | Cold responsive gene |
| PCA | Principal component analysis |
| DEG | Differentially expressed gene |
| qPCR | Quantitative real-time PCR |
| GO | Gene ontology |
| CID | Cold-induced damage |
| WGCNA | Weighted gene coexpression network analysis |
| KEGG | Kyoto encyclopedia of genes and genomes |
| GSEA | Gene set enrichment analyses |
| TPM | Transcript per million |
| GEO | Gene expression omnibus |
| MCC | Maximal clique centrality |
References
- Donaldson, M.R.; Cooke, S.J.; Patterson, D.A.; Macdonald, J.S. Cold shock and fish. J. Fish. Biol. 2008, 73, 1491–1530. [Google Scholar] [CrossRef]
- Somero, G.N.; Hochachka, P.W. Biochemical Adaptation to Environment. Am. Zool. 1971, 11, 159–167. [Google Scholar] [CrossRef]
- Los, D.A.; Murata, N. Membrane fluidity and its roles in the perception of environmental signals. Biochim. Biophys. Acta 2004, 1666, 142–157. [Google Scholar] [CrossRef] [PubMed]
- Eckerle, L.G.; Lucassen, M.; Hirse, T.; Portner, H.O. Cold induced changes of adenosine levels in common eelpout (Zoarces viviparus): A role in modulating cytochrome c oxidase expression. J. Exp. Biol. 2008, 211, 1262–1269. [Google Scholar] [CrossRef]
- Ou, J.X.; Ball, J.M.; Luan, Y.Z.; Zhao, T.T.; Miyagishima, K.J.; Xu, Y.F.; Zhou, H.Z.; Chen, J.G.; Merriman, D.K.; Xie, Z.; et al. iPSCs from a Hibernator Provide a Platform for Studying Cold Adaptation and Its Potential Medical Applications. Cell 2018, 173, 851–863. [Google Scholar] [CrossRef] [PubMed]
- Hattori, K.; Ishikawa, H.; Sakauchi, C.; Takayanagi, S.; Naguro, I.; Ichijo, H. Cold stress-induced ferroptosis involves the ASK1-p38 pathway. EMBO Rep. 2017, 18, 2067–2078. [Google Scholar] [CrossRef] [PubMed]
- Kassahn, K.S.; Crozier, R.H.; Portner, H.O.; Caley, M.J. Animal performance and stress: Responses and tolerance limits at different levels of biological organisation. Biol. Rev. 2009, 84, 277–292. [Google Scholar] [CrossRef]
- van den Burg, E.H.; Peeters, R.R.; Verhoye, M.; Meek, J.; Flik, G.; Van der Linden, A. Brain responses to ambient temperature fluctuations in fish: Reduction of blood volume and initiation of a whole-body stress response. J. Neurophysiol. 2005, 93, 2849–2855. [Google Scholar] [CrossRef] [PubMed]
- Black, M.C.; Millsap, D.S.; Mccarthy, J.F. Effects of Acute Temperature-Change on Respiration and Toxicant Uptake by Rainbow-Trout, Salmo-Gairdneri (Richardson). Physiol. Zool. 1991, 64, 145–168. [Google Scholar] [CrossRef]
- Abram, Q.H.; Dixon, B.; Katzenback, B.A. Impacts of Low Temperature on the Teleost Immune System. Biology 2017, 6, 39. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Gui, L.; Liu, M.; Li, W.; Hu, P.; Duarte, D.F.C.; Niu, H.; Chen, L. Transcriptomic responses to low temperature stress in the Nile tilapia, Oreochromis niloticus. Fish. Shellfish Immunol. 2019, 84, 1145–1156. [Google Scholar] [CrossRef] [PubMed]
- Hurst, T.P.; Conover, D.O. Winter mortality of young-of-the-year Hudson River striped bass (Morone saxatilis): Size-dependent patterns and effects on recruitment. Can. J. Fish. Aquat. Sci. 1998, 55, 1122–1130. [Google Scholar] [CrossRef]
- Somero, G.N. The cellular stress response and temperature: Function, regulation, and evolution. J. Exp. Zool. Part A 2020, 333, 379–397. [Google Scholar] [CrossRef]
- Hurst, T.P. Causes and consequences of winter mortality in fishes. J. Fish. Biol. 2007, 71, 315–345. [Google Scholar] [CrossRef]
- Schulte, P.M. What is environmental stress? Insights from fish living in a variable environment. J. Exp. Biol. 2014, 217, 23–34. [Google Scholar] [CrossRef]
- Johnston, I.A.; Dunn, J. Temperature acclimation and metabolism in ectotherms with particular reference to teleost fish. Symp. Soc. Exp. Biol. 1987, 41, 67–93. [Google Scholar] [PubMed]
- Long, Y.; Song, G.; Yan, J.; He, X.; Li, Q.; Cui, Z. Transcriptomic characterization of cold acclimation in larval zebrafish. BMC Genom. 2013, 14, 612. [Google Scholar] [CrossRef]
- Long, Y.; Li, X.; Li, F.; Ge, G.; Liu, R.; Song, G.; Li, Q.; Qiao, Z.; Cui, Z. Transcriptional Programs Underlying Cold Acclimation of Common Carp (Cyprinus carpio L.). Front. Genet. 2020, 11, 556418. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Qiang, J.; Yang, H.; Xu, P.; Zhu, Z.X.; Yang, R.Q. Changes in the fatty acid composition and regulation of antioxidant enzymes and physiology of juvenile genetically improved farmed tilapia Oreochromis niloticus (L.), subjected to short-term low temperature stress. J. Biol. 2015, 53, 90–97. [Google Scholar] [CrossRef]
- Mommsen, T.P.; Vijayan, M.M.; Moon, T.W. Cortisol in teleosts: Dynamics, mechanisms of action, and metabolic regulation. Rev. Fish. Biol. Fish. 1999, 9, 211–268. [Google Scholar] [CrossRef]
- Gracey, A.Y.; Fraser, E.J.; Li, W.Z.; Fang, Y.X.; Taylor, R.R.; Rogers, J.; Brass, A.; Cossins, A.R. Coping with cold: An integrative, multitissue analysis of the transcriptome of a poikilothermic vertebrate. Proc. Natl. Acad. Sci. USA 2004, 101, 16970–16975. [Google Scholar] [CrossRef] [PubMed]
- Jayasundara, N.; Tomanek, L.; Dowd, W.W.; Somero, G.N. Proteomic analysis of cardiac response to thermal acclimation in the eurythermal goby fish Gillichthys mirabilis. J. Exp. Biol. 2015, 218, 1359–1372. [Google Scholar] [CrossRef]
- Wen, X.; Zhang, X.Y.; Hu, Y.D.; Xu, J.J.; Wang, T.; Yin, S.W. iTRAQ-based quantitative proteomic analysis of Takifugu fasciatus liver in response to low-temperature stress. J. Proteom. 2019, 201, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.J.; Long, Y.; Zhou, T.; Ren, J.; Li, Q.; Song, G.L.; Cui, Z.B. Dynamic Phosphoproteome Profiling of Zebrafish Embryonic Fibroblasts during Cold Acclimation. Proteomics 2020, 20, e1900257. [Google Scholar] [CrossRef]
- Melis, R.; Sanna, R.; Braca, A.; Bonaglini, E.; Cappuccinelli, R.; Slawski, H.; Roggio, T.; Uzzau, S.; Anedda, R. Molecular details on gilthead sea bream (Sparus aurata) sensitivity to low water temperatures from H-1 NMR metabolomics. Comp. Biochem. Phys. A 2017, 204, 129–136. [Google Scholar] [CrossRef]
- Mele, M.; Ferreira, P.G.; Reverter, F.; DeLuca, D.S.; Monlong, J.; Sammeth, M.; Young, T.R.; Goldmann, J.M.; Pervouchine, D.D.; Sullivan, T.J.; et al. Human genomics. The human transcriptome across tissues and individuals. Science 2015, 348, 660–665. [Google Scholar] [CrossRef]
- Ge, G.; Long, Y.; Shi, L.; Ren, J.; Yan, J.; Li, C.; Li, Q.; Cui, Z. Transcriptomic profiling revealed key signaling pathways for cold tolerance and acclimation of two carp species. BMC Genom. 2020, 21, 539. [Google Scholar] [CrossRef]
- Li, B.J.; Zhu, Z.X.; Qin, H.; Meng, Z.N.; Lin, H.R.; Xia, J.H. Genome-Wide Characterization of Alternative Splicing Events and Their Responses to Cold Stress in Tilapia. Front. Genet. 2020, 11, 244. [Google Scholar] [CrossRef] [PubMed]
- Healy, T.M.; Schulte, P.M. Patterns of alternative splicing in response to cold acclimation in fish. J. Exp. Biol. 2019, 222, jeb193516. [Google Scholar] [CrossRef]
- Hu, P.; Liu, M.; Zhang, D.; Wang, J.; Niu, H.; Liu, Y.; Wu, Z.; Han, B.; Zhai, W.; Shen, Y.; et al. Global identification of the genetic networks and cis-regulatory elements of the cold response in zebrafish. Nucleic Acids Res. 2015, 43, 9198–9213. [Google Scholar] [CrossRef]
- Liang, L.Q.; Chang, Y.M.; He, X.L.; Tang, R. Transcriptome Analysis to Identify Cold-Responsive Genes in Amur Carp (Cyprinus carpio haematopterus). PLoS ONE 2015, 10, e0130526. [Google Scholar] [CrossRef]
- Hung, I.C.; Hsiao, Y.C.; Sun, H.S.; Chen, T.M.; Lee, S.J. MicroRNAs regulate gene plasticity during cold shock in zebrafish larvae. BMC Genom. 2016, 17, 922. [Google Scholar] [CrossRef] [PubMed]
- Qiang, J.; Cui, Y.T.; Tao, F.Y.; Bao, W.J.; He, J.; Li, X.H.; Xu, P.; Sun, L.Y. Physiological response and microRNA expression profiles in head kidney of genetically improved farmed tilapia (GIFT, Oreochromis niloticus) exposed to acute cold stress. Sci. Rep. UK 2018, 8, 172. [Google Scholar] [CrossRef]
- Fujiwara, Y.; Denlinger, D.L. p38 MAPK is a likely component of the signal transduction pathway triggering rapid cold hardening in the flesh fly Sarcophaga crassipalpis. J. Exp. Biol. 2007, 210, 3295–3300. [Google Scholar] [CrossRef]
- Luo, Z.Z.; Huang, W.; Wang, G.X.; Sun, H.M.; Chen, X.Y.; Luo, P.; Liu, J.S.; Hu, C.Q.; Li, H.; Shu, H. Identification and characterization of p38(MAPK) in response to acute cold stress in the gill of Pacific white shrimp (Litopenaeus vannamei). Aquacult. Rep. 2020, 17, 100365. [Google Scholar] [CrossRef]
- Xu, B.; Lian, S.; Guo, J.R.; Wang, J.F.; Zhang, L.P.; Li, S.Z.; Yang, H.M. Activation of the MAPK signaling pathway induces upregulation of pro-apoptotic proteins in the hippocampi of cold stressed adolescent mice. Neurosci. Lett. 2019, 699, 97–102. [Google Scholar] [CrossRef]
- Lopez-Olmeda, J.F.; Sanchez-Vazquez, F.J. Thermal biology of zebrafish (Danio rerio). J. Biol. 2011, 36, 91–104. [Google Scholar] [CrossRef]
- Chou, M.Y.; Hsiao, C.D.; Chen, S.C.; Chen, I.W.; Liu, S.T.; Hwang, P.P. Effects of hypothermia on gene expression in zebrafish gills: Upregulation in differentiation and function of ionocytes as compensatory responses. J. Exp. Biol. 2008, 211, 3077–3084. [Google Scholar] [CrossRef] [PubMed]
- Martinet, W.; Timmermans, J.P.; De Meyer, G.R. Methods to assess autophagy in situ--transmission electron microscopy versus immunohistochemistry. Methods Enzym. 2014, 543, 89–114. [Google Scholar]
- Burattini, S.; Falcieri, E. Analysis of cell death by electron microscopy. Methods Mol. Biol. 2013, 1004, 77–89. [Google Scholar] [PubMed]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Henderson, Y.C.; Chen, Y.Y.; Frederick, M.J.; Lai, S.Y.; Clayman, G.L. MEK Inhibitor PD0325901 Significantly Reduces the Growth of Papillary Thyroid Carcinoma Cells in vitro and In vivo. Mol. Cancer 2010, 9, 1968–1976. [Google Scholar] [CrossRef]
- Long, Y.; Li, L.C.; Li, Q.; He, X.Z.; Cui, Z.B. Transcriptomic Characterization of Temperature Stress Responses in Larval Zebrafish. PLoS ONE 2012, 7, e37209. [Google Scholar] [CrossRef] [PubMed]
- Clark, K.J.; Boczek, N.J.; Ekker, S.C. Stressing zebrafish for behavioral genetics. Rev. Neurosci. 2011, 22, 49–62. [Google Scholar] [CrossRef] [PubMed]
- Francis, A.; Baynosa, R. Ischaemia-reperfusion injury and hyperbaric oxygen pathways: A review of cellular mechanisms. Diving Hyperb. Med. 2017, 47, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, M.; Cahill, G.M. Light-dependent development of circadian gene expression in transgenic zebrafish. PLoS Biol. 2005, 3, 313–323. [Google Scholar] [CrossRef] [PubMed]
- Caratozzolo, M.F.; Micale, L.; Turturo, M.G.; Cornacchia, S.; Fusco, C.; Marzano, F.; Augello, B.; D’Erchia, A.M.; Guerrini, L.; Pesole, G.; et al. TRIM8 modulates p53 activity to dictate cell cycle arrest. Cell Cycle 2012, 11, 511–523. [Google Scholar] [CrossRef]
- Ohoka, N.; Yoshii, S.; Hattori, T.; Onozaki, K.; Hayashi, H. TRB3, a novel ER stress-inducible gene, is induced via ATF4-CHOP pathway and is involved in cell death. EMBO J. 2005, 24, 1243–1255. [Google Scholar] [CrossRef]
- Qian, N.; Frank, D.; OKeefe, D.; Dao, D.; Zhao, L.; Yuan, L.; Wang, Q.; Keating, M.; Walsh, C.; Tycko, B. The IPL gene on chromosome 11p15.5 is imprinted in humans and mice and is similar to TDAG51, implicated in Fas expression and apoptosis. Hum. Mol. Genet. 1997, 6, 2021–2029. [Google Scholar] [CrossRef]
- Zhang, X.; Li, C.F.; Zhang, L.; Wu, C.Y.; Han, L.X.; Jin, G.X.; Rezaeian, A.H.; Han, F.; Liu, C.F.; Xu, C.; et al. TRAF6 Restricts p53 Mitochondrial Translocation, Apoptosis, and Tumor Suppression. Mol. Cell 2016, 64, 803–814. [Google Scholar] [CrossRef]
- Chen, D.; Zhang, J.B.; Li, M.; Rayburn, E.R.; Wang, H.; Zhang, R.W. RYBP stabilizes p53 by modulating MDM2. EMBO Rep. 2009, 10, 166–172. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Hitchler, M.J.; Sun, W.; Sarsour, E.H.; Goswami, P.C.; Klingelhutz, A.J.; Domann, F.E. AP-2alpha Inhibits c-MYC Induced Oxidative Stress and Apoptosis in HaCaT Human Keratinocytes. J. Oncol. 2009, 2009, 780874. [Google Scholar] [CrossRef] [PubMed]
- Alva, N.; Palomeque, J.; Carbonell, T. Oxidative stress and antioxidant activity in hypothermia and rewarming: Can RONS modulate the beneficial effects of therapeutic hypothermia? Oxid Med. Cell Longev. 2013, 2013, 957054. [Google Scholar] [CrossRef] [PubMed]
- Wen, W.; Pillai-Kastoori, L.; Wilson, S.G.; Morris, A.C. Sox4 regulates choroid fissure closure by limiting Hedgehog signaling during ocular morphogenesis. Dev. Biol. 2015, 399, 139–153. [Google Scholar] [CrossRef] [PubMed]
- Hartel, F.V.; Holl, M.; Arshad, M.; Aslam, M.; Gunduz, D.; Weyand, M.; Micoogullari, M.; Abdallah, Y.; Piper, H.M.; Noll, T. Transient hypoxia induces ERK-dependent anti-apoptotic cell survival in endothelial cells. Am. J. Physiol. Cell Ph. 2010, 298, C1501–C1509. [Google Scholar] [CrossRef] [PubMed]
- Cagnol, S.; Chambard, J.C. ERK and cell death: Mechanisms of ERK-induced cell death—Apoptosis, autophagy and senescence. FEBS J. 2010, 277, 2–21. [Google Scholar] [CrossRef]
- Teets, N.M.; Yi, S.X.; Lee, R.E.; Denlinger, D.L. Calcium signaling mediates cold sensing in insect tissues. Proc. Natl. Acad. Sci. USA 2013, 110, 9154–9159. [Google Scholar] [CrossRef] [PubMed]
- Long, Y.; Li, Q.; Zhong, S.; Wang, Y.; Cui, Z. Molecular characterization and functions of zebrafish ABCC2 in cellular efflux of heavy metals. Comp. Biochem. Physiol. C Toxicol. Pharm. 2011, 153, 381–391. [Google Scholar] [CrossRef]
- Hagedorn, M.; Carter, V.L. Zebrafish reproduction: Revisiting in vitro fertilization to increase sperm cryopreservation success. PLoS ONE 2011, 6, e21059. [Google Scholar] [CrossRef]
- Long, Y.; Liu, R.; Song, G.; Li, Q.; Cui, Z. Establishment and characterization of a cold-sensitive neural cell line from the brain of tilapia (Oreochromis niloticus). J. Fish. Biol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef] [PubMed]
- Soneson, C.; Love, M.I.; Robinson, M.D. Differential analyses for RNA-seq: Transcript-level estimates improve gene-level inferences. F1000Res 2015, 4, 1521. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Kelly, R.; Fang, H.; Tong, W. ArrayTrack: A free FDA bioinformatics tool to support emerging biomedical research—An update. Hum. Genom. 2010, 4, 428–434. [Google Scholar] [CrossRef]
- de Hoon, M.J.; Imoto, S.; Nolan, J.; Miyano, S. Open source clustering software. Bioinformatics 2004, 20, 1453–1454. [Google Scholar] [CrossRef]
- Page, R.D. TreeView: An application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 1996, 12, 357–358. [Google Scholar]
- Venny Oliveros, J.C. An interactive tool for comparing lists with Venn’s diagrams. In Anonymous, 2007. [Google Scholar]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Maere, S.; Heymans, K.; Kuiper, M. BiNGO: A Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 2005, 21, 3448–3449. [Google Scholar] [CrossRef] [PubMed]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.H.; Pages, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [PubMed]
- Chin, C.H.; Chen, S.H.; Wu, H.H.; Ho, C.W.; Ko, M.T.; Lin, C.Y. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014, 8, S11. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ren, J.; Long, Y.; Liu, R.; Song, G.; Li, Q.; Cui, Z. Characterization of Biological Pathways Regulating Acute Cold Resistance of Zebrafish. Int. J. Mol. Sci. 2021, 22, 3028. https://doi.org/10.3390/ijms22063028
Ren J, Long Y, Liu R, Song G, Li Q, Cui Z. Characterization of Biological Pathways Regulating Acute Cold Resistance of Zebrafish. International Journal of Molecular Sciences. 2021; 22(6):3028. https://doi.org/10.3390/ijms22063028
Chicago/Turabian StyleRen, Jing, Yong Long, Ran Liu, Guili Song, Qing Li, and Zongbin Cui. 2021. "Characterization of Biological Pathways Regulating Acute Cold Resistance of Zebrafish" International Journal of Molecular Sciences 22, no. 6: 3028. https://doi.org/10.3390/ijms22063028
APA StyleRen, J., Long, Y., Liu, R., Song, G., Li, Q., & Cui, Z. (2021). Characterization of Biological Pathways Regulating Acute Cold Resistance of Zebrafish. International Journal of Molecular Sciences, 22(6), 3028. https://doi.org/10.3390/ijms22063028





