Advanced Spheroid, Tumouroid and 3D Bioprinted In-Vitro Models of Adult and Paediatric Glioblastoma
Abstract
1. Introduction
2. The Tumour Microenvironment
2.1. Glioma Stem Cells
2.2. Hypoxia
2.3. Extracellular Matrix
2.4. Tumour Interactions with Non-Tumour Cells
2.5. Tumour Microtubules
2.6. Mechanical Properties
| Sample Size, Age Range (y) | Range of Excitation Frequency (Hz) | Complex Modulus (rad) | Phase Angle | Ref. | ||||
|---|---|---|---|---|---|---|---|---|
| GBM | NAWM | GBM | NAWM | |||||
| 9, 60–80 | 30–60 | 1.10 ± 0.29 | 1.81 ± 0.23 | 0.65 ± 0.04 | 0.62 ± 0.19 | 0.36 ± 0.10 | 0.54 ± 0.15 | [54] |
| 6, 25–68 | 60 | 1.7 ± 0.5 | 3.3 ± 0.7 | — | — | — | — | [63] |
| 11, 42–86 | 30–60 | 1.37 ± 0.26 | 1.64 ± 0.21 | 0.64 ± 0.10 | 0.85 ± 0.22 | 0.44 ± 0.07 | 0.70 ± 0.11 | [61] |
| 22, 18–86 | 30–60 | 1.32 ± 0.26 | 1.54 ± 0.27 | 0.58 ± 0.07 | 0.88 ± 0.19 | 0.37 ± 0.08 | 0.66 ± 0.15 | [65] |
| 3, 53–69 | 45 | 1.24 ± 0.31 | 2.11 ± 0.31 | 0.41 ± 0.06 | 0.59 ± 0.09 | 0.30 ± 0.04 | 0.74 ± 0.19 | [60] |
3. Traditional In-Vitro Models of High Grade Glioma
3.1. Three-Dimensional In-Vitro Models of High Grade Glioma
3.2. Free Spheroid and Tumouroid Models
3.3. Matrix-Supported Spheroid and Tumouroid Models
4. In-Vitro Models of Healthy Brain
5. Cerebral Organoid/Glioblastoma Co-Culture
6. Bioprinted Organoid/Tumouroid Models of High-Grade Glioma
6.1. Addition of Stromal Components to 3D Bioprinted High-Grade Glioma Models
6.2. Future Trends in 3D Bioprinting
7. Conclusions and Future Outlook
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| 2D | Two-dimensional |
| Three-dimensional (3D) | Three-dimensional |
| ASCs | Adult stem cells |
| AUKRA | Aurora A kinase |
| BBB | Blood–brain barrier |
| CSCs | Cancer stem cells |
| ECM | Extracellular matrix |
| EGFR | Epidermal growth factor receptor |
| GA-MSCs | Glioma associated mesenchymal stem cells |
| GAGs | Glycosaminoglyca |
| GBM | Glioblastoma |
| GelMA | Gelatine methacrylate |
| GICs | Glioma initiating cells |
| GSCs | Glioma stem cells |
| HA | Hyaluronic acid |
| HESC | Human embryonic stem cell |
| HGG | High-grade glioma |
| HIFs | Hypoxia inducible factors |
| HUVECs | Human umbilical vascular endothelial cells |
| IFF | Interstitial fluid flow |
| IPSCs | Induced pluripotent stem cells |
| MMRE | Multifrequency MRE |
| MRI | Magnetic resonance imaging |
| NAWM | Normal-appearing white matter |
| OPCs | Oligodendroglial precursor cells |
| PDX | Patient-derived xenograft |
| PEG | Polyethylene glycol |
| PGBM | Paediatric GBM |
| RBM | Reconstituted basement membrane |
| RCCS | Rotary cell culture system |
| TME | Tumour microenvironment |
| TMs | Tumour microtubules |
| TMZ | Temozolomide |
References
- Zhang, I.; Beus, M.; Stochaj, U.; Le, P.U.; Zorc, B.; Rajić, Z.; Petrecca, K.; Maysinger, D. Inhibition of Glioblastoma Cell Proliferation, Invasion, and Mechanism of Action of a Novel Hydroxamic Acid Hybrid Molecule. Cell Death Discov. 2018, 4, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Hermida, M.A.; Kumar, J.D.; Schwarz, D.; Laverty, K.G.; Di Bartolo, A.; Ardron, M.; Bogomolnijs, M.; Clavreul, A.; Brennan, P.M.; Wiegand, U.K.; et al. Three Dimensional in Vitro Models of Cancer: Bioprinting Multilineage Glioblastoma Models. Adv. Biol. Regul. 2020, 75, 100658. [Google Scholar] [CrossRef] [PubMed]
- Jacob, F.; Salinas, R.D.; Zhang, D.Y.; Nguyen, P.T.; Schnoll, J.G.; Wong, S.Z.H.; Thokala, R.; Sheikh, S.; Saxena, D.; Prokop, S.; et al. A Patient-Derived Glioblastoma Organoid Model and Biobank Recapitulates Inter- and Intra-Tumoral Heterogeneity. Cell 2020, 180, 188–204. [Google Scholar] [CrossRef] [PubMed]
- Freeman, C.R.; Farmer, J.P. Pediatric Brain Stem Gliomas: A Review. Int. J. Radiat. Oncol. Biol. Phys. 1998, 40, 265–271. [Google Scholar] [CrossRef]
- Hargrave, D.; Bartels, U.; Bouffet, E. Diffuse Brainstem Glioma in Children: Critical Review of Clinical Trials. Lancet Oncol. 2006, 7, 241–248. [Google Scholar] [CrossRef]
- Janssens, G.O.; Gidding, C.E.; Van Lindert, E.J.; Oldenburger, F.R.; Erasmus, C.E.; Schouten-Meeteren, A.Y.; Kaanders, J.H. The role of Hypofractionation Radiotherapy for Diffuse Intrinsic Brainstem Glioma in Children: A Pilot Study. Int. J. Radiat. Oncol. Biol. Phys. 2009, 73, 722–726. [Google Scholar] [CrossRef]
- Vanan, M.I.; Eisenstat, D.D. Management of High-grade Gliomas in the Pediatric Patient: Past, Present, and Future. Neuro-Oncol. Pract. 2014, 1, 145–157. [Google Scholar] [CrossRef]
- Fangusaro, J.; Warren, K.E. Unclear Standard of Care for Pediatric High Grade Glioma Patients. J. Neuro-Oncol. 2013, 113, 341–342. [Google Scholar] [CrossRef] [PubMed]
- Cohen, K.J.; Pollack, I.F.; Zhou, T.; Buxton, A.; Holmes, E.J.; Burger, P.C.; Brat, D.J.; Rosenblum, M.K.; Hamilton, R.L.; Lavey, R.S.; et al. Temozolomide in the Treatment of High-grade Gliomas in Children: A Report from the Children’s Oncology Group. Neuro-Oncology 2011, 13, 317–323. [Google Scholar] [CrossRef]
- Jansen, M.; Van Vuurden, D.; Vandertop, W.; Kaspers, G. Diffuse Intrinsic Pontine Gliomas: A Systematic Update on Clinical Trials and Biology. Cancer Treat. Rev. 2012, 38, 27–35. [Google Scholar] [CrossRef]
- Linkous, A.; Balamatsias, D.; Snuderl, M.; Edwards, L.; Miyaguchi, K.; Milner, T.; Reich, B.; Cohen-Gould, L.; Storaska, A.; Nakayama, Y.; et al. Modeling Patient-Derived Glioblastoma with Cerebral Organoids. Cell Rep. 2019, 26, 3203–3211. [Google Scholar] [CrossRef] [PubMed]
- Farrell, K.; Mahajan, G.; Srinivasan, P.; Lee, M.Y.; Kothapalli, C.R. Pediatric Glioblastoma Cells Inhibit Neurogenesis and Promote Astrogenesis, Phenotypic Transformation and Migration of Human Neural Progenitor Cells Within Cocultures. Exp. Cell Res. 2018, 362, 159–171. [Google Scholar] [CrossRef]
- Martínez-Vélez, N.; Garcia-Moure, M.; Marigil, M.; González-Huarriz, M.; Puigdelloses, M.; Pérez-Larraya, J.G.; Zalacaín, M.; Marrodán, L.; Varela-Guruceaga, M.; Laspidea, V.; et al. The Oncolytic Virus Delta-24-RGD Elicits an Antitumor Effect in Pediatric Glioma and DIPG Mouse Models. Nat. Commun. 2019, 10, 2235. [Google Scholar] [CrossRef]
- Gritsenko, P.G.; Ilina, O.; Friedl, P. Interstitial Guidance of Cancer Invasion. J. Pathol. 2012, 226, 185–199. [Google Scholar] [CrossRef] [PubMed]
- Gritsenko, P.; Leenders, W.; Friedl, P. Recapitulating -like Plasticity of Glioma Cell Invasion Along Blood Vessels and in Astrocyte-rich Stroma. Histochem. Cell Biol. 2017, 148, 395–406. [Google Scholar] [CrossRef] [PubMed]
- Osswald, M.; Jung, E.; Sahm, F.; Solecki, G.; Venkataramani, V.; Blaes, J.; Weil, S.; Horstmann, H.; Wiestler, B.; Syed, M.; et al. Brain Tumour Cells Interconnect to a Functional and Resistant Network. Nature 2015, 528, 93–98. [Google Scholar] [CrossRef]
- Gritsenko, P.G.; Atlasy, N.; Dieteren, C.E.; Navis, A.C.; Venhuizen, J.H.; Veelken, C.; Schubert, D.; Acker-Palmer, A.; Westerman, B.A.; Wurdinger, T.; et al. p120-catenin-dependent Collective Brain Infiltration by Glioma Cell Networks. Nat. Cell Biol. 2020, 22, 97–107. [Google Scholar] [CrossRef]
- Goranci-Buzhala, G.; Mariappan, A.; Gabriel, E.; Ramani, A.; Ricci-Vitiani, L.; Buccarelli, M.; D’Alessandris, Q.G.; Pallini, R.; Gopalakrishnan, J. Rapid and Efficient Invasion Assay of Glioblastoma in Human Brain Organoids. Cell Rep. 2020, 31, 107738. [Google Scholar] [CrossRef] [PubMed]
- Caretti, V.; Zondervan, I.; Meijer, D.H.; Idema, S.; Vos, W.; Hamans, B.; Bugiani, M.; Hulleman, E.; Wesseling, P.; Vandertop, W.P.; et al. Monitoring of Tumor Growth and Post-irradiation Recurrence in a Diffuse Intrinsic Pontine Glioma Mouse Model. Brain Pathol. 2011, 21, 441–451. [Google Scholar] [CrossRef]
- Vinci, M.; Burford, A.; Molinari, V.; Kessler, K.; Popov, S.; Clarke, M.; Taylor, K.R.; Pemberton, H.N.; Lord, C.J.; Gutteridge, A.; et al. Functional Diversity and Cooperativity Between Subclonal Populations of Pediatric Glioblastoma and Diffuse Intrinsic Pontine Glioma Cells. Nat. Med. 2018, 24, 1204–1215. [Google Scholar] [CrossRef]
- Bhaduri, A.; Di Lullo, E.; Jung, D.; Müller, S.; Crouch, E.E.; Espinosa, C.S.; Ozawa, T.; Alvarado, B.; Spatazza, J.; Cadwell, C.R.; et al. Outer Radial Glia-like Cancer Stem Cells Contribute to Heterogeneity of Glioblastoma. Cell Stem Cell 2020, 26, 48–63. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, S.; Louis, D.N.; Curry, W.T.; Batchelor, T.T.; Dietrich, J. Diagnostic and Therapeutic Avenues for Glioblastoma: No Longer a Dead End? Nat. Rev. Clin. Oncol. 2013, 10, 14. [Google Scholar] [CrossRef]
- Bayir, E.; Sendemir, A.; Missirlis, Y.F. Mechanobiology of Cells and Cell Systems, Such as Organoids. Biophys. Rev. 2019, 11, 721–728. [Google Scholar] [CrossRef]
- Hoffman, M.; Gillmor, A.H.; Kunz, D.J.; Johnston, M.J.; Nikolic, A.; Narta, K.; Zarrei, M.; King, J.; Ellestad, K.; Dang, N.H.; et al. Intratumoral Genetic and Functional Heterogeneity in Pediatric Glioblastoma. Cancer Res. 2019, 79, 2111–2123. [Google Scholar] [CrossRef] [PubMed]
- Dobson, T.H.; Gopalakrishnan, V. Preclinical Models of Pediatric Brain Tumors—Forging Ahead. Bioengineering 2018, 5, 81. [Google Scholar] [CrossRef]
- Figueroa, J.; Phillips, L.M.; Shahar, T.; Hossain, A.; Gumin, J.; Kim, H.; Bean, A.J.; Calin, G.A.; Fueyo, J.; Walters, E.T.; et al. Exosomes from Glioma-Associated Mesenchymal Stem Cells Increase the Tumorigenicity of Glioma Stem-Like Cells via Transfer of miR-1587. Cancer Res. 2017, 77, 5808–5819. [Google Scholar] [CrossRef]
- Hossain, A.; Gumin, J.; Gao, F.; Figueroa, J.; Shinojima, N.; Takezaki, T.; Priebe, W.; Villarreal, D.; Kang, S.G.; Joyce, C.; et al. Mesenchymal Stem Cells Isolated from Human Gliomas Increase Proliferation and Maintain Stemness of Glioma Stem Cells Through the IL-6/gp130/STAT3 Pathway. Stem Cells 2015, 33, 2400–2415. [Google Scholar] [CrossRef]
- Gillespie, S.; Monje, M. An Active Role for Neurons in Glioma Progression: Making Sense of Scherer’s structures. Neuro-Oncology 2018, 20, 1292–1299. [Google Scholar] [CrossRef]
- Osswald, M.; Solecki, G.; Wick, W.; Winkler, F. A Malignant Cellular Network in Gliomas: Potential Clinical Implications. Neuro-Oncology 2016, 18, 479–485. [Google Scholar] [CrossRef] [PubMed]
- Osswald, M.; Jung, E.; Wick, W.; Winkler, F. Tunneling Nanotube-Like Structures in Brain Tumors. Cancer Rep. 2019, 2, e1181. [Google Scholar] [CrossRef]
- Singh, S.K.; Hawkins, C.; Clarke, I.D.; Squire, J.A.; Bayani, J.; Hide, T.; Henkelman, R.M.; Cusimano, M.D.; Dirks, P.B. Identification of Human Brain Tumour Initiating Cells. Nature 2004, 432, 396–401. [Google Scholar] [CrossRef]
- Hubert, C.G.; Rivera, M.; Spangler, L.C.; Wu, Q.; Mack, S.C.; Prager, B.C.; Couce, M.; McLendon, R.E.; Sloan, A.E.; Rich, J.N. A Three-dimensional Organoid Culture System Derived from Human Glioblastomas Recapitulates the Hypoxic Gradients and Cancer Stem Cell Heterogeneity of Tumors Found. Cancer Res. 2016, 76, 2465–2477. [Google Scholar] [CrossRef]
- Wenger, A.; Larsson, S.; Danielsson, A.; Elbæk, K.J.; Kettunen, P.; Tisell, M.; Sabel, M.; Lannering, B.; Nordborg, C.; Schepke, E.; et al. Stem Cell Cultures Derived from Pediatric Brain Tumors Accurately Model the Originating Tumors. Oncotarget 2017, 8, 18626. [Google Scholar] [CrossRef] [PubMed]
- Aziz-Bose, R.; Monje, M. Diffuse Intrinsic Pontine Glioma: Molecular Landscape and Emerging Therapeutic Targets. Curr. Opin. Oncol. 2019, 31, 522–530. [Google Scholar] [CrossRef]
- Surowiec, R.K.; Ferris, S.F.; Apfelbaum, A.; Espinoza, C.; Mehta, R.K.; Monchamp, K.; Sirihorachai, V.R.; Bedi, K.; Ljungman, M.; Galban, S. Transcriptomic Analysis of Diffuse Intrinsic Pontine Glioma (DIPG) Identifies a Targetable ALDH-positive Subset of Highly Tumorigenic Cancer Stem-like Cells. Mol. Cancer Res. 2020, 19, 223–239. [Google Scholar] [CrossRef]
- Jung, E.; Alfonso, J.; Osswald, M.; Monyer, H.; Wick, W.; Winkler, F. Emerging Intersections Between Neuroscience and Glioma Biology. Nat. Neurosci. 2019, 22, 1951–1960. [Google Scholar] [CrossRef] [PubMed]
- Agnihotri, S.; Burrell, K.; Buczkowicz, P.; Remke, M.; Golbourn, B.; Chornenkyy, Y.; Gajadhar, A.; Fernandez, N.A.; Clarke, I.D.; Barszczyk, M.S.; et al. ATM Regulates 3-Methylpurine-DNA Glycosylase and Promotes Therapeutic Resistance to Alkylating Agents. Cancer Discov. 2014, 4, 1198–1213. [Google Scholar] [CrossRef]
- Martínez-Ramos, C.; Lebourg, M. Three-Dimensional Constructs Using Hyaluronan Cell Carrier as a Tool for the Study of Cancer stem Cells. J. Biomed. Mater. Res. Part B Appl. Biomater. 2015, 103, 1249–1257. [Google Scholar] [CrossRef]
- Rath, B.H.; Wahba, A.; Camphausen, K.; Tofilon, P.J. Coculture with Astrocytes Reduces the Radiosensitivity of Glioblastoma Stem-like Cells and Identifies Additional Targets for Radiosensitization. Cancer Med. 2015, 4, 1705–1716. [Google Scholar] [CrossRef]
- Vartanian, A.; Singh, S.K.; Agnihotri, S.; Jalali, S.; Burrell, K.; Aldape, K.D.; Zadeh, G. GBM’s Multifaceted Landscape: Highlighting Regional and Microenvironmental Heterogeneity. Neuro-Oncology 2014, 16, 1167–1175. [Google Scholar] [CrossRef] [PubMed]
- Mudassar, F.; Shen, H.; O’Neill, G.; Hau, E. Targeting Tumor Hypoxia and Mitochondrial Metabolism with Anti-parasitic Drugs to ImproveRadiation Response in High-grade Gliomas. J. Exp. Clin. Cancer Res. 2020, 39, 1–17. [Google Scholar] [CrossRef]
- Blandin, A.F.; Durand, A.; Litzler, M.; Tripp, A.; Guérin, É.; Ruhland, E.; Obrecht, A.; Keime, C.; Fuchs, Q.; Reita, D.; et al. Hypoxic Environment and Paired Hierarchical 3D and 2D Models of Pediatric H3. 3-mutated Gliomas Recreate the Patient Tumor Complexity. Cancers 2019, 11, 1875. [Google Scholar] [CrossRef]
- Yeom, K.W.; Lober, R.M.; Nelson, M.D.; Panigrahy, A.; Blüml, S. Citrate Concentrations Increase with Hypoperfusion in Pediatric Diffuse Intrinsic Pontine Glioma. J. Neuro-Oncol. 2015, 122, 383–389. [Google Scholar] [CrossRef]
- Xiao, W.; Sohrabi, A.; Seidlits, S.K. Integrating the Glioblastoma Microenvironment into Engineered Experimental Models. Future Sci. OA 2017, 3, FSO189. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Kumar, S. CD44-Mediated Adhesion to Hyaluronic Acid Contributes to Mechanosensing and Invasive Motility. Mol. Cancer Res. 2014, 12, 1416–1429. [Google Scholar] [CrossRef]
- Wolf, K.J.; Chen, J.; Coombes, J.D.; Aghi, M.K.; Kumar, S. Dissecting and Rebuilding the Glioblastoma Microenvironment with Engineered Materials. Nat. Rev. Mater. 2019, 4, 651–668. [Google Scholar] [CrossRef]
- Xiao, W.; Zhang, R.; Sohrabi, A.; Ehsanipour, A.; Sun, S.; Liang, J.; Walthers, C.M.; Ta, L.; Nathanson, D.A.; Seidlits, S.K. Brain-mimetic 3D Culture Platforms Allow Investigation of Cooperative Effects of Extracellular Matrix Features on Therapeutic Resistance in Glioblastoma. Cancer Res. 2018, 78, 1358–1370. [Google Scholar] [CrossRef] [PubMed]
- Heinrich, M.A.; Bansal, R.; Lammers, T.; Zhang, Y.S.; Michel Schiffelers, R.; Prakash, J. Three Dimensional Bioprinted Mini-Brain: A Glioblastoma Model to Study Cellular Interactions and Therapeutics. Adv. Mater. 2019, 31, 1806590. [Google Scholar] [CrossRef] [PubMed]
- Venkatesh, H.S.; Morishita, W.; Geraghty, A.C.; Silverbush, D.; Gillespie, S.M.; Arzt, M.; Tam, L.T.; Espenel, C.; Ponnuswami, A.; Ni, L.; et al. Electrical and Synaptic Integration of Glioma into Neural Circuits. Nature 2019, 573, 539–545. [Google Scholar] [CrossRef]
- Franze, K. The Mechanical Control of Nervous System Development. Development 2013, 140, 3069–3077. [Google Scholar] [CrossRef]
- Bouchonville, N.; Meyer, M.; Gaude, C.; Gay, E.; Ratel, D.; Nicolas, A. AFM Mapping of the Elastic Properties of Brain Tissue Reveals kPa m- 1 Gradients of Rigidity. Soft Matter 2016, 12, 6232–6239. [Google Scholar] [CrossRef]
- MacManus, D.B.; Pierrat, B.; Murphy, J.G.; Gilchrist, M.D. Region and Species Dependent Mechanical Properties of Adolescent and Young Adult Brain Tissue. Sci. Rep. 2017, 7, 13729. [Google Scholar] [CrossRef] [PubMed]
- Northcott, J.M.; Dean, I.S.; Mouw, J.K.; Weaver, V.M. Feeling Stress: The Mechanics of Cancer Progression and Aggression. Front. Cell Dev. Biol. 2018, 6, 17. [Google Scholar] [CrossRef]
- Streitberger, K.J.; Lilaj, L.; Schrank, F.; Braun, J.; Hoffmann, K.T.; Reiss-Zimmermann, M.; Käs, J.A.; Sack, I. How Tissue Fluidity Influences Brain Tumor Progression. Proc. Natl. Acad. Sci. USA 2020, 117, 128–134. [Google Scholar] [CrossRef] [PubMed]
- Jugé, L.; Petiet, A.; Lambert, S.A.; Nicole, P.; Chatelin, S.; Vilgrain, V.; Van Beers, B.E.; Bilston, L.E.; Sinkus, R. Microvasculature Alters the Dispersion Properties of Shear Waves—A Multi-Frequency MR Elastography Study. NMR Biomed. 2015, 28, 1763–1771. [Google Scholar] [CrossRef]
- Green, M.A.; Bilston, L.E.; Sinkus, R. In Vivo Brain Viscoelastic Properties Measured by Magnetic Resonance Elastography. NMR Biomed. Int. J. Devoted Dev. Appl. Magn. Reson. Vivo 2008, 21, 755–764. [Google Scholar]
- Bunevicius, A.; Schregel, K.; Sinkus, R.; Golby, A.; Patz, S. MR elastography of brain tumors. NeuroImage Clin. 2020, 25, 102109. [Google Scholar] [CrossRef]
- Venkatesh, S.K.; Yin, M.; Ehman, R.L. Magnetic Resonance Elastography of Liver: Technique, Analysis, and Clinical Applications. J. Magn. Reson. Imaging 2013, 37, 544–555. [Google Scholar] [CrossRef]
- Sack, I.; Beierbach, B.; Wuerfel, J.; Klatt, D.; Hamhaber, U.; Papazoglou, S.; Martus, P.; Braun, J. The Impact of Aging and Gender on Brain Viscoelasticity. Neuroimage 2009, 46, 652–657. [Google Scholar] [CrossRef] [PubMed]
- Simon, M.; Guo, J.; Papazoglou, S.; Scholand-Engler, H.; Erdmann, C.; Melchert, U.; Bonsanto, M.; Braun, J.; Petersen, D.; Sack, I.; et al. Non-invasive Characterization of Intracranial Tumors by Magnetic Resonance Elastography. New J. Phys. 2013, 15, 085024. [Google Scholar] [CrossRef]
- Reiss-Zimmermann, M.; Streitberger, K.J.; Sack, I.; Braun, J.; Arlt, F.; Fritzsch, D.; Hoffmann, K.T. High Resolution Imaging of Viscoelastic Properties of Intracranial Tumours by Multi-Frequency Magnetic Resonance Elastography. Clin. Neuroradiol. 2015, 25, 371–378. [Google Scholar] [CrossRef]
- Sakai, N.; Takehara, Y.; Yamashita, S.; Ohishi, N.; Kawaji, H.; Sameshima, T.; Baba, S.; Sakahara, H.; Namba, H. Shear Stiffness of 4 Common Intracranial Tumors Measured Using MR Elastography: Comparison with Intraoperative Consistency Grading. Am. J. Neuroradiol. 2016, 37, 1851–1859. [Google Scholar] [CrossRef]
- Pepin, K.M.; McGee, K.P.; Arani, A.; Lake, D.S.; Glaser, K.J.; Manduca, A.; Parney, I.F.; Ehman, R.L.; Huston, J. MR Elastography Analysis of Glioma Stiffness and IDH1-Mutation Status. Am. J. Neuroradiol. 2018, 39, 31–36. [Google Scholar] [CrossRef]
- Hughes, J.D.; Fattahi, N.; Van Gompel, J.; Arani, A.; Meyer, F.; Lanzino, G.; Link, M.J.; Ehman, R.; Huston, J. Higher-Resolution Magnetic Resonance Elastography in Meningiomas to Determine Intratumoral Consistency. Neurosurgery 2015, 77, 653–659. [Google Scholar] [CrossRef]
- Streitberger, K.J.; Reiss-Zimmermann, M.; Freimann, F.B.; Bayerl, S.; Guo, J.; Arlt, F.; Wuerfel, J.; Braun, J.; Hoffmann, K.T.; Sack, I. High-Resolution Mechanical Imaging of Glioblastoma by Multifrequency Magnetic Resonance Elastography. PLoS ONE 2014, 9, e110588. [Google Scholar] [CrossRef]
- Fernandez-Fuente, G.; Mollinedo, P.; Grande, L.; Vazquez-Barquero, A.; Fernandez-Luna, J.L. Culture Dimensionality Influences the Resistance of Glioblastoma Stem-like Cells to Multikinase Inhibitors. Mol. Cancer Ther. 2014, 13, 1664–1672. [Google Scholar] [CrossRef]
- Lee, J.; Kotliarova, S.; Kotliarov, Y.; Li, A.; Su, Q.; Donin, N.M.; Pastorino, S.; Purow, B.W.; Christopher, N.; Zhang, W.; et al. Tumor Stem Cells Derived from Glioblastomas Cultured in bFGF and EGF More Closely Mirror the Phenotype and Genotype of Primary Tumors than do Serum-Cultured Cell Llines. Cancer Cell 2006, 9, 391–403. [Google Scholar] [CrossRef] [PubMed]
- Stringer, B.W.; Day, B.W.; D’Souza, R.C.; Jamieson, P.R.; Ensbey, K.S.; Bruce, Z.C.; Lim, Y.C.; Goasdoué, K.; Offenhäuser, C.; Akgül, S.; et al. A Reference Collection of Patient-derived Cell Line and Xenograft Models of Proneural, Classical and Mesenchymal Glioblastoma. Sci. Rep. 2019, 9, 4902. [Google Scholar] [CrossRef] [PubMed]
- Kogiso, M.; Qi, L.; Braun, F.K.; Injac, S.G.; Zhang, L.; Du, Y.; Zhang, H.; Lin, F.Y.; Zhao, S.; Lindsay, H.; et al. Concurrent Inhibition of Neurosphere and Monolayer Cells of Pediatric Glioblastoma by Aurora A Inhibitor MLN8237 Predicted Survival Extension in PDOX Models. Clin. Cancer Res. 2018, 24, 2159–2170. [Google Scholar] [CrossRef] [PubMed]
- Kingsmore, K.M.; Logsdon, D.K.; Floyd, D.H.; Peirce, S.M.; Purow, B.W.; Munson, J.M. Interstitial Flow Differentially Increases Patient-Derived Glioblastoma Stem Cell Invasion via CXCR4, CXCL12, and CD44-mediated Mechanisms. Integr. Biol. 2016, 8, 1246–1260. [Google Scholar] [CrossRef]
- Miroshnikova, Y.A.; Mouw, J.K.; Barnes, J.M.; Pickup, M.W.; Lakins, J.N.; Kim, Y.; Lobo, K.; Persson, A.I.; Reis, G.F.; McKnight, T.R.; et al. Tissue Mechanics Promote IDH1-dependent HIF1α–Tenascin C Feedback to Regulate Glioblastoma Aggression. Nat. Cell Biol. 2016, 18, 1336–1345. [Google Scholar] [CrossRef]
- Marumoto, T.; Zhang, D.; Saya, H. Aurora-A—A Guardian of Poles. Nat. Rev. Cancer 2005, 5, 42–50. [Google Scholar] [CrossRef]
- Smith, S.J.; Wilson, M.; Ward, J.H.; Rahman, C.V.; Peet, A.C.; Macarthur, D.C.; Rose, F.R.; Grundy, R.G.; Rahman, R. Recapitulation of Tumor Heterogeneity and Molecular Signatures in a 3D Brain Cancer Model with Decreased Sensitivity to Histone Deacetylase Inhibition. PLoS ONE 2012, 7, e52335. [Google Scholar] [CrossRef]
- Cockle, J.V.; Brüning-Richardson, A.; Scott, K.J.; Thompson, J.; Kottke, T.; Morrison, E.; Ismail, A.; Carcaboso, A.M.; Rose, A.; Selby, P.; et al. Oncolytic Herpes Simplex Virus Inhibits Pediatric Brain Tumor Migration and Invasion. Mol. Ther. Oncolytics 2017, 5, 75–86. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, J.; Pao, G.M.; Shokhirev, M.N.; Verma, I.M. Glioblastoma Model Using Human Cerebral Organoids. Cell Rep. 2018, 23, 1220–1229. [Google Scholar] [CrossRef] [PubMed]
- Azzarelli, R. Organoid Models of Glioblastoma to Study Brain Tumor Stem Cells. Front. Cell Dev. Biol. 2020, 8, 220. [Google Scholar] [CrossRef] [PubMed]
- Nayernia, Z.; Turchi, L.; Cosset, E.; Peterson, H.; Dutoit, V.; Dietrich, P.Y.; Tirefort, D.; Chneiweiss, H.; Lobrinus, J.A.; Krause, K.H.; et al. The Relationship Between Brain Tumor Cell Invasion of Engineered Neural Tissues and Features of Glioblastoma. Biomaterials 2013, 34, 8279–8290. [Google Scholar] [CrossRef]
- Fan, Y.; Nguyen, D.T.; Akay, Y.; Xu, F.; Akay, M. Engineering a Brain Cancer Chip for High-Throughput Drug Screening. Sci. Rep. 2016, 6, 25062. [Google Scholar] [CrossRef]
- Souza, G.R.; Molina, J.R.; Raphael, R.M.; Ozawa, M.G.; Stark, D.J.; Levin, C.S.; Bronk, L.F.; Ananta, J.S.; Mandelin, J.; Georgescu, M.M.; et al. Three-Dimensional Tissue Culture Based on Magnetic Cell Levitation. Nat. Nanotechnol. 2010, 5, 291–296. [Google Scholar] [CrossRef]
- Weeber, F.; van de Wetering, M.; Hoogstraat, M.; Dijkstra, K.K.; Krijgsman, O.; Kuilman, T.; Gadellaa-van Hooijdonk, C.G.; van der Velden, D.L.; Peeper, D.S.; Cuppen, E.P.; et al. Preserved Genetic Diversity in Organoids Cultured from Biopsies of Human Colorectal Cancer Metastases. Proc. Natl. Acad. Sci. USA 2015, 112, 13308–13311. [Google Scholar] [CrossRef]
- Singh, S.K.; Clarke, I.D.; Terasaki, M.; Bonn, V.E.; Hawkins, C.; Squire, J.; Dirks, P.B. Identification of a Cancer Stem Cell in Human Brain Tumors. Cancer Res. 2003, 63, 5821–5828. [Google Scholar] [PubMed]
- Wang, Z.; Hu, P.; Tang, F.; Lian, H.; Chen, X.; Zhang, Y.; He, X.; Liu, W.; Xie, C. HDAC6 Promotes Cell Proliferation and Confers Resistance to Temozolomide in Glioblastoma. Cancer Lett. 2016, 379, 134–142. [Google Scholar] [CrossRef]
- Bar, E.E.; Chaudhry, A.; Lin, A.; Fan, X.; Schreck, K.; Matsui, W.; Piccirillo, S.; Vescovi, A.L.; DiMeco, F.; Olivi, A.; et al. Cyclopamine-mediated Hedgehog Pathway Inhibition Depletes Stem-like Cancer Cells in Glioblastoma. Stem Cells 2007, 25, 2524–2533. [Google Scholar] [CrossRef] [PubMed]
- Yu, K.K.H.; Taylor, J.T.; Pathmanaban, O.N.; Youshani, A.S.; Beyit, D.; Dutko-Gwozdz, J.; Benson, R.; Griffiths, G.; Peers, I.; Cueppens, P.; et al. High Content Screening of Patient-derived Cell Lines Highlights the Potential of Non-standard Chemotherapeutic Agents for the Treatment of Glioblastoma. PLoS ONE 2018, 13, e0193694. [Google Scholar] [CrossRef] [PubMed]
- Hemmati, H.D.; Nakano, I.; Lazareff, J.A.; Masterman-Smith, M.; Geschwind, D.H.; Bronner-Fraser, M.; Kornblum, H.I. Cancerous Stem Cells Can Arise from Pediatric Brain Tumors. Proc. Natl. Acad. Sci. USA 2003, 100, 15178–15183. [Google Scholar] [CrossRef]
- Badekila, A.K.; Kini, S.; Jaiswal, A.K. Fabrication Techniques of Biomimetic Scaffolds in Three-dimensional Cell Culture: A Review. J. Cell. Physiol. 2021, 236, 741–762. [Google Scholar] [CrossRef] [PubMed]
- Ozturk, M.S.; Lee, V.K.; Dai, G.; Intes, X. Longitudinal Volumetric Assessment of Glioblastoma Brain Tumors in 3D Bio-Printed Environment by Mesoscopic Fluorescence Molecular Tomography. In Proceedings of the Cancer Imaging and Therapy 2016, Fort Lauderdale, FL, USA, 25–28 April 2016; pp. 3–46. [Google Scholar]
- Magno, V.; Meinhardt, A.; Werner, C. Polymer Hydrogels to Guide Organotypic and Organoid Cultures. Adv. Funct. Mater. 2020, 30, 2000097. [Google Scholar] [CrossRef]
- Xiao, W.; Wang, S.; Zhang, R.; Sohrabi, A.; Yu, Q.; Liu, S.; Ehsanipour, A.; Liang, J.; Bierman, R.D.; Nathanson, D.A.; et al. Bioengineered Scaffolds for 3D Culture Demonstrate Extracellular Matrix-mediated Mechanisms of Chemotherapy Resistance in Glioblastoma. Matrix Biol. 2019, 85–86, 128–146. [Google Scholar] [CrossRef]
- Xu, J.; Margol, A.; Asgharzadeh, S.; Erdreich-Epstein, A. Pediatric Brain Tumor Cell Lines. J. Cell. Biochem. 2015, 116, 218–224. [Google Scholar] [CrossRef]
- Dutta, D.; Heo, I.; Clevers, H. Disease Modeling in Stem Cell-Derived 3D Organoid Systems. Trends Mol. Med. 2017, 23, 393–410. [Google Scholar] [CrossRef] [PubMed]
- Ho, B.X.; Pek, N.M.Q.; Soh, B.S. Disease Modeling Using 3D Organoids Derived From Human Induced Pluripotent Stem Cells. Int. J. Mol. Sci. 2018, 19, 936. [Google Scholar] [CrossRef] [PubMed]
- Chhibber, T.; Bagchi, S.; Lahooti, B.; Verma, A.; Al-Ahmad, A.; Paul, M.K.; Pendyala, G.; Jayant, R.D. CNS Organoids: An Innovative Tool for Neurological Disease Modeling and Drug Neurotoxicity Screening. Drug Discov. Today 2020, 25, 456–465. [Google Scholar] [CrossRef] [PubMed]
- Papaioannou, M.D.; Sangster, K.; Sajid, R.S.; Djuric, U.; Diamandis, P. Cerebral Organoids: Emerging Humanoid Models of Glioblastoma. Acta Neuropathol. Commun. 2020, 8, 209. [Google Scholar] [CrossRef]
- Lancaster, M.A.; Renner, M.; Martin, C.A.; Wenzel, D.; Bicknell, L.S.; Hurles, M.E.; Homfray, T.; Penninger, J.M.; Jackson, A.P.; Knoblich, J.A. Cerebral Organoids Model Human Brain Development and Microcephaly. Nature 2013, 501, 373–379. [Google Scholar] [CrossRef]
- Bian, S.; Repic, M.; Guo, Z.; Kavirayani, A.; Burkard, T.; Bagley, J.A.; Krauditsch, C.; Knoblich, J.A. Genetically Engineered Cerebral Organoids Model Brain Tumor Formation. Nat. Methods 2018, 15, 631–639. [Google Scholar] [CrossRef]
- Qian, X.; Song, H.; Ming, G.l. Brain Organoids: Advances, Applications and Challenges. Development 2019, 146, dev166074. [Google Scholar] [CrossRef]
- Berger, E.; Magliaro, C.; Paczia, N.; Monzel, A.S.; Antony, P.; Linster, C.L.; Bolognin, S.; Ahluwalia, A.; Schwamborn, J.C. Millifluidic Culture Improves Human Midbrain Organoid Vitality and Differentiation. Lab Chip 2018, 18, 3172–3183. [Google Scholar] [CrossRef] [PubMed]
- Sarker, F.A.; Prior, V.G.; Bax, S.; O’Neill, G.M. Forcing a Growth Factor Response–Tissue-Stiffness Modulation of Integrin Signaling and Crosstalk with Growth Factor Receptors. J. Cell Sci. 2020, 133, jcs242461. [Google Scholar] [CrossRef]
- An, Z.; Aksoy, O.; Zheng, T.; Fan, Q.W.; Weiss, W.A. Epidermal Growth Factor Receptor and EGFRvIII in Glioblastoma: Signaling Pathways and Targeted Therapies. Oncogene 2018, 37, 1561–1575. [Google Scholar] [CrossRef] [PubMed]
- Dababneh, A.B.; Ozbolat, I.T. Bioprinting Technology: A Current State-of-the-art Review. J. Manuf. Sci. Eng. 2014, 136, 061016. [Google Scholar] [CrossRef]
- Cui, X.; Gao, G.; Yonezawa, T.; Dai, G. Human cartilage tissue fabrication using three-dimensional inkjet printing technology. J. Vis. Exp. JoVE 2014, 88, 51294. [Google Scholar] [CrossRef]
- Wang, W.; Li, G.; Huang, Y. Modeling of bubble expansion-induced cell mechanical profile in laser-assisted cell direct writing. J. Manuf. Sci. Eng. 2009, 131, 051013. [Google Scholar] [CrossRef]
- Sun, H.; Jia, Y.; Dong, H.; Dong, D.; Zheng, J. Combining Additive Manufacturing with Microfluidics: An Emerging Method for Developing Novel Organs-on-Chips. Curr. Opin. Chem. Eng. 2020, 28, 1–9. [Google Scholar] [CrossRef]
- Gungor-Ozkerim, P.S.; Inci, I.; Zhang, Y.S.; Khademhosseini, A.; Dokmeci, M.R. Bioinks for 3D Bioprinting: An Overview. Biomater. Sci. 2018, 6, 915–946. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Yao, R.; Ouyang, L.; Ding, H.; Zhang, T.; Zhang, K.; Cheng, S.; Sun, W. Three-dimensional Printing of Hela Cells for Cervical Tumor Model. Biofabrication 2014, 6, 035001. [Google Scholar] [CrossRef] [PubMed]
- Lee, V.K.; Dai, G.; Zou, H.; Yoo, S.S. Generation of 3-D glioblastoma-Vascular Niche using 3-D Bioprinting. In Proceedings of the IEEE 2015 41st Annual Northeast Biomedical Engineering Conference (Nebec), Troy, NY, USA, 17–19 April 2015; pp. 1–2. [Google Scholar]
- Mirani, B.; Pagan, E.; Shojaei, S.; Duchscherer, J.; Toyota, B.D.; Ghavami, S.; Akbari, M. A 3D Bioprinted Hydrogel Mesh Loaded with All-Trans Retinoic Acid for Treatment of Glioblastoma. Eur. J. Pharmacol. 2019, 854, 201–212. [Google Scholar] [CrossRef]
- Ouyang, L.; Highley, C.B.; Rodell, C.B.; Sun, W.; Burdick, J.A. 3D Printing of Shear-Thinning Hyaluronic Acid Hydrogels with Secondary Cross-Linking. ACS Biomater. Sci. Eng. 2016, 2, 1743–1751. [Google Scholar] [CrossRef] [PubMed]
- Suri, S.; Schmidt, C.E. Cell-Laden Hydrogel Constructs of Hyaluronic Acid, Collagen, and Laminin for Neural Tissue Engineering. Tissue Eng. Part A 2010, 16, 1703–1716. [Google Scholar] [CrossRef]
- Yi, H.G.; Jeong, Y.H.; Kim, Y.; Choi, Y.J.; Moon, H.E.; Park, S.H.; Kang, K.S.; Bae, M.; Jang, J.; Youn, H.; et al. A Bioprinted Human-Glioblastoma-on-a-Chip for the Identification of Patient-Specific Responses to Chemoradiotherapy. Nat. Biomed. Eng. 2019, 3, 509–519. [Google Scholar] [CrossRef]
- Datta, P.; Dey, M.; Ataie, Z.; Unutmaz, D.; Ozbolat, I.T. 3D Bioprinting for Reconstituting the Cancer Microenvironment. NPJ Precis. Oncol. 2020, 4, 1–13. [Google Scholar] [CrossRef]
- Pham, M.T.; Pollock, K.M.; Rose, M.D.; Cary, W.A.; Stewart, H.R.; Zhou, P.; Nolta, J.A.; Waldau, B. Generation of Human Vascularized Brain Organoids. Neuroreport 2018, 29, 588. [Google Scholar] [CrossRef]
- Mansour, A.; Gage, F.H.; Ozkan, A.; Kumar, W.; Basak, A.N.; Macklis, J.D.; Pappalardo, Z.; Ohlemscher, S.; Chien, C.C. In Vivo Brain Organoid Model for Generation of Vascularized and Functional PSC-Derived Human Brain Organoids. Neuroscience 2000, 6, 163. [Google Scholar]
- Tomasina, C.; Bodet, T.; Mota, C.; Moroni, L.; Camarero-Espinosa, S. Bioprinting Vasculature: Materials, Cells and Emergent Techniques. Materials 2019, 12, 2701. [Google Scholar] [CrossRef]
- Lee, C.S.; Leong, K.W. Advances in Microphysiological Blood-brain Barrier (BBB) Models Towards Drug Delivery. Curr. Opin. Biotechnol. 2020, 66, 78–87. [Google Scholar] [CrossRef] [PubMed]
- Lim, K.S.; Schon, B.S.; Mekhileri, N.V.; Brown, G.C.; Chia, C.M.; Prabakar, S.; Hooper, G.J.; Woodfield, T.B. New Visible-Light Photoinitiating System for Improved Print Fidelity in Gelatin-Based Bioinks. ACS Biomater. Sci. Eng. 2016, 2, 1752–1762. [Google Scholar] [CrossRef]
- Lee, C.; Abelseth, E.; de la Vega, L.; Willerth, S.M. Bioprinting a Novel Glioblastoma Tumor Model Using a Fibrin-Based Bioink for Drug Screening. Mater. Today Chem. 2019, 12, 78–84. [Google Scholar] [CrossRef]
- Jahromi, M.A.M.; Abdoli, A.; Rahmanian, M.; Bardania, H.; Bayandori, M.; Basri, S.M.M.; Kalbasi, A.; Aref, A.R.; Karimi, M.; Hamblin, M.R. Microfluidic Brain-on-a-chip: Perspectives for mimicking neural system disorders. Mol. Neurobiol. 2019, 56, 8489–8512. [Google Scholar] [CrossRef] [PubMed]
- Murphy, S.V.; De Coppi, P.; Atala, A. Opportunities and Challenges of Translational 3D Bioprinting. Nat. Biomed. Eng. 2020, 4, 370–380. [Google Scholar] [CrossRef] [PubMed]
- Mao, H.; Yang, L.; Zhu, H.; Wu, L.; Ji, P.; Yang, J.; Gu, Z. Recent Advances and Challenges in Materials for 3D Bioprinting. Prog. Nat. Sci. Mater. Int. 2020, 30, 618–634. [Google Scholar] [CrossRef]
- Brown, T.E.; Marozas, I.A.; Anseth, K.S. Amplified Photodegradation of Cell-Laden Hydrogels via an Addition–Fragmentation Chain Transfer Reaction. Adv. Mater. 2017, 29, 1605001. [Google Scholar] [CrossRef]
- Bakarich, S.E.; Gorkin III, R.; Panhuis, M.I.H.; Spinks, G.M. 4D Printing with Mechanically Robust, Thermally Actuating Hhydrogels. Macromol. Rapid Commun. 2015, 36, 1211–1217. [Google Scholar] [CrossRef]
- Tomaskovic-Crook, E.; Crook, J.M. 3D Bioprinting Electrically Conductive Bioink with Human Neural Stem Cells for Human Neural Tissues. In 3D Bioprinting; Springer: New York, NY, USA, 2020; pp. 159–170. [Google Scholar]
- Zhang, Y.S.; Zhang, Y.N.; Zhang, W. Cancer-on-a-Chip Systems at the Frontier of Nanomedicine. Drug Discov. Today 2017, 22, 1392–1399. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Barker, J.; Zhou, C.; Li, W.; Zhang, J.; Lin, B.; Foltz, G.; Küblbeck, J.; Honkakoski, P. Towards Personalized Medicine with a Three-dimensional Micro-scale Perfusion-based Two-chamber Tissue Model System. Biomaterials 2012, 33, 4353–4361. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Oh, S.; Hoang, H.H.; Nguyen, D.T.T.; Lim, W.; Shin, T.H.; Lee, G.; Park, S. Recapitulation of Cancer Stem Cell Niches in Glioblastoma on 3D Microfluidic Cell Culture Devices Under Gravity-driven Perfusion. J. Ind. Eng. Chem. 2018, 62, 352–361. [Google Scholar] [CrossRef]

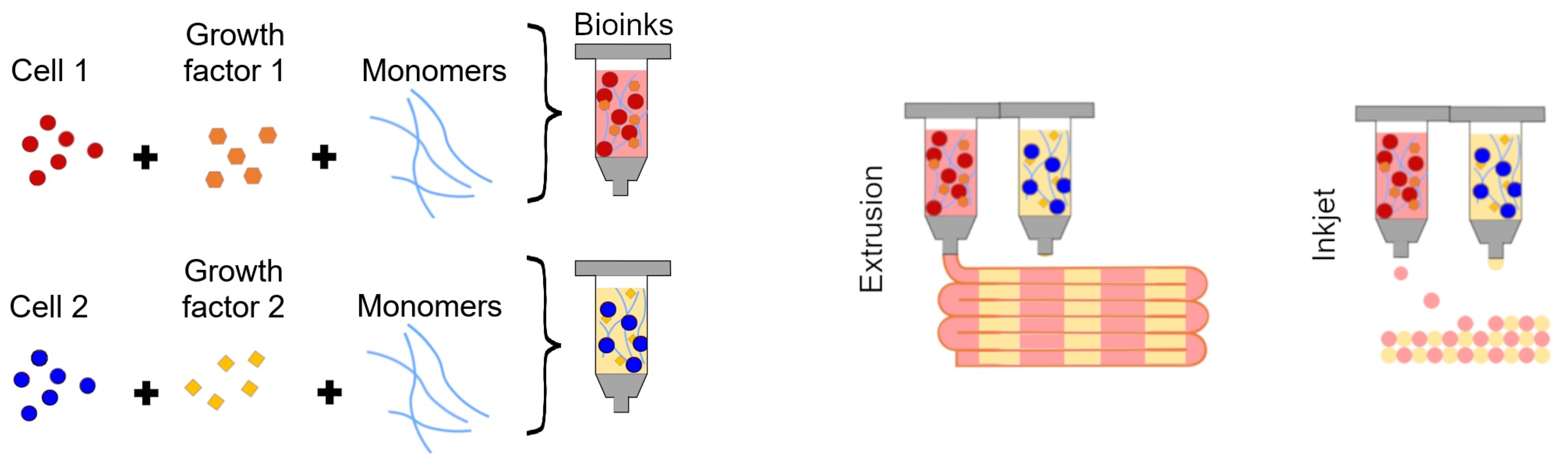
| Model Type | Definition |
|---|---|
| Glioma spheroid (GS) (with serum) | Dense conglomerate of cells cultured in serum—growth of CSCs not specifically promoted. |
| Glioma tumouroid (GT) | Tumour organoids generated by growing primary tumour material in suspension under defined media conditions in the absence of serum, CSCs specifically promoted and cellular heterogeneity maintained. |
| Brain organoid (BO) | Derived from stem cells under specific media and growth conditions to promote tissue lineage differentiation, displays some functionality and morphological features of model organ. |
| GS/BO and GT/BO | Glioma spheroid or tumouroid co-cultured with a BO. |
| Free | Single cells/spheroid/tumouroid suspended in liquid medium. |
| Matrix-supported | Single cells/spheroid/organoid encapsulated in a 3D matrix. |
| Model Type | Cell Origin | Culture Method | Findings | Ref. |
|---|---|---|---|---|
| Adult GBM | ||||
| Free tumouroid | Cerebral organoid generated from hESC cell line H1. | Oncogenesis transduced with oncogene and knockdown of p53. | Tumouroids can be generated from cerebral organoids via gene manipulation. | [75] |
| Free tumouroid | Dissociated GBM specimens. | Suspended in serum-free media. | Tumouroids recapitulated the morphology and expression profile of parent GBM tumours. | [3] |
| Free tumouroid co-culture | GA-MSCs and CSCs were isolated from surgical specimens of GBM stroma and GBM, respectively. | Dissociated and resuspended in liquid differentiation media. | Stromal GA-MSCs excrete exosomes that increased proliferation of GSC xenografts and decreased median survival of the host animals when pre-treated with stromal GA-MSCs-derived exosomes. | [26] |
| Free tumouroid/ spheroid | Patient-derived GSCs/ cell line U87 | Non-adherent plates. | All patient-derived tumouroids from primary GSCs were Nestin and Sox2 positive. Chemotherapeutics were effective only on 3D U87 spheroids. Tumouroids from the one recurrent cell line were the most drug-resistant. TMZ efficacy was patient-specific. | [84] |
| Paediatric GBM | ||||
| Ex-supported tumouroid (passaged in PDX models then extracted) | Specimens of pGBM | Xenografts of human pGBM patients with therapy-naive, recurrent and lethal disease were extracted, minced and enriched for CSCs. | An AUKRA inhibitor was most effective on therapy-naive tumouroids, followed by recurrent ex-xenografted tumouroids. | [69] |
| Free tumouroid | Tumour specimens from six pGBM patients | Stem cell population expanded via specialised media. | EGFR and PDGFRA amplification and deletion of RB1, CDKN2A/B & PTEN was observed. | [33] |
| Free tumouroid | Dissociated pGBM specimens from two patients | Suspended in serum-free media. | Stemness markers nestin, CD133, Sox2, melk, PSP and bmi-1 were expressed. | [85] |
| Free tumouroid | Dissociated pGBM specimens from 14 patients | Suspended in neural stem-cell media. | Stemness markers CD133 and Nestin were expressed and self-renewal was retained even when secondary tumouroids were formed from a single cell. | [81] |
| Model Type | Cell Origin | Culture Method | Findings | Ref. |
|---|---|---|---|---|
| Matrix -supported spheroid | GBM lines E98, E468 & U-251MG | Spheroids formed with hanging drop and implanted in nude rats, rat brain slices, rBM-based hydrogel layers or 3-layers of astrocytes. Hyaluronic acid was added to media. | Migration on brain slices was through blood vessels. Spheroids on rBM hydrogel and astrocyte layers recapitulated some migratory patterns seen in live rat brains. Higher HA concentration in media induced more rapid migration. | [15] |
| Matrix -supported spheroid | GBM cell line U251N | Hanging drop then embedded in collagen gel. | TMZ was effective in dose- and time-dependent manner | [1] |
| Matrix -supported spheroid | Patient-derived cell lines K301, GBM6, GS024 & GS025 | Tumouroids were formed in suspension, dissociated, then transferred to HA-based hydrogel in a microfluidic chip. | Higher HA induced proliferation and drug resistance. | [89] |
| Matrix- supported tumouroid | Patient-derived CSCs. | Low-attachment plates and neurobasal media then encapsulation in HA/collagen hydrogel. Interstitial pressure was applied by deferentially filling a Millipore insert in a cell culture well. | Increased flow through the channel induced patient-specific increase in migration between 1.3 and 1.5-fold. With knockdown of CXR4, CXCL12 and CD44, a flow-induced increase in migration was neutralised. | [70] |
| Model Type | Cell Origin | Culture Method | Findings | Ref |
|---|---|---|---|---|
| Brain organoid | hESC cell line H9 | Differentiation media | Organoids were transduced to invoke oncogenesis. The number of modified, malignant cells surpassed healthy organoid cells over weeks. | [75] |
| Brain organoid | hESC cell lines H1, H6 or H9 | Matrigel-coated plates & differentiation media | A primitive ventricular system and neural rosettes were formed & a proliferative zone of neural stem cells was present. | [11] |
| Brain organoid | iPSCs | Differentiation media & transfer to orbital shaker or millifluidic device | Millifluidic media exchange successfully reduced size of necrotic and hypoxic regions. No overall size difference was observed. | [98] |
| Brain organoid | hESCs | Low-attachment plates & differentiation media | Induction of common GBM genes with electroporation resulted in malignant cells overtaking healthy organoid cells within a month. | [96] |
| Cancerous Constituent | Culture Method | Healthy Brain Constituent | Culture Method | Findings | Ref. |
|---|---|---|---|---|---|
| Tumouroid | Dissociated primary CSCs cultured inlow-attachment plates with differentiation media | Brain organoid | hESC cell line H1 cultured inlow-attachment plates & differentiation media | Radial migration of tumouroid cells. Modification of ECM related expression similar to in-vivo. | [77] |
| Spheroid | SK2176 GBM cell-line cultured inlow-attachment plates | Brain organoid | hESC cell line H1 cultured in differentiation media | Spontaneous attachment and invasion of tumour cells into cerebral organoid. 30% of organoid volume was invaded after 24 days. Degree of invasiveness in model correlated with lethality of orthotopically xenografted tumouroids. | [75] |
| GSC cell line insuspension | Co-culture | Brain organoid | hESC cell line H1, H6 or H9 culture inMatrigel-coated plates with differentiation media | Co-cultures were more resistant to chemo-therapeutic agents and radiation versus 2D cultures. EGFR levels of parent tissue were recapitulated in 3D co-cultures and absent in 2D analogues. | [11] |
| Transfection of 18 GBM-like gene mutations/ amplifications | Oncogenesis of organoid via electroporation | Cerebral organoid | Generated from EBs with differentiation media | GBM can be initiated by selective gene manipulation. Increased invasiveness, higher expression of invasion-related genes and lower expression of tumour-inhibitive genes were observed in gene-altered cells. | [96] |
| Model Type | Cells Used | Gel Material and Organisation | Findings | Ref. |
|---|---|---|---|---|
| Bioprinted matrix -supported co-culture | GBM cell line U87MG, GSC lines G166, G144 & G7 monocyte cell line MM6 | RGD-alginate + <250 mg/L HA or collagen I. Central tumouroid was printed then surrounded by astroma-like cell-laden gel construct. | Printed GBM cells remained viable (>90%) for months and CSCs retained stemness. Temozolomide IC50 doubled for printed spheroids compared to 2D co-cultures. GBM cells printed alongside fibroblasts were more resistant to TMZ. | [2] |
| Bioprinted matrix -supported co-culture | GBM cell line GL261 & macrophage cell line RAW 264.7 | GelMA was used as both GBM and stroma-like bioink to create a GBM tumour model enclosed by amacrophage-laden gel construct. | Shear-thinning GelMA decreased printing-related cell death. Macrophages migrated towards GBM cells in co-culture and GBM cells had 15-fold increases inGBM-specific markers compared to 3D and 2D mono-culture. | [48] |
| Model Type | Gel Material and Layout | Findings | Ref. |
|---|---|---|---|
| 3D GBM- vascular niche with patient- derived CSCs co-cultured with HUVECs | A straight fluidic vascular channel was printed with collagen I and lined with HUVECs. CSCs were seeded adjacent to the microvessel. | At the highest concentration of laminin (100 µg/mL), CSCs migrated 1.5× further than inthegel containing 10 µg/mL of laminin. | [107] |
| GBM-on-a- chip with continuous cell line U-87 and patient- derived line co-cultured with HUVECs | A circular fluidic vascular channel was printed in collagen and abioink developed from decellularised porcine brain ECM. GBM-laden hydrogel was printed in the centre of a ring of collagen gel containing HUVECs. This was surrounded again by amicrochannel with an outer boundary printed in gas permeable silicone. | GBM cells grew in dense spheres with ananoxia-normoxia gradient and peripheral pseudopalisading cells. Cells in the intermediate region excreted factors leading to microvessel formation in the periphery. In porcine brain-derived gel, angiogenesis, proliferation and expression of pro-angiogenic genes and ECM remodelling proteins increased. All patient-derived cells in co-culture with HUVECs exhibited a dose-dependent response to TMZ but those on-chip recapitulated clinical therapy resistance, unlike the same cells cultured in 2D and 3D monoculture. Following multiple treatments, GBM cells extracted from patients with a longer survival exhibited decreased metabolic activity even after treatment ceased, whereas the metabolic activity increased after treatment ceased in the cells originating from patients with a shorter survival. | [111] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Orcheston-Findlay, L.; Bax, S.; Utama, R.; Engel, M.; Govender, D.; O’Neill, G. Advanced Spheroid, Tumouroid and 3D Bioprinted In-Vitro Models of Adult and Paediatric Glioblastoma. Int. J. Mol. Sci. 2021, 22, 2962. https://doi.org/10.3390/ijms22062962
Orcheston-Findlay L, Bax S, Utama R, Engel M, Govender D, O’Neill G. Advanced Spheroid, Tumouroid and 3D Bioprinted In-Vitro Models of Adult and Paediatric Glioblastoma. International Journal of Molecular Sciences. 2021; 22(6):2962. https://doi.org/10.3390/ijms22062962
Chicago/Turabian StyleOrcheston-Findlay, Louise, Samuel Bax, Robert Utama, Martin Engel, Dinisha Govender, and Geraldine O’Neill. 2021. "Advanced Spheroid, Tumouroid and 3D Bioprinted In-Vitro Models of Adult and Paediatric Glioblastoma" International Journal of Molecular Sciences 22, no. 6: 2962. https://doi.org/10.3390/ijms22062962
APA StyleOrcheston-Findlay, L., Bax, S., Utama, R., Engel, M., Govender, D., & O’Neill, G. (2021). Advanced Spheroid, Tumouroid and 3D Bioprinted In-Vitro Models of Adult and Paediatric Glioblastoma. International Journal of Molecular Sciences, 22(6), 2962. https://doi.org/10.3390/ijms22062962






