Reduced Granule Cell Proliferation and Molecular Dysregulation in the Cerebellum of Lysosomal Acid Phosphatase 2 (ACP2) Mutant Mice
Abstract
1. Introduction
2. Results
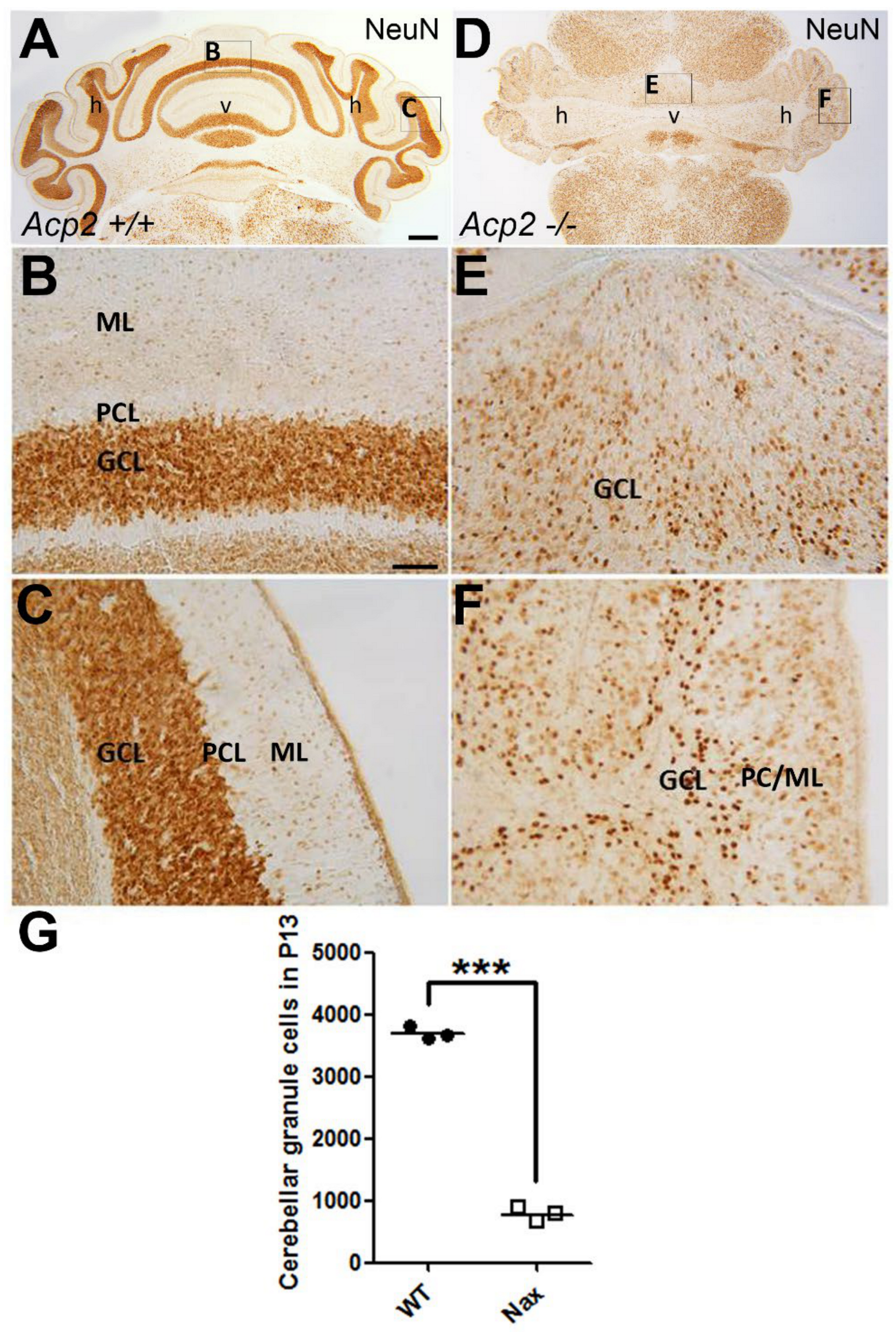
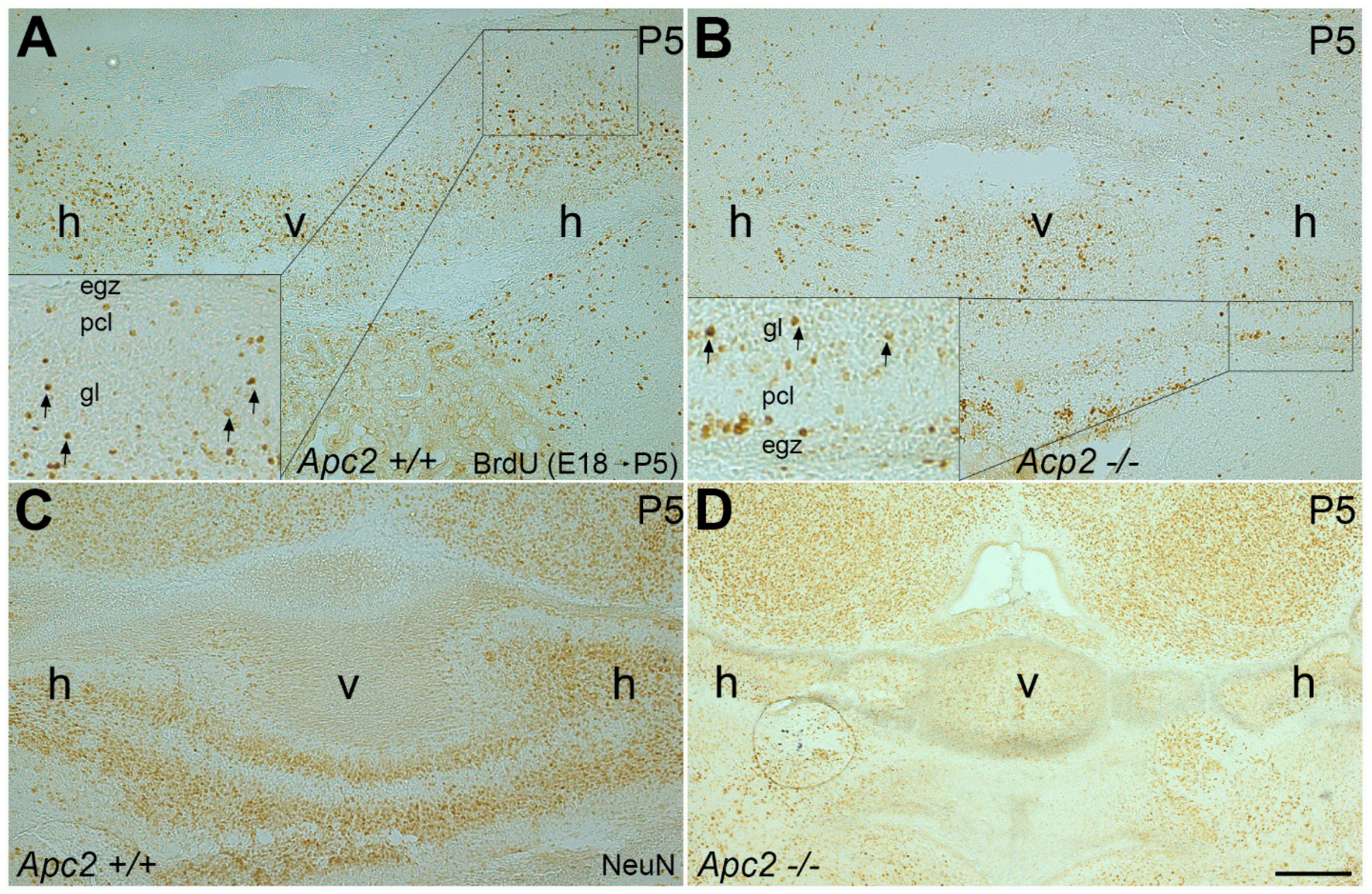
2.1. The Number of Cerebellar GCs Is Significantly Decreased in the nax Cerebellum
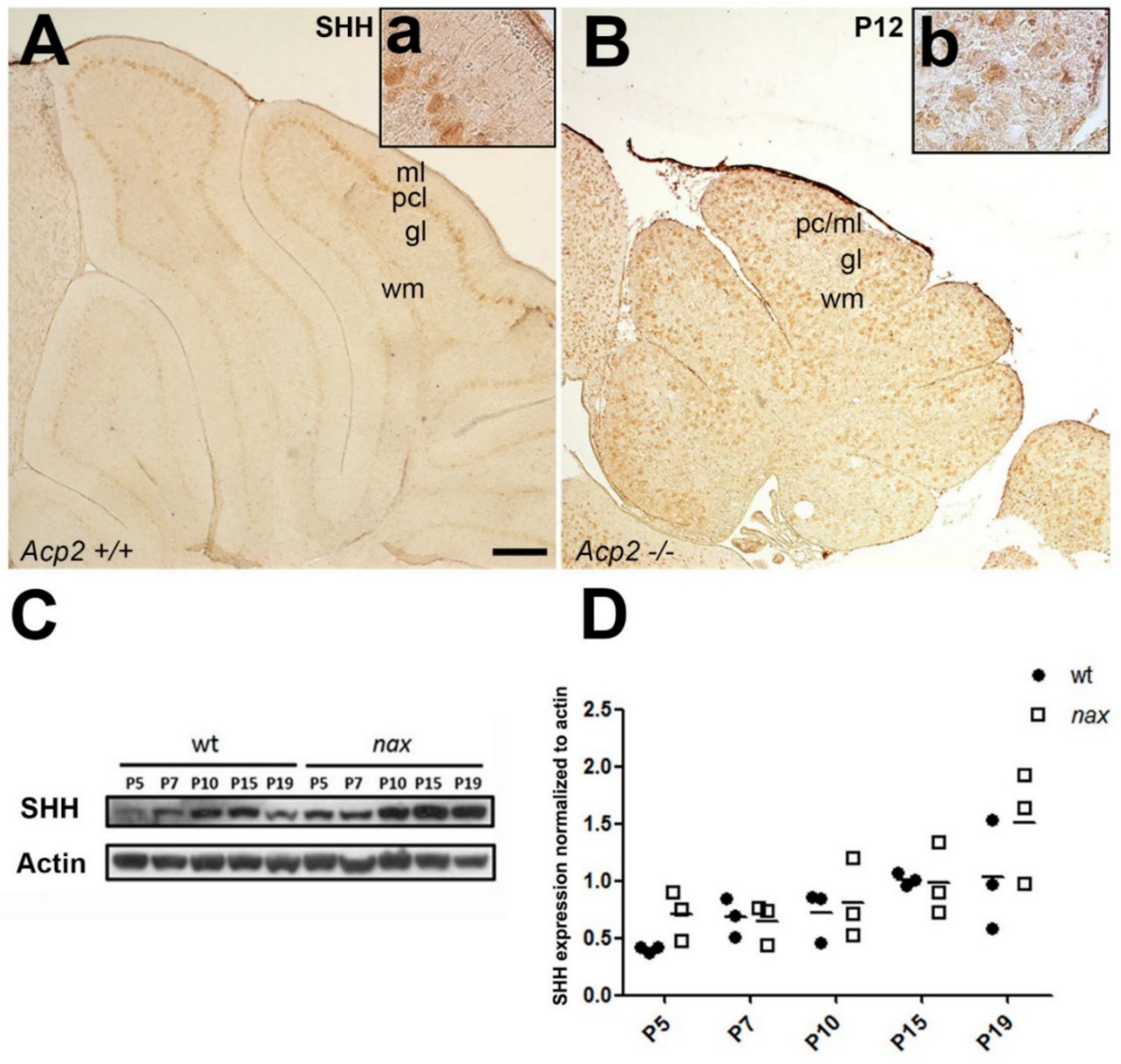
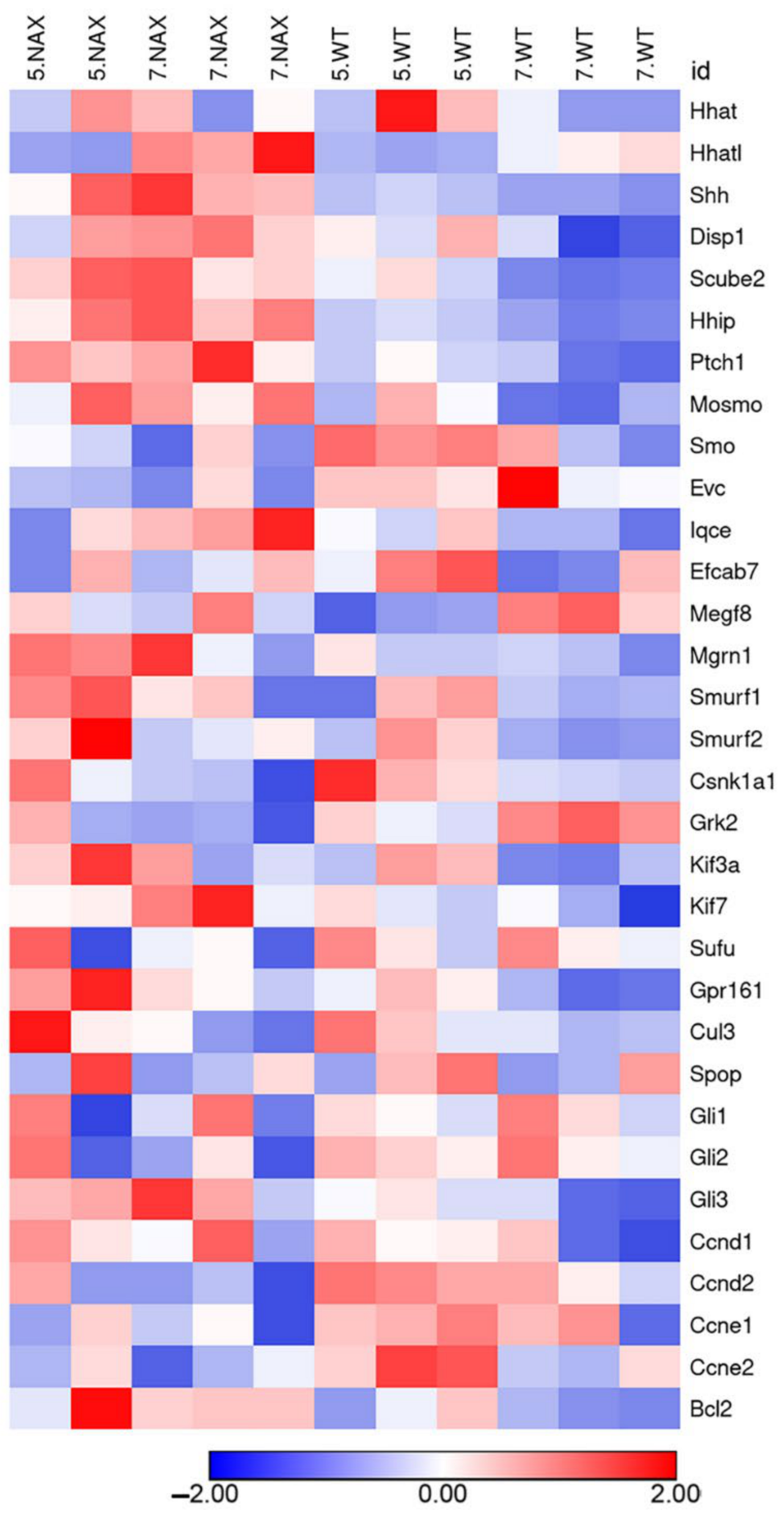
2.2. Is the SHH Signaling Pathway Impaired in the nax Cerebellum?
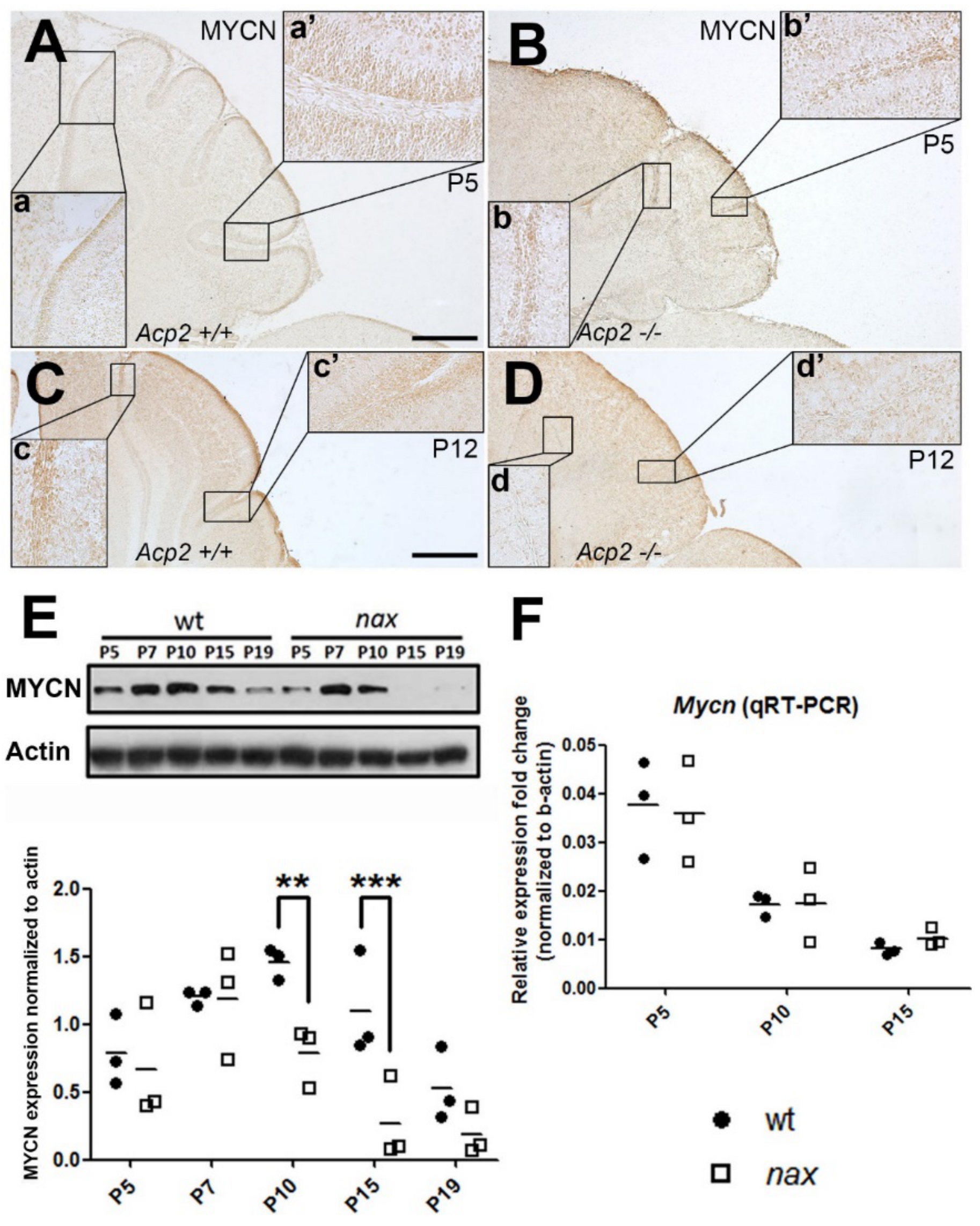
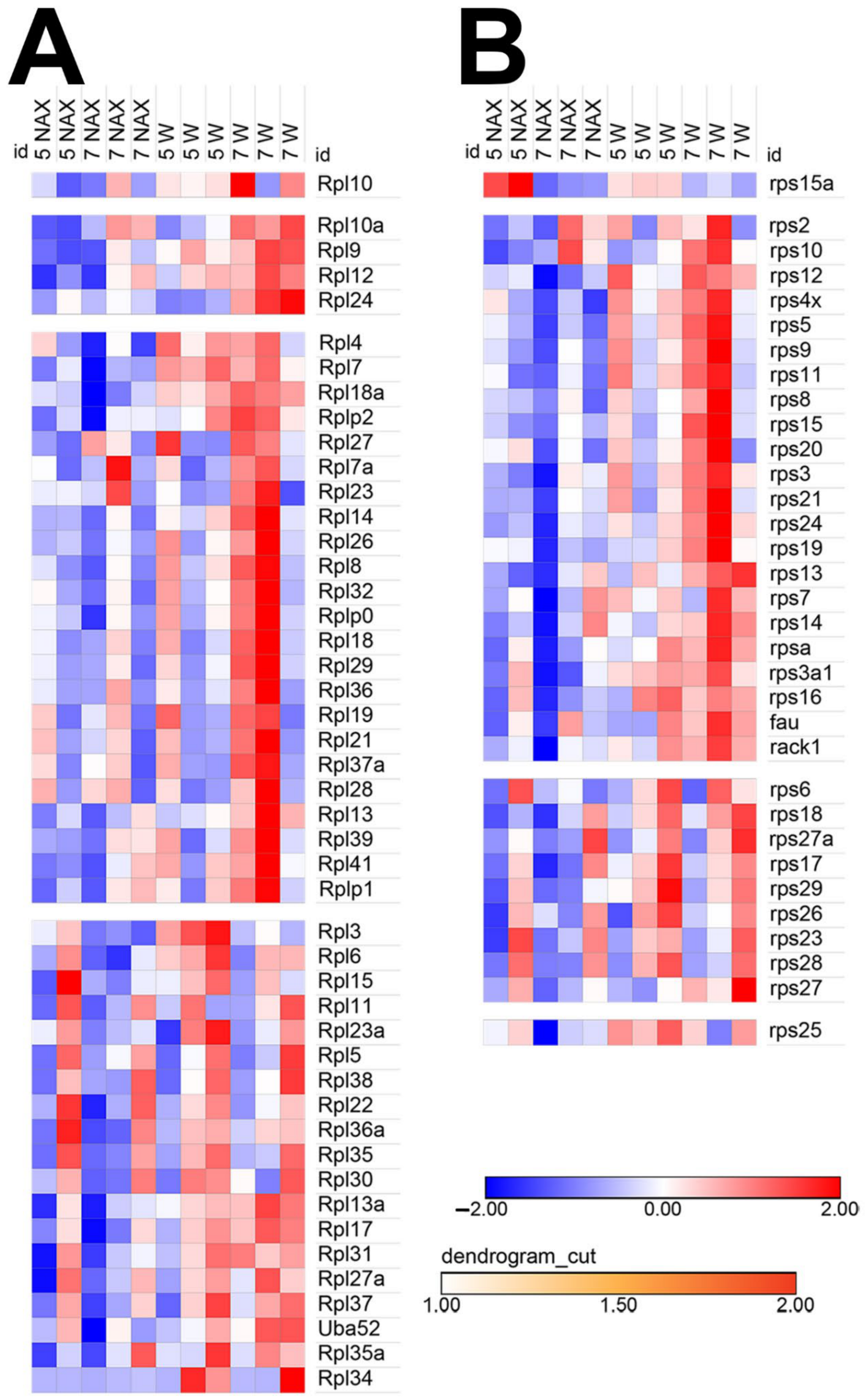
2.3. MYCN Expression Patterns Are Altered in the nax Cerebellum
3. Discussion
4. Materials and Methods
4.1. Animal Model
4.2. Genotyping
4.3. Cryosection
4.4. Immunohistochemistry
4.5. Bromodeoxyuridine (BrdU) Incorporation Assay
4.6. Western Blotting
4.7. RNA Isolation and Analyses
4.8. Gene Expression Analyses
4.9. Mouse Embryonic Fibroblast (MEF) Culture
4.10. Proteasome Activity Assay
4.11. Imaging
4.12. Cell Counting
4.13. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Acp2 | Lysosomal acid phosphatase 2 |
| BOC | Brother of Codon |
| CDON | Codon |
| CNS | central nervous system |
| DAB | diaminobenzidine |
| E | embryonic day |
| EGL | external granule cell layer |
| EGZ | external germinal zone |
| GCL | granule cell layer |
| GFAP | glial fibrillary acidic protein |
| Gli | glioma-associated oncogene homolog |
| IHC | immunohistochemistry |
| MEF | Mouse Embryonic Fibroblast |
| ML | molecular layer |
| nax | naked and ataxia |
| PAX6 | paired box protein 6 |
| PCL | Purkinje cell layer |
| PFA | paraformaldehyde |
| PTCH1 | Patched 1 |
| SHH | sonic hedgehog |
| SMO | smoothened |
| WB | Western blotting |
| wt | wild type |
References
- Steinlin, M. Cerebellar Disorders in Childhood: Cognitive Problems. Cerebellum 2008, 7, 607–610. [Google Scholar] [CrossRef]
- Thach, W.T. On the mechanism of cerebellar contributions to cognition. Cerebellum 2007, 6, 163–167. [Google Scholar] [CrossRef]
- Wolf, U.; Rapoport, M.J.; Schweizer, T.A. Evaluating the affective component of the cerebellar cognitive affective syndrome. J. Neuropsychiatry Clin. Neurosci. 2009, 21, 245–253. [Google Scholar] [CrossRef]
- Marzban, H.; Del Bigio, M.R.; Alizadeh, J.; Ghavami, S.; Zachariah, R.M.; Rastegar, M. Cellular commitment in the developing cer-ebellum. Front. Cell. Neurosci. 2014, 8, 450. [Google Scholar]
- Llinas, R.; Negrello, M.N. Cerebellum. Scholarpedia 2015, 10, 4606. [Google Scholar] [CrossRef]
- Azevedo, F.A.C.; Carvalho, L.R.B.; Grinberg, L.T.; Farfel, J.M.; Ferretti, R.E.L.; Leite, R.E.P.; Filho, W.J.; Lent, R.; Herculano-Houzel, S. Equal numbers of neuronal and nonneuronal cells make the human brain an isometrically scaled-up primate brain. J. Comp. Neurol. 2009, 513, 532–541. [Google Scholar] [CrossRef] [PubMed]
- Consalez, G.G.; Goldowitz, D.; Casoni, F.; Hawkes, R. Origins, Development, and Compartmentation of the Granule Cells of the Cerebellum. Front. Neural Circuits 2021, 14. [Google Scholar] [CrossRef] [PubMed]
- Machold, R.; Fishell, G. Math1 Is Expressed in Temporally Discrete Pools of Cerebellar Rhombic-Lip Neural Progenitors. Neuron 2005, 48, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Yeung, J.; Ha, T.J.; Swanson, U.J.; Choi, K.; Tong, Y.; Goldowitz, D. Wls provides a new compartmental view of the rhombic lip in mouse cerebellar development. J. Neurosci. 2014, 34, 12527–12537. [Google Scholar] [CrossRef]
- Yacubova, E.; Komuro, H. Cellular and molecular mechanisms of cerebellar granule cell migration. Cell Biophys. 2002, 37, 213–234. [Google Scholar] [CrossRef]
- Espinosa, J.S.; Luo, L. Timing Neurogenesis and Differentiation: Insights from Quantitative Clonal Analyses of Cerebellar Granule Cells. J. Neurosci. 2008, 28, 2301–2312. [Google Scholar] [CrossRef] [PubMed]
- Legué, E.; Riedel, E.; Joyner, A.L. Clonal analysis reveals granule cell behaviors and compartmentalization that determine the folded morphology of the cerebellum. Development 2015, 142, 1661–1671. [Google Scholar] [CrossRef]
- Nakashima, K.; Umeshima, H.; Kengaku, M. Cerebellar granule cells are predominantly generated by terminal symmetric divisions of granule cell precursors. Dev. Dyn. 2015, 244, 748–758. [Google Scholar] [CrossRef]
- Zanin, J.P.; Verpeut, J.L.; Li, Y.; Shiflett, M.W.; Wang, S.S.-H.; Santhakumar, V.; Friedman, W.J. The p75NTR Influences Cerebellar Circuit Development and Adult Behavior via Regulation of Cell Cycle Duration of Granule Cell Progenitors. J. Neurosci. 2019, 39, 9119–9129. [Google Scholar] [CrossRef] [PubMed]
- Aguilar, A.; Meunier, A.; Strehl, L.; Martinovic, J.; Bonniere, M.; Attie-Bitach, T.; Encha-Razavi, F.; Spassky, N. Analysis of human samples reveals impaired SHH-dependent cerebellar development in Joubert syndrome/Meckel syndrome. Proc. Natl. Acad. Sci. USA 2012, 109, 16951–16956. [Google Scholar] [CrossRef]
- Haldipur, P.; Bharti, U.; Govindan, S.; Sarkar, C.; Iyengar, S.; Gressens, P.; Mani, S. Expression of Sonic Hedgehog During Cell Proliferation in the Human Cerebellum. Stem Cells Dev. 2012, 21, 1059–1068. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.J.; Lindsey, S.; Graves, B.; Yoo, S.; Olson, J.M.; Langhans, S.A. Sonic Hedgehog-Induced Histone Deacetylase Activation Is Required for Cerebellar Granule Precursor Hyperplasia in Medulloblastoma. PLoS ONE 2013, 8, e71455. [Google Scholar] [CrossRef]
- Lewis, P.M.; Gritli-Linde, A.; Smeyne, R.; Kottmann, A.; McMahon, A.P. Sonic hedgehog signaling is required for expansion of granule neuron precursors and patterning of the mouse cerebellum. Dev. Biol. 2004, 270, 393–410. [Google Scholar] [CrossRef] [PubMed]
- Delogu, A.; Sellers, K.; Zagoraiou, L.; Bocianowska-Zbrog, A.; Mandal, S.; Guimera, J.; Rubenstein, J.L.; Sugden, D.; Jessell, T.; Lumsden, A. Subcortical Visual Shell Nuclei Targeted by ipRGCs Develop from a Sox14+-GABAergic Progenitor and Require Sox14 to Regulate Daily Activity Rhythms. Neuron 2012, 75, 648–662. [Google Scholar] [CrossRef]
- Vuong, T.A.; Leem, Y.-E.; Kim, B.-G.; Cho, H.; Lee, S.-J.; Bae, G.-U.; Kang, J.-S. A Sonic hedgehog coreceptor, BOC regulates neuronal differentiation and neurite outgrowth via interaction with ABL and JNK activation. Cell. Signal. 2017, 30, 30–40. [Google Scholar] [CrossRef]
- Delloye-Bourgeois, C.; Rama, N.; Brito, J.; Le Douarin, N.; Mehlen, P. Sonic Hedgehog promotes the survival of neural crest cells by limiting apoptosis induced by the dependence receptor CDON during branchial arch development. Biochem. Biophys. Res. Commun. 2014, 452, 655–660. [Google Scholar] [CrossRef]
- Xavier, G.M.; Seppala, M.; Papageorgiou, S.N.; Fan, C.-M.; Cobourne, M.T. Genetic interactions between the hedgehog co-receptors Gas1 and Boc regulate cell proliferation during murine palatogenesis. Oncotarget 2016, 7, 79233–79246. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wechsler-Reya, R.J.; Scott, M.P. Control of Neuronal Precursor Proliferation in the Cerebellum by Sonic Hedgehog. Neuron 1999, 22, 103–114. [Google Scholar] [CrossRef]
- Corrales, J.D.; Rocco, G.L.; Blaess, S.; Guo, Q.; Joyner, A.L. Spatial pattern of sonic hedgehog signaling through Gli genes during cerebellum development. Development 2004, 131, 5581–5590. [Google Scholar] [CrossRef] [PubMed]
- Altaba, A.R.I. Gli proteins encode context-dependent positive and negative functions: Implications for development and disease. Development 1999, 126, 3205–3216. [Google Scholar]
- Ruppert, J.M.; Kinzler, K.W.; Wong, A.J.; Bigner, S.H.; Kao, F.T.; Law, M.L.; Seuanez, H.N.; O’Brien, S.J.; Vogelstein, B. The GLI-Kruppel family of human genes. Mol. Cell. Biol. 1988, 8, 3104–3113. [Google Scholar] [CrossRef]
- Mimeault, M.; Batra, S.K. Frequent Deregulations in the Hedgehog Signaling Network and Cross-Talks with the Epidermal Growth Factor Receptor Pathway Involved in Cancer Progression and Targeted Therapies. Pharmacol. Rev. 2010, 62, 497–524. [Google Scholar] [CrossRef]
- Carballo, G.B.; Honorato, J.R.; De Lopes, G.P.F.; Spohr, T.C.L.D.S.E. A highlight on Sonic hedgehog pathway. Cell Commun. Signal. 2018, 16, 1–15. [Google Scholar] [CrossRef]
- Kenney, A.M. Nmyc upregulation by sonic hedgehog signaling promotes proliferation in developing cerebellar granule neuron precursors. Development 2003, 130, 15–28. [Google Scholar] [CrossRef]
- Knoepfler, P.S.; Cheng, P.F.; Eisenman, R.N. N-myc is essential during neurogenesis for the rapid expansion of progenitor cell populations and the inhibition of neuronal differentiation. Genes Dev. 2002, 16, 2699–2712. [Google Scholar] [CrossRef]
- Hatton, B.A.; Knoepfler, P.S.; Kenney, A.M.; Rowitch, D.H.; De Alborán, I.M.; Olson, J.M.; Eisenman, R.N. N-myc Is an Essential Downstream Effector of Shh Signaling during both Normal and Neoplastic Cerebellar Growth. Cancer Res. 2006, 66, 8655–8661. [Google Scholar] [CrossRef] [PubMed]
- Pogoriler, J.; Millen, K.; Utset, M.; Du, W. Loss of cyclin D1 impairs cerebellar development and suppresses medulloblastoma formation. Development 2006, 133, 3929–3937. [Google Scholar] [CrossRef] [PubMed]
- Otero, J.J.; Kalaszczynska, I.; Michowski, W.; Wong, M.; Gygli, P.E.; Gokozan, H.N.; Griveau, A.; Odajima, J.; Czeisler, C.; Catacutan, F.P.; et al. Cerebellar cortical lamination and foliation require cyclin A2. Dev. Biol. 2014, 385, 328–339. [Google Scholar] [CrossRef] [PubMed]
- Ceccarelli, M.; Micheli, L.; D’Andrea, G.; De Bardi, M.; Scheijen, B.; Ciotti, M.; Leonardi, L.; Luvisetto, S.; Tirone, F. Altered cerebellum development and impaired motor coordination in mice lacking the Btg1 gene: Involvement of cyclin D1. Dev. Biol. 2015, 408, 109–125. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lübke, T.; Lobel, P.; Sleat, D.E. Proteomics of the lysosome. Biochim. Biophys. Acta (BBA) Bioenerg. 2009, 1793, 625–635. [Google Scholar] [CrossRef]
- Saftig, P.; Klumperman, J. Lysosome biogenesis and lysosomal membrane proteins: Trafficking meets function. Nat. Rev. Mol. Cell Biol. 2009, 10, 623–635. [Google Scholar] [CrossRef]
- Mannan, A.U.; Roussa, E.; Kraus, C.; Rickmann, M.; Maenner, J.; Nayernia, K.; Krieglstein, K.; Engel, W. Mutation in the gene encoding lysosomal acid phosphatase (Acp2) causes cerebellum and skin malformation in mouse. Neurogenetics 2004, 5, 229–238. [Google Scholar] [CrossRef]
- Bailey, K.; Balaei, M.R.; Mehdizadeh, M.; Marzban, H. Spatial and Temporal Expression of Lysosomal Acid Phosphatase 2 (ACP2) Reveals Dynamic Patterning of the Mouse Cerebellar Cortex. Cerebellum 2013, 12, 870–881. [Google Scholar] [CrossRef] [PubMed]
- Bailey, K.; Balaei, M.R.; Mannan, A.; Del Bigio, M.R.; Marzban, H. Purkinje Cell Compartmentation in the Cerebellum of the Lysosomal Acid Phosphatase 2 Mutant Mouse (Nax–Naked-Ataxia Mutant Mouse). PLoS ONE 2014, 9, e94327. [Google Scholar] [CrossRef]
- Canterini, S.; Dragotto, J.; Dardis, A.; Zampieri, S.; De Stefano, M.E.; Mangia, F.; Erickson, R.P.; Fiorenza, M.T. Shortened primary cilium length and dysregulated Sonic hedgehog signaling in Niemann-Pick C1 disease. Hum. Mol. Genet. 2017, 26, 2277–2289. [Google Scholar] [CrossRef]
- Yue, S.; Tang, L.-Y.; Tang, Y.; Tang, Y.; Shen, Q.-H.; Ding, J.; Chen, Y.; Zhang, Z.; Yu, T.-T.; E Zhang, Y.; et al. Requirement of Smurf-mediated endocytosis of Patched1 in sonic hedgehog signal reception. eLife 2014, 3. [Google Scholar] [CrossRef] [PubMed]
- Roussel, M.F.; Hatten, M.E. Cerebellum. Curr. Top. Dev. Biol. 2011, 94, 235–282. [Google Scholar]
- Ma, M.; Wu, W.; Li, Q.; Li, J.; Sheng, Z.; Shi, J.; Zhang, M.; Yang, H.; Wang, Z.; Sun, R.; et al. N-myc is a key switch regulating the proliferation cycle of postnatal cerebellar granule cell progenitors. Sci. Rep. 2015, 5, 12740. [Google Scholar] [CrossRef]
- Mallet, J.; Huchet, M.; Pougeois, R.; Changeux, J.-P. Anatomical, physiological and biochemical studies on the cerebellum from mutant mice. III. Protein differences associated with the weaver, staggerer and nervous mutations. Brain Res. 1976, 103, 291–312. [Google Scholar] [CrossRef]
- Rahimi-Balaei, M.; Bergen, H.; Kong, J.; Marzban, H. Neuronal Migration During Development of the Cerebellum. Front. Cell. Neurosci. 2018, 12. [Google Scholar] [CrossRef]
- Beachy, P.A.; Hymowitz, S.G.; Lazarus, R.A.; Leahy, D.J.; Siebold, C. Interactions between Hedgehog proteins and their binding partners come into view. Genes Dev. 2010, 24, 2001–2012. [Google Scholar] [CrossRef]
- Zhao, X.; Heng, J.I.-T.; Guardavaccaro, D.; Jiang, R.; Pagano, M.; Guillemot, F.; Iavarone, A.; Lasorella, A. The HECT-domain ubiquitin ligase Huwe1 controls neural differentiation and proliferation by destabilizing the N-Myc oncoprotein. Nat. Cell Biol. 2008, 10, 643–653. [Google Scholar] [CrossRef]
- Vriend, J.; Ghavami, S.; Marzban, H. The role of the ubiquitin proteasome system in cerebellar development and medulloblastoma. Mol. Brain 2015, 8, 1–14. [Google Scholar] [CrossRef]
- Boon, K.; Caron, H.N.; Van Asperen, R.; Valentijn, L.; Hermus, M.-C.; Van Sluis, P.; Roobeek, I.; Weis, I.; Voûte, P.A.; Schwab, M.; et al. N-myc enhances the expression of a large set of genes functioning in ribosome biogenesis and protein synthesis. EMBO J. 2001, 20, 1383–1393. [Google Scholar] [CrossRef] [PubMed]
- Corrales, J.D.; Blaess, S.; Mahoney, E.M.; Joyner, A.L. The level of sonic hedgehog signaling regulates the complexity of cerebellar foliation. Development 2006, 133, 1811–1821. [Google Scholar] [CrossRef] [PubMed]
- Dahmane, N.; Altaba, A.R.I. Sonic hedgehog regulates the growth and patterning of the cerebellum. Development 1999, 126, 3089–3100. [Google Scholar] [PubMed]
- Wallace, V.A. Purkinje-cell-derived Sonic hedgehog regulates granule neuron precursor cell proliferation in the developing mouse cerebellum. Curr. Biol. 1999, 9, 445–448. [Google Scholar] [CrossRef]
- Chuang, P.-T.; McMahon, A.P. Vertebrate Hedgehog signalling modulated by induction of a Hedgehog-binding protein. Nat. Cell Biol. 1999, 397, 617–621. [Google Scholar] [CrossRef]
- Wang, H.; Ge, G.; Uchida, Y.; Luu, B.; Ahn, S. Gli3 Is Required for Maintenance and Fate Specification of Cortical Progenitors. J. Neurosci. 2011, 31, 6440–6448. [Google Scholar] [CrossRef] [PubMed]
- Jacob, J.; Briscoe, J. Gli proteins and the control of spinal-cord patterning. EMBO Rep. 2003, 4, 761–765. [Google Scholar] [CrossRef]
- Borday, C.; Cabochette, P.; Parain, K.; Mazurier, N.; Janssens, S.; Tran, H.T.; Sekkali, B.; Bronchain, O.; Vleminckx, K.; Locker, M.; et al. Antagonistic cross-regulation between Wnt and Hedgehog signalling pathways controls post-embryonic retinal proliferation. Development 2012, 139, 3499–3509. [Google Scholar] [CrossRef]
- Ocasio, J.K.; Bates, R.D.P.; Rapp, C.D.; Gershon, T.R. GSK-3 modulates SHH-driven proliferation in postnatal cerebellar neurogenesis and medulloblastoma. Development 2019, 146, dev177550. [Google Scholar] [CrossRef]
- Anne, S.L.; Govek, E.-E.; Ayrault, O.; Kim, J.H.; Zhu, X.; Murphy, D.A.; Van Aelst, L.; Roussel, M.F.; Hatten, M.E. WNT3 Inhibits Cerebellar Granule Neuron Progenitor Proliferation and Medulloblastoma Formation via MAPK Activation. PLoS ONE 2013, 8, e81769. [Google Scholar] [CrossRef]
- Hu, M.C.; Mo, R.; Bhella, S.; Wilson, C.W.; Chuang, P.-T.; Hui, C.-C.; Rosenblum, N.D. GLI3-dependent transcriptional repression of Gli1, Gli2 and kidney patterning genes disrupts renal morphogenesis. Development 2006, 133, 569–578. [Google Scholar] [CrossRef]
- Charron, J.; A Malynn, B.; Fisher, P.; Stewart, V.; Jeannotte, L.; Goff, S.P.; Robertson, E.J.; Alt, F.W. Embryonic lethality in mice homozygous for a targeted disruption of the N-myc gene. Genes Dev. 1992, 6, 2248–2257. [Google Scholar] [CrossRef]
- Cohn, S.L.; Salwen, H.; Quasney, M.W.; Ikegaki, N.; Cowan, J.M.; Herst, C.V.; Kennett, R.H.; Rosen, S.T.; A DiGiuseppe, J.; Brodeur, G.M. Prolonged N-myc protein half-life in a neuroblastoma cell line lacking N-myc amplification. Oncogene 1990, 5, 1821–1827. [Google Scholar] [PubMed]
- Sharma, T.; Schwalbe, E.C.; Williamson, D.; Sill, M.; Hovestadt, V.; Mynarek, M.; Rutkowski, S.; Robinson, G.W.; Gajjar, A.; Cavalli, F.; et al. Second-generation molecular subgrouping of medulloblastoma: An international meta-analysis of Group 3 and Group 4 subtypes. Acta Neuropathol. 2019, 138, 309–326. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.Y.; Erkek, S.; Tong, Y.; Yin, L.; Federation, A.J.; Zapatka, M.; Haldipur, P.; Kawauchi, D.; Risch, T.; Warnatz, H.-J.; et al. Active medulloblastoma enhancers reveal subgroup-specific cellular origins. Nat. Cell Biol. 2016, 530, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Oliver, T.G.; Grasfeder, L.L.; Carroll, A.L.; Kaiser, C.; Gillingham, C.L.; Lin, S.M.; Wickramasinghe, R.; Scott, M.P.; Wechsler-Reya, R.J. Transcriptional profiling of the Sonic hedgehog response: A critical role for N-myc in proliferation of neuronal precursors. Proc. Natl. Acad. Sci. USA 2003, 100, 7331–7336. [Google Scholar] [CrossRef]
- Zhao, X.; Arca, D.D.; Lim, W.K.; Brahmachary, M.; Carro, M.S.; Ludwig, T.; Cardo, C.C.; Guillemot, F.; Aldape, K.; Califano, A.; et al. The N-Myc-DLL3 Cascade Is Suppressed by the Ubiquitin Ligase Huwe1 to Inhibit Proliferation and Promote Neurogenesis in the Developing Brain. Dev. Cell 2009, 17, 210–221. [Google Scholar] [CrossRef]
- Mehdizadeh, M.; Ashtari, N.; Jiao, X.; Balaei, M.R.; Marzban, A.; Qiyami-Hour, F.; Kong, J.; Ghavami, S.; Marzban, H. Alteration of the Dopamine Receptors’ Expression in the Cerebellum of the Lysosomal Acid Phosphatase 2 Mutant (Naked–Ataxia (NAX)) Mouse. Int. J. Mol. Sci. 2020, 21, 2914. [Google Scholar] [CrossRef]
- Zhao, Z.; Lee, R.T.H.; Pusapati, G.V.; Iyu, A.; Rohatgi, R.; Ingham, P.W. An essential role for Grk2 in Hedgehog signalling downstream of Smoothened. EMBO Rep. 2016, 17, 739–752. [Google Scholar] [CrossRef]
- Garcia-Lopez, J.; Kumar, R.; Smith, K.S.; Northcott, P.A. Deconstructing Sonic Hedgehog Medulloblastoma: Molecular Subtypes, Drivers, and Beyond. Trends Genet. 2021, 37, 235–250. [Google Scholar] [CrossRef]
- Matsuo, S.; Takahashi, M.; Inoue, K.; Tamura, K.; Irie, K.; Kodama, Y.; Nishikawa, A.; Yoshida, M. Thickened area of external granular layer and Ki-67 positive focus are early events of medulloblastoma in Ptch1+/− mice. Exp. Toxicol. Pathol. 2013, 65, 863–873. [Google Scholar] [CrossRef]
- Iriana, S.; Asha, K.; Repak, M.; Sharma-Walia, N. Hedgehog Signaling: Implications in Cancers and Viral Infections. Int. J. Mol. Sci. 2021, 22, 1042. [Google Scholar] [CrossRef]
- Balaei, M.R.; Jiao, X.; Ashtari, N.; Afsharinezhad, P.; Ghavami, S.; Marzban, H. Cerebellar Expression of the Neurotrophin Receptor p75 in Naked-Ataxia Mutant Mouse. Int. J. Mol. Sci. 2016, 17, 115. [Google Scholar] [CrossRef]
- Emao, S.; Exiong, G.; Ezhang, L.; Edong, H.; Eliu, B.; Cohen, N.A.; Cohen, A.S. Verification of the Cross Immunoreactivity of A60, a Mouse Monoclonal Antibody against Neuronal Nuclear Protein. Front. Neuroanat. 2016, 10, 54. [Google Scholar] [CrossRef]
- Moreno, N.; Joven, A.; Morona, R.; Bandín, S.; López, J.M.; González, A. Conserved localization of Pax6 and Pax7 transcripts in the brain of representatives of sarcopterygian vertebrates during development supports homologous brain regionalization. Front. Neuroanat. 2014, 8. [Google Scholar] [CrossRef] [PubMed]
- Eberhardt, A.; Hansen, J.N.; Koster, J.; Jr, L.T.L.; Wang, S.; Livingstone, E.; Qian, K.; Valentijn, L.J.; Zheng, Y.G.; Schor, N.F.; et al. Protein arginine methyltransferase 1 is a novel regulator of MYCN in neuroblastoma. Oncotarget 2016, 7, 63629–63639. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinform. 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. The Subread aligner: Fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013, 41, e108. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2013, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Eissa, N.; Hussein, H.; Rabbi, M.F.; Munyaka, P.M.; Khafipour, A.; Bernstein, C.N.; Ghia, J.-E. Tu1832 Stability of Reference Genes for Messenger RNA Quantification by Real-Time PCR in Mouse Dextran Sodium Sulfate Experimental Colitis. Gastroenterol. 2016, 150, S955–S956. [Google Scholar] [CrossRef]
- Eissa, N.; Kermarrec, L.; Hussein, H.; Bernstein, C.N.; Ghia, J.-E. Appropriateness of reference genes for normalizing messenger RNA in mouse 2,4-dinitrobenzene sulfonic acid (DNBS)-induced colitis using quantitative real time PCR. Sci. Rep. 2017, 7, 42427. [Google Scholar] [CrossRef] [PubMed]
- Abu-El-Rub, E.; Sequiera, G.L.; Sareen, N.; Yan, W.; Moudgil, M.; Sabbir, M.G.; Dhingra, S. Hypoxia-induced 26S proteasome dysfunction increases immunogenicity of mesenchymal stem cells. Cell Death Dis. 2019, 10, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Abu-El-Rub, E.; Sareen, N.; Yan, W.; Alagarsamy, K.N.; Rafieerad, A.; Srivastava, A.; Desiderio, V.; Dhingra, S. Hypoxia-induced shift in the phenotype of proteasome from 26S toward immunoproteasome triggers loss of immunoprivilege of mesenchymal stem cells. Cell Death Dis. 2020, 11, 1–12. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiao, X.; Rahimi Balaei, M.; Abu-El-Rub, E.; Casoni, F.; Pezeshgi Modarres, H.; Dhingra, S.; Kong, J.; Consalez, G.G.; Marzban, H. Reduced Granule Cell Proliferation and Molecular Dysregulation in the Cerebellum of Lysosomal Acid Phosphatase 2 (ACP2) Mutant Mice. Int. J. Mol. Sci. 2021, 22, 2994. https://doi.org/10.3390/ijms22062994
Jiao X, Rahimi Balaei M, Abu-El-Rub E, Casoni F, Pezeshgi Modarres H, Dhingra S, Kong J, Consalez GG, Marzban H. Reduced Granule Cell Proliferation and Molecular Dysregulation in the Cerebellum of Lysosomal Acid Phosphatase 2 (ACP2) Mutant Mice. International Journal of Molecular Sciences. 2021; 22(6):2994. https://doi.org/10.3390/ijms22062994
Chicago/Turabian StyleJiao, Xiaodan, Maryam Rahimi Balaei, Ejlal Abu-El-Rub, Filippo Casoni, Hassan Pezeshgi Modarres, Sanjiv Dhingra, Jiming Kong, Giacomo G. Consalez, and Hassan Marzban. 2021. "Reduced Granule Cell Proliferation and Molecular Dysregulation in the Cerebellum of Lysosomal Acid Phosphatase 2 (ACP2) Mutant Mice" International Journal of Molecular Sciences 22, no. 6: 2994. https://doi.org/10.3390/ijms22062994
APA StyleJiao, X., Rahimi Balaei, M., Abu-El-Rub, E., Casoni, F., Pezeshgi Modarres, H., Dhingra, S., Kong, J., Consalez, G. G., & Marzban, H. (2021). Reduced Granule Cell Proliferation and Molecular Dysregulation in the Cerebellum of Lysosomal Acid Phosphatase 2 (ACP2) Mutant Mice. International Journal of Molecular Sciences, 22(6), 2994. https://doi.org/10.3390/ijms22062994







