m5U54 tRNA Hypomodification by Lack of TRMT2A Drives the Generation of tRNA-Derived Small RNAs
Abstract
1. Introduction
2. Results
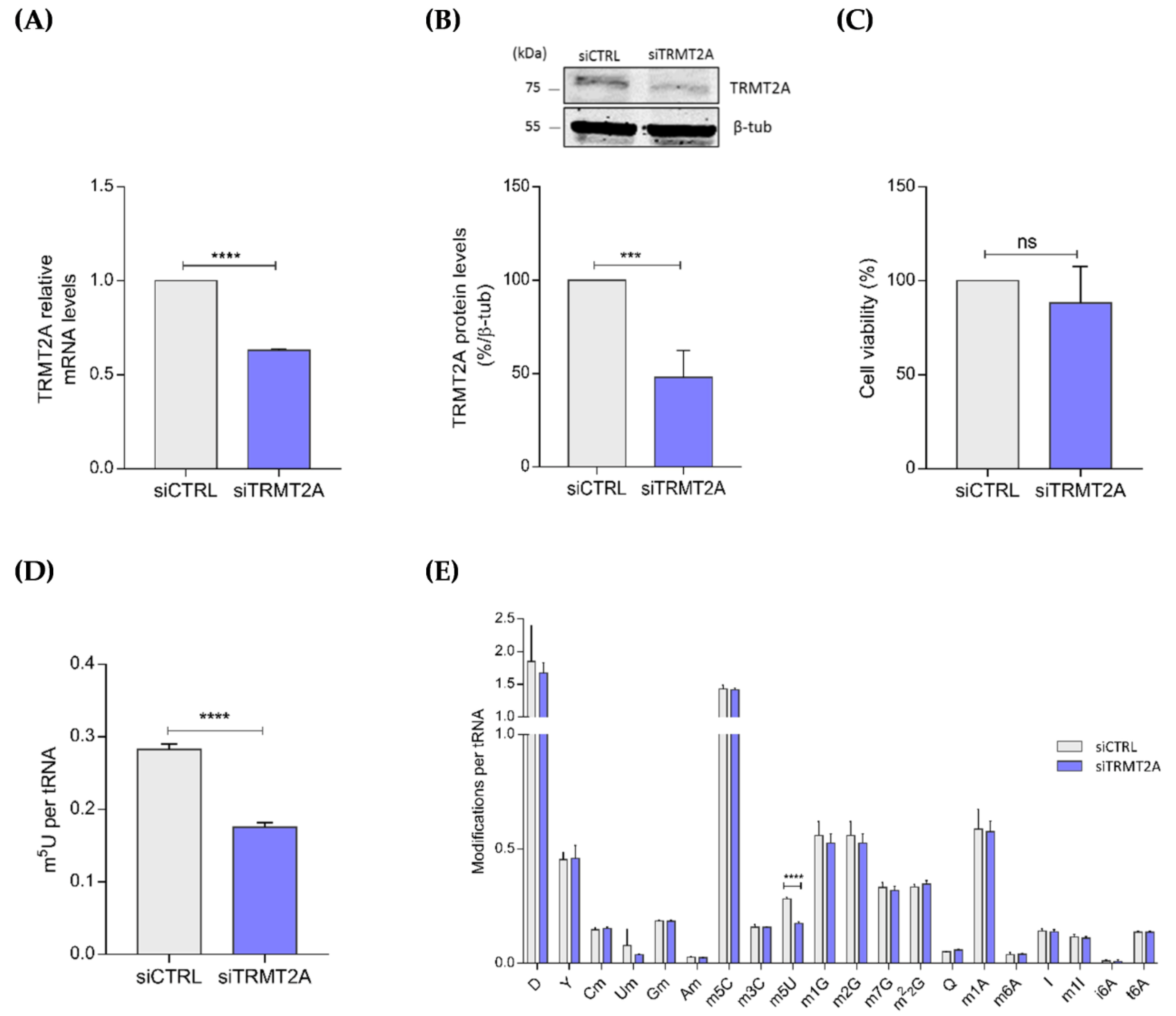
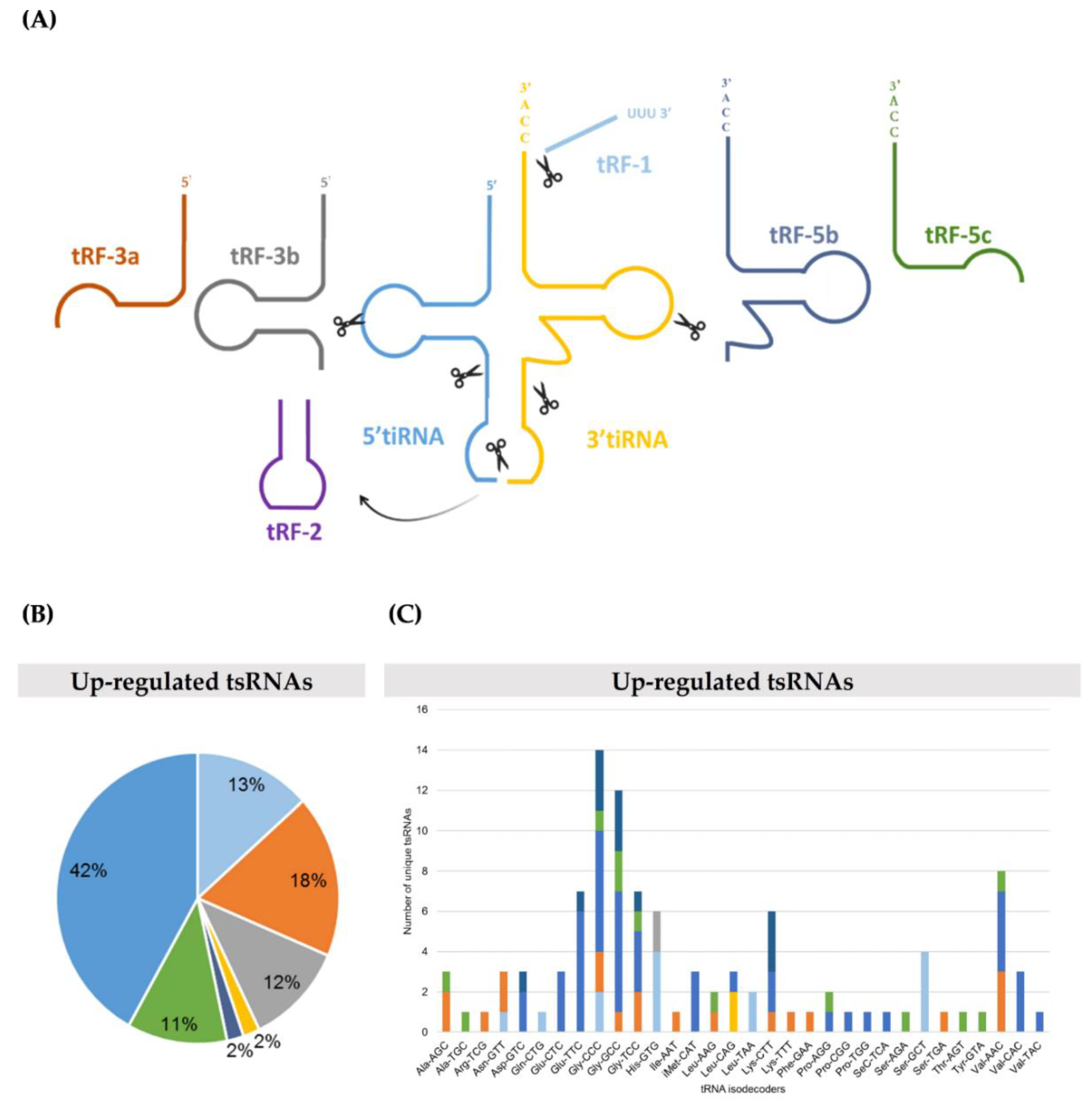
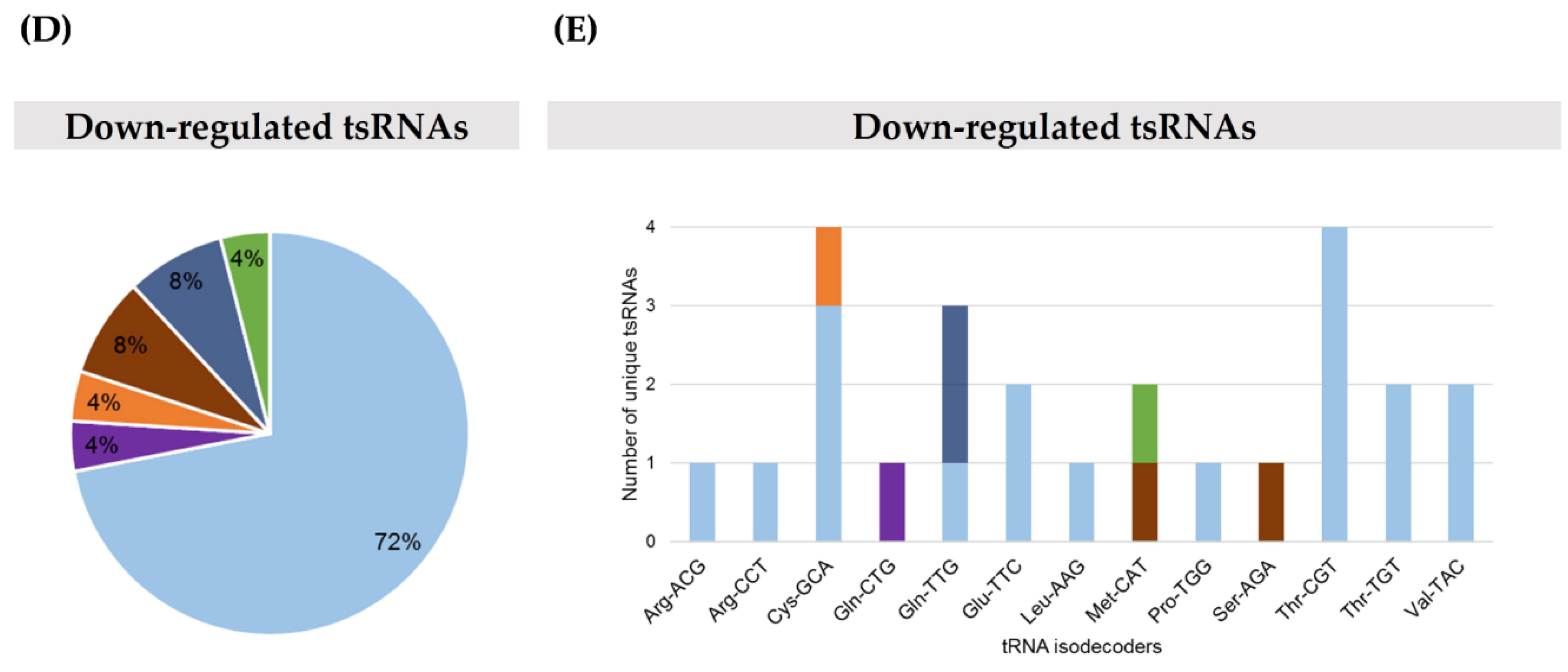
2.1. TRMT2A Silencing Induces tRNA Hypomodification and the Formation of a Panoply of tsRNAs
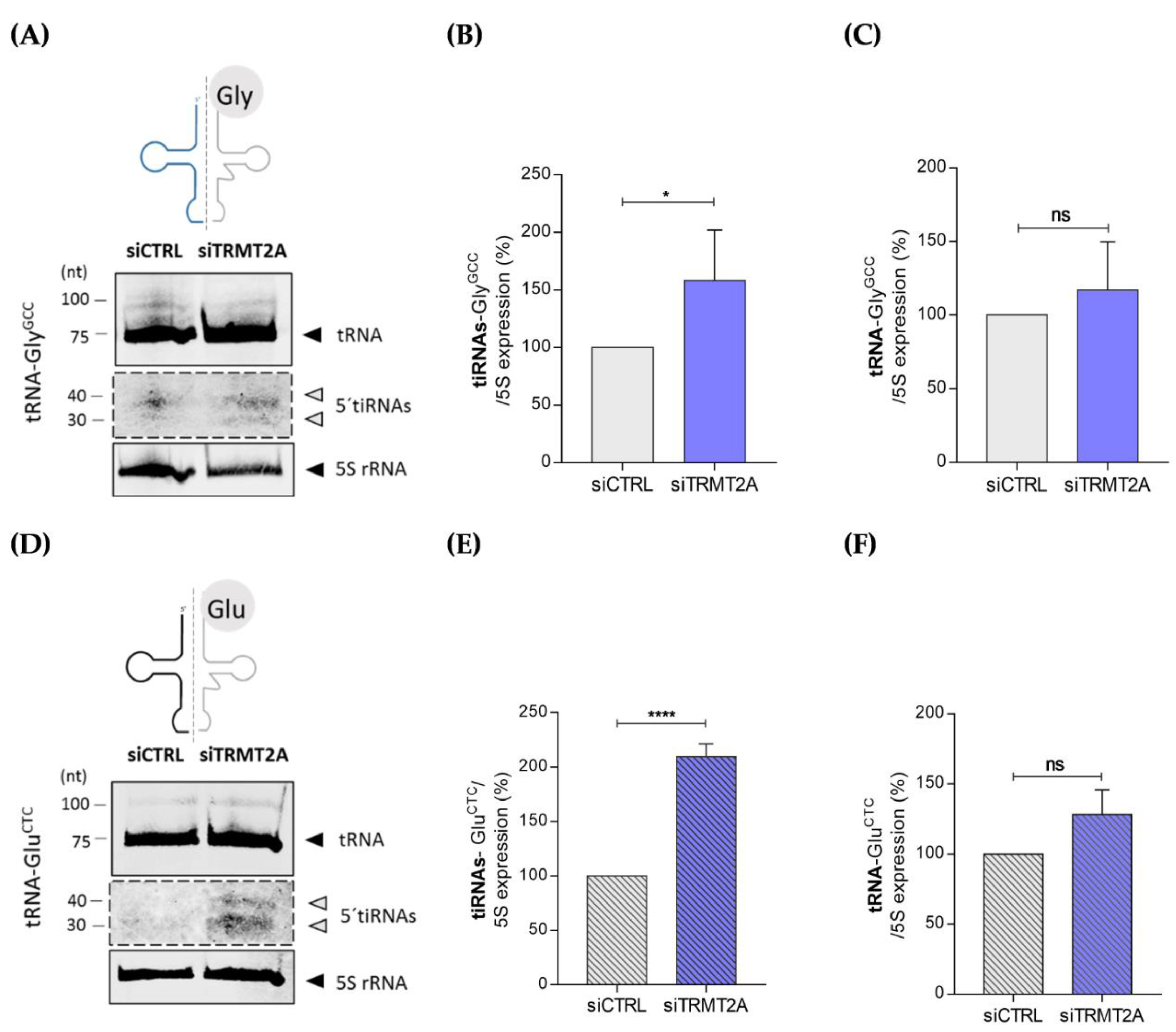
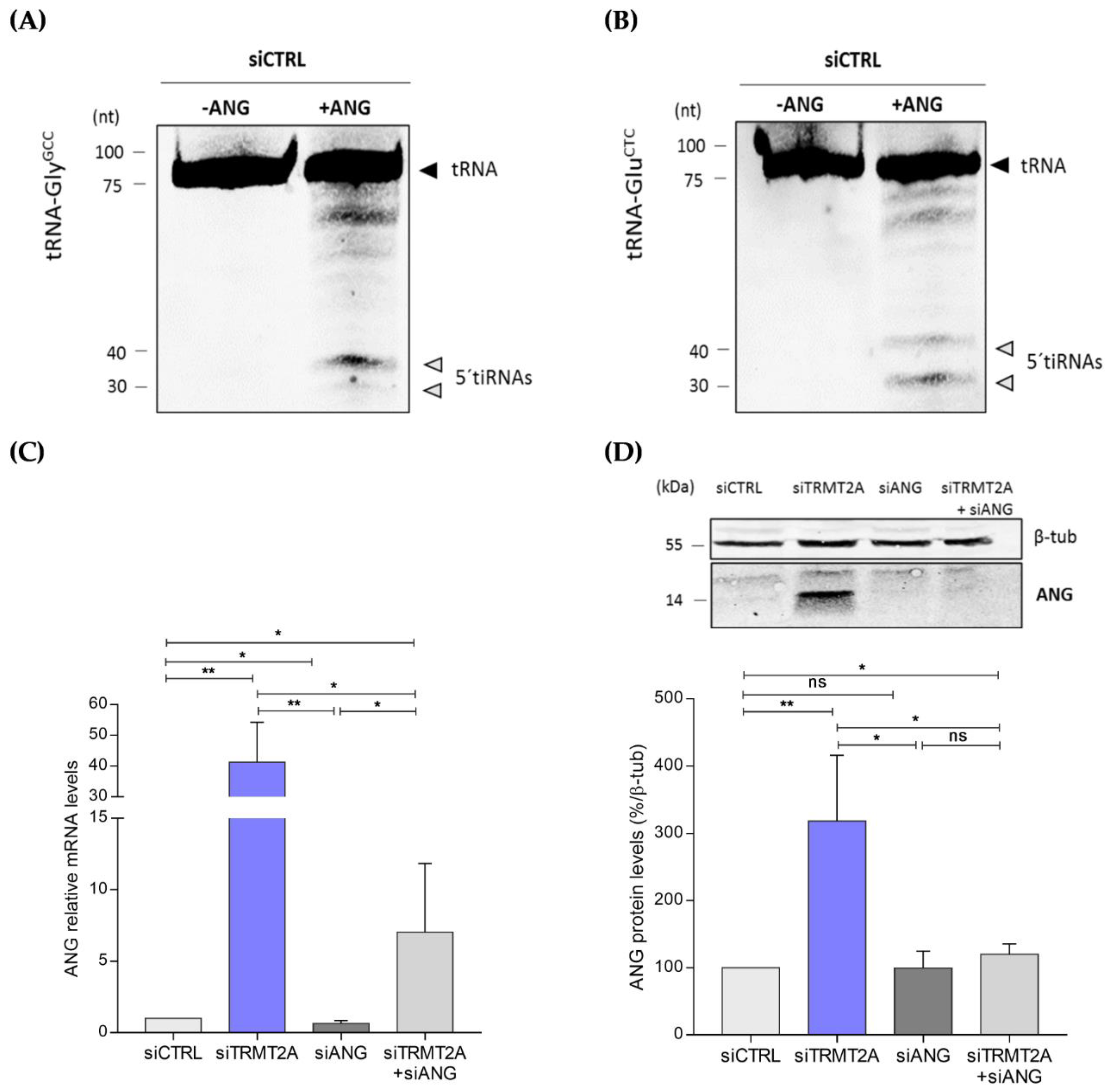
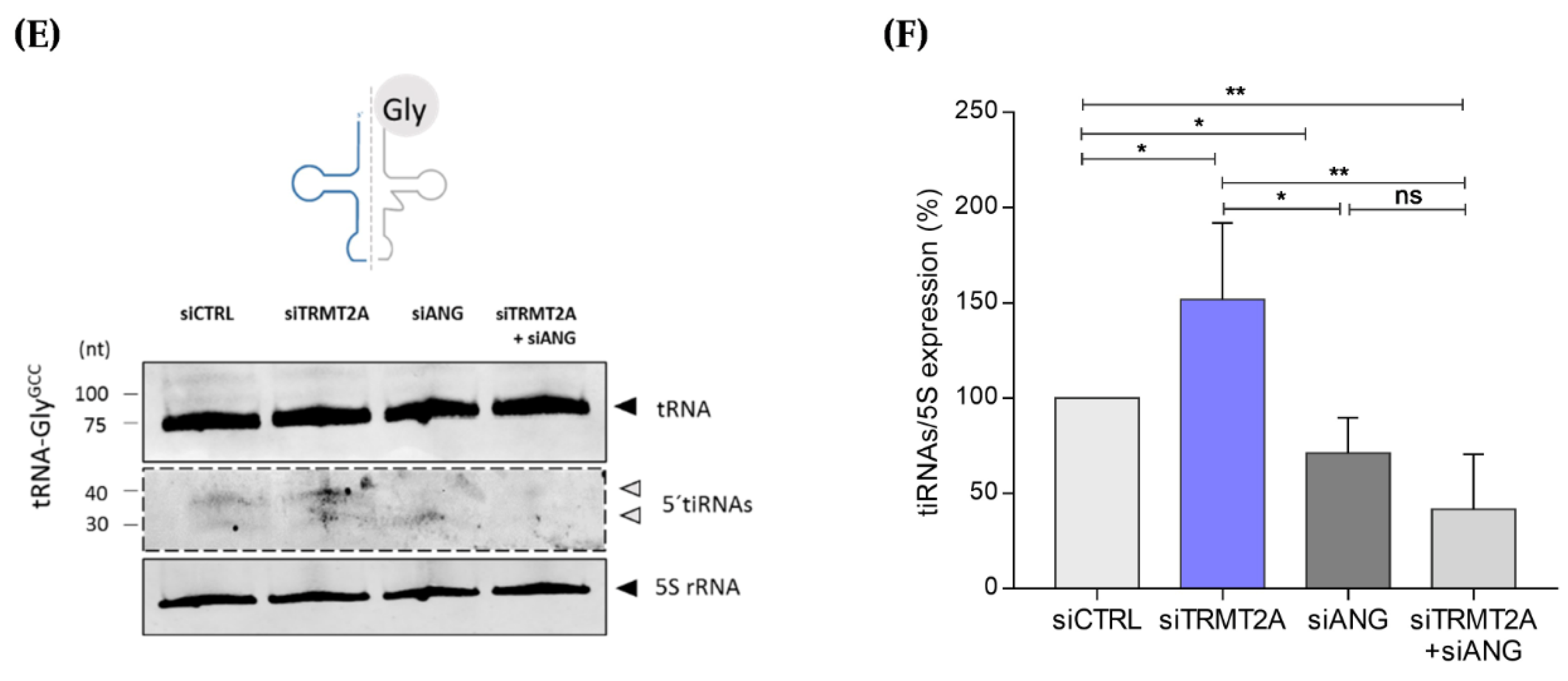
2.2. m5U Hypomodification Induces ANG Dependent 5ʹtiRNA-GlyGCC and 5′tiRNA-GluCTC Formation
2.3. TRMT2A Silencing Affects Protein Synthesis Rate and Elicits the Endoplasmic Reticulum Stress Response
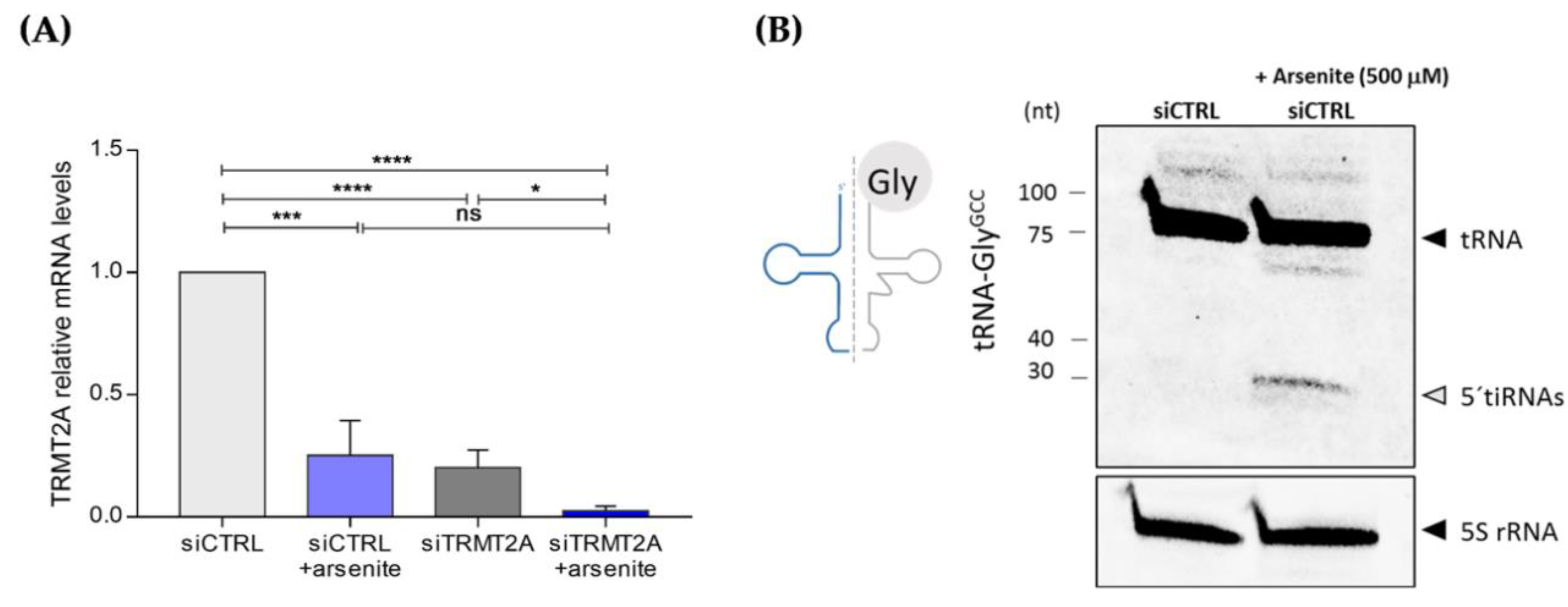
2.4. Oxidative Stress Induces TRMT2A Down-Regulation and tiRNA Generation
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Reverse Transfection with siRNA
4.3. RNA Extraction
4.4. cDNA Synthesis and qPCR
4.5. Quantification of m5U Modification by LC-MS/MS
4.6. Human tRF and tiRNA Sequencing-ArrayStar
4.7. Total Protein Extraction
4.8. Western Blotting
4.9. Northern Blotting
4.10. Cellular Viability Assay
4.11. Cellular Proliferation Assay
4.12. SUnSET Method
4.13. Gene Expression Analysis Using Oligonucleotide Microarrays
4.14. Gene Ontology (GO) Analysis
4.15. Stress Experiments
4.16. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ANG | Angiogenin |
| DEGs | Differentially expressed genes |
| FC | Fold Change |
| FDR | False discovery rate |
| GO | Gene ontology |
| hpt | Hours post-transfection |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| LC-MS/MS | Liquid chromatography coupled mass spectrometry |
| m1A | 1-methyladenosine |
| m5C | Cytosine-5 methylation |
| m5U | 5-methyluridine |
| MTT | 3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide |
| ncRNAs | Small non-coding RNAs |
| NGS | Next generation sequencing |
| nts | Nucleotides |
| qPCR | Quantitative polymerase chain reaction |
| tiRNAs | tRNA-derived stress-induced RNAs |
| tRFs | tRNA-derived fragments |
| tRNAs | Transfer RNAs |
| tsRNAs | tRNA-derived small RNAs formation |
References
- Kirchner, S.; Ignatova, Z. Emerging roles of tRNA in adaptive translation, signalling dynamics and disease. Nat. Rev. Genet. 2015, 16, 98–112. [Google Scholar] [CrossRef]
- Jackman, J.E.; Alfonzo, J.D. Transfer RNA modifications: Nature’s combinatorial chemistry playground. Wiley Interdiscip. Rev. RNA 2013, 4, 35–48. [Google Scholar] [CrossRef] [PubMed]
- Pereira, M.; Francisco, S.; Varanda, A.S.; Santos, M.; Santos, M.A.S.; Soares, A.R. Impact of tRNA modifications and tRNA-modifying enzymes on proteostasis and human disease. Int. J. Mol. Sci. 2018, 19, 3738. [Google Scholar] [CrossRef] [PubMed]
- Ny, T.; Björk, G.R. Cloning and restriction mapping of the trmA gene coding for transfer ribonucleic acid (5-methyluridine)-methyltransferase in Escherichia coli K-12. J. Bacteriol. 1980, 142, 371–379. [Google Scholar] [CrossRef]
- Nordlund, M.E.; Johansson, M.J.O.; Von Pawel-Rammingen, U.; Byström, A.S. Identification of the TRM2 gene encoding the tRNA (m5U54) methyltransferase of Saccharomyces cerevisiae. RNA 2000, 6, 844–860. [Google Scholar] [CrossRef] [PubMed]
- Carter, J.-M.; Emmett, W.; Mozos, I.R.; Kotter, A.; Helm, M.; Ule, J.; Hussain, S. FICC-Seq: A method for enzyme-specified profiling of methyl-5-uridine in cellular RNA. Nucleic Acids Res. 2019, 47, e113. [Google Scholar] [CrossRef] [PubMed]
- Powell, C.A.; Minczuk, M. TRMT2B is responsible for both tRNA and rRNA m5U-methylation in human mitochondria. RNA Biol. 2020, 17, 451–462. [Google Scholar] [CrossRef]
- Yao, L.J.; James, T.L.; Kealey, J.T.; Santi, D.V.; Schmitz, U. The dynamic NMR structure of the TψC-loop: Implications for the specificity of tRNA methylation. J. Biomol. NMR 1997, 9, 229–244. [Google Scholar] [CrossRef] [PubMed]
- Sengupta, R.; Vainauskas, S.; Yarian, C.; Sochacka, E.; Malkiewicz, A.; Guenther, R.H.; Koshlap, K.M.; Agris, P.F. Modified constructs of the tRNA TΨC domain to probe substrate conformational requirements of m1A58 and m5U54 tRNA methyltransferases. Nucleic Acids Res. 2000, 28, 1374–1380. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kumar, P.; Kuscu, C.; Dutta, A. Biogenesis and function of transfer RNA-Related fragments (tRFs). Trends Biochem. Sci. 2016, 41, 679–689. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Xu, Z.; Sheng, J. tRNA-derived small RNA: A Novel regulatory small non-coding RNA. Genes (Basel) 2018, 9, 246. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, P.; Emara, M.M.; Villen, J.; Gygi, S.P.; Anderson, P. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol. Cell 2011, 43, 613–623. [Google Scholar] [CrossRef]
- Yamasaki, S.; Ivanov, P.; Hu, G.; Anderson, P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J. Cell Biol. 2009, 185, 35–42. [Google Scholar] [CrossRef]
- Lee, Y.S.; Shibata, Y.; Malhotra, A.; Dutta, A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev. 2009, 23, 2639–2649. [Google Scholar] [CrossRef] [PubMed]
- Goodarzi, H.; Liu, X.; Fish, L.; Tavazoie, S.F.; Goodarzi, H.; Liu, X.; Nguyen, H.C.B.; Zhang, S.; Fish, L.; Tavazoie, S.F. Endogenous tRNA-derived fragments suppress breast cancer progression via YBX1 displacement. Cell 2015, 161, 790–802. [Google Scholar] [CrossRef]
- Schaffer, A.E.; Eggens, V.R.C.; Caglayan, A.O.; Reuter, M.S.; Scott, E.; Coufal, N.G.; Silhavy, J.L.; Xue, Y.; Kayserili, H.; Yasuno, K.; et al. CLP1 founder mutation links tRNA splicing and maturation to cerebellar development and neurodegeneration. Cell 2014, 157, 651–663. [Google Scholar] [CrossRef]
- Deng, J.; Ptashkin, R.N.; Chen, Y.; Cheng, Z.; Liu, G.; Phan, T.; Deng, X.; Zhou, J.; Lee, I.; Lee, Y.S.; et al. Respiratory syncytial virus utilizes a tRNA fragment to suppress antiviral responses through a novel targeting mechanism. Mol. Ther. 2015, 23, 1622–1629. [Google Scholar] [CrossRef] [PubMed]
- Kuscu, C.; Kumar, P.; Kiran, M.; Su, Z.; Malik, A.; Dutta, A. tRNA fragments (tRFs) guide Ago to regulate gene expression post-transcriptionally in a Dicer-independent manner. RNA 2018, 24, 1093–1105. [Google Scholar] [CrossRef] [PubMed]
- Soares, A.R.; Fernandes, N.; Reverendo, M.; Araújo, H.R.; Oliveira, J.L.; Moura, G.M.R.; Santos, M.A.S. Conserved and highly expressed tRNA derived fragments in zebrafish. BMC Mol. Biol. 2015, 16, 22. [Google Scholar] [CrossRef] [PubMed]
- Gebetsberger, J.; Zywicki, M.; Künzi, A.; Polacek, N. tRNA-derived fragments target the ribosome and function as regulatory non-coding RNA in Haloferax volcanii. Archaea 2012, 260909. [Google Scholar] [CrossRef]
- Kim, H.K.; Fuchs, G.; Wang, S.; Wei, W.; Zhang, Y.; Park, H.; Roy-chaudhuri, B.; Li, P.; Xu, J.; Chu, K.; et al. A transfer-RNA-derived small RNA regulates ribosome biogenesis. Nature 2017, 552, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Thompson, D.M.; Lu, C.; Green, P.J.; Parker, R. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA 2008, 14, 2095–2103. [Google Scholar] [CrossRef]
- Elkordy, A.; Mishima, E.; Niizuma, K.; Akiyama, Y.; Fujimura, M.; Tominaga, T.; Abe, T. Stress-induced tRNA cleavage and tiRNA generation in rat neuronal PC12 cells. J. Neurochem. 2018, 146, 560–569. [Google Scholar] [CrossRef]
- Lyons, S.M.; Achorn, C.; Kedersha, N.L.; Anderson, P.J.; Ivanov, P. YB-1 regulates tiRNA-induced stress granule formation but not translational repression. Nucleic Acids Res. 2016, 44, 6949–6960. [Google Scholar] [CrossRef]
- Yu, M.; Lu, B.; Zhang, J.; Ding, J.; Liu, P.; Lu, Y. tRNA-derived RNA fragments in cancer: Current status and future perspectives. J. Hematol. Oncol. 2020, 13, 121. [Google Scholar] [CrossRef] [PubMed]
- Nunes, A.; Ribeiro, D.R.; Marques, M.; Santos, M.A.S.; Ribeiro, D.; Soares, A.R. Emerging roles of tRNAs in RNA virus infections. Trends Biochem. Sci. 2020, 45, 794–805. [Google Scholar] [CrossRef]
- Jia, Y.; Tan, W.; Zhou, Y. Transfer RNA-derived small RNAs: Potential applications as novel biomarkers for disease diagnosis and prognosis. Ann. Transl. Med. 2020, 8, 1092. [Google Scholar] [CrossRef] [PubMed]
- Pliatsika, V.; Loher, P.; Magee, R.; Telonis, A.G.; Londin, E.; Shigematsu, M.; Kirino, Y.; Rigoutsos, I. MINTbase v2.0: A comprehensive database for tRNA-derived fragments that includes nuclear and mitochondrial fragments from all The Cancer Genome Atlas projects. Nucleic Acids Res. 2018, 46, D152–D159. [Google Scholar] [CrossRef]
- Kumar, P.; Mudunuri, S.B.; Anaya, J.; Dutta, A. tRFdb: A database for transfer RNA fragments. Nucleic Acids Res. 2015, 43, D141–D145. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, M.; Pollex, T.; Hanna, K.; Tuorto, F.; Meusburger, M.; Helm, M.; Lyko, F. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. 2010, 24, 1590–1595. [Google Scholar] [CrossRef]
- Blanco, S.; Dietmann, S.; Flores, J.V.; Hussain, S.; Kutter, C.; Humphreys, P.; Lukk, M.; Lombard, P.; Treps, L.; Popis, M.; et al. Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J. 2014, 33, 2020–2039. [Google Scholar] [CrossRef]
- Blanco, S.; Bandiera, R.; Popis, M.; Hussain, S.; Lombard, P.; Aleksic, J.; Sajini, A.; Tanna, H.; Cortés-Garrido, R.; Gkatza, N.; et al. Stem cell function and stress response are controlled by protein synthesis. Nature 2016, 534, 335–340. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Qi, M.; Shen, B.; Luo, G.; Wu, Y.; Li, J.; Lu, Z.; Zheng, Z.; Dai, Q.; Wang, H. Transfer RNA demethylase ALKBH3 promotes cancer progression via induction of tRNA-derived small RNAs. Nucleic Acids Res. 2019, 47, 2533–2545. [Google Scholar] [CrossRef] [PubMed]
- Kellner, S.; Ochel, A.; Thüring, K.; Spenkuch, F.; Neumann, J.; Sharma, S.; Entian, K.D.; Schneider, D.; Helm, M. Absolute and relative quantification of RNA modifications via biosynthetic isotopomers. Nucleic Acids Res. 2014, 42, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Hagelskamp, F.; Borland, K.; Ramos, J.; Hendrick, A.G.; Fu, D.; Kellner, S. Broadly applicable oligonucleotide mass spectrometry for the analysis of RNA writers and erasers in vitro. Nucleic Acids Res. 2020, 48, e41. [Google Scholar] [CrossRef]
- Torres, A.G.; Reina, O.; Attolini, C.S.-O.; de Pouplana, L.R. Differential expression of human tRNA genes drives the abundance of tRNA-derived fragments. Proc. Natl. Acad. Sci. USA 2019, 116, 8451–8456. [Google Scholar] [CrossRef]
- Thomas, S.P.; Hoang, T.T.; Ressler, V.T.; Raines, R.T. Human angiogenin is a potent cytotoxin in the absence of ribonuclease inhibitor. RNA 2018, 24, 1018–1027. [Google Scholar] [CrossRef]
- Drino, A.; Oberbauer, V.; Troger, C.; Janisiw, E.; Anrather, D.; Hartl, M.; Kaiser, S.; Kellner, S.; Schaefer, M.R. Production and purification of endogenously modified tRNA-derived small RNAs. RNA Biol. 2020, 17, 1104–1115. [Google Scholar] [CrossRef]
- Fu, H.; Feng, J.; Liu, Q.; Sun, F.; Tie, Y.; Zhu, J.; Xing, R.; Sun, Z.; Zheng, X. Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett. 2009, 583, 437–442. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. ClusterProfiler: An R package for comparing biological themes among gene clusters. OMICS J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Soares, A.R.; Santos, M. Discovery and function of transfer RNA-derived fragments and their role in disease. Wiley Interdiscip. Rev. RNA 2017, 8, 1–13. [Google Scholar] [CrossRef]
- Schmidt, E.K.; Clavarino, G.; Ceppi, M.; Pierre, P. SUnSET, a nonradioactive method to monitor protein synthesis. Nat. Methods 2009, 6, 275–277. [Google Scholar] [CrossRef]
- Saikia, M.; Krokowski, D.; Guan, B.-J.; Ivanov, P.; Parisien, M.; Hu, G.; Anderson, P.; Pan, T.; Hatzoglou, M. Genome-wide identification and quantitative analysis of cleaved tRNA fragments induced by cellular stress. J. Biol. Chem. 2012, 287, 42708–42725. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Chen, Y.; Ren, Y.; Zhou, J.; Ren, J.; Lee, I.; Bao, X. A tRNA-derived RNA Fragment plays an important role in the mechanism of arsenite-induced cellular responses. Sci. Rep. 2018, 8, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Machnicka, M.A.; Olchowik, A.; Grosjean, H.; Bujnicki, J.M. Distribution and frequencies of post-transcriptional modifications in tRNAs. RNA Biol. 2014, 11, 1619–1629. [Google Scholar] [CrossRef]
- Gkatza, N.A.; Castro, C.; Harvey, R.F.; Heiß, M.; Popis, M.C.; Blanco, S.; Bornelöv, S.; Sajini, A.A.; Gleeson, J.G.; Griffin, J.L.; et al. Cytosine-5 RNA methylation links protein synthesis to cell metabolism. PLoS Biol. 2019, 17, e3000297. [Google Scholar] [CrossRef] [PubMed]
- Tuorto, F.; Liebers, R.; Musch, T.; Schaefer, M.; Hofmann, S.; Kellner, S.; Frye, M.; Helm, M.; Stoecklin, G.; Lyko, F. RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat. Struct. Mol. Biol. 2012, 19. [Google Scholar] [CrossRef] [PubMed]
- Akiyama, Y.; Kharel, P.; Abe, T.; Anderson, P.; Ivanov, P. Isolation and initial structure-functional characterization of endogenous tRNA-derived stress-induced RNAs. RNA Biol. 2020, 17, 1116–1124. [Google Scholar] [CrossRef]
- Huber, S.; Leonardi, A.; Dedon, P.; Begley, T. The versatile roles of the tRNA epitranscriptome during cellular responses to toxic exposures and environmental stress. Toxics 2019, 7, 17. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.T.Y.; Dyavaiah, M.; DeMott, M.S.; Taghizadeh, K.; Dedon, P.C.; Begley, T.J. A Quantitative systems approach reveals dynamic control of tRNA modifications during cellular stress. PLoS Genet. 2010, 6, e1001247. [Google Scholar] [CrossRef]
- Chan, C.T.Y.; Pang, Y.L.J.; Deng, W.; Babu, I.R.; Dyavaiah, M.; Begley, T.J.; Dedon, P.C. Reprogramming of tRNA modifications controls the oxidative stress response by codon-biased translation of proteins. Nat. Commun. 2012, 3, 937. [Google Scholar] [CrossRef]
- Hicks, D.G.; Janarthanan, B.R.; Vardarajan, R.; Kulkarni, S.A.; Khoury, T.; Dim, D.; Budd, G.T.; Yoder, B.J.; Tubbs, R.; Schreeder, M.T.; et al. The expression of TRMT2A, a novel cell cycle regulated protein, identifies a subset of breast cancer patients with HER2 over-expression that are at an increased risk of recurrence. BMC Cancer 2010, 10, 108. [Google Scholar] [CrossRef]
- Jehn, J.; Treml, J.; Wulsch, S.; Ottum, B.; Erb, V.; Hewel, C.; Kooijmans, R.N.; Wester, L.; Fast, I.; Rosenkranz, D. 5’ tRNA halves are highly expressed in the primate hippocampus and might sequence-specifically regulate gene expression. RNA 2020, 26, 694–707. [Google Scholar] [CrossRef]
- Borland, K.; Diesend, J.; Ito-Kureha, T.; Heissmeyer, V.; Hammann, C.; Buck, A.H.; Michalakis, S.; Kellner, S. Production and application of stable isotope-labeled internal standards for RNA modification analysis. Genes 2019, 10, 26. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Carlson, M. hgfocus.db: Affymetrix Human Genome Focus Array annotation data (chip hgfocus), R package version 3.2.3; Bioconductor: Buffalo, NY, USA, 2016. [Google Scholar] [CrossRef]
- Merico, D.; Isserlin, R.; Stueker, O.; Emili, A.; Bader, G.D. Enrichment map: A network-based method for gene-set enrichment visualization and interpretation. PLoS ONE 2010, 5, e13984. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Kucera, M.; Isserlin, R.; Arkhangorodsky, A.; Bader, G.D. AutoAnnotate: A Cytoscape app for summarizing networks with semantic annotations. F1000Research 2016, 5, 1717. [Google Scholar] [CrossRef]
- Morris, J.H.; Apeltsin, L.; Newman, A.M.; Baumbach, J.; Wittkop, T.; Su, G.; Bader, G.D.; Ferrin, T.E. ClusterMaker: A multi-algorithm clustering plugin for Cytoscape. BMC Bioinform. 2011, 12. [Google Scholar] [CrossRef]
- Oesper, L.; Merico, D.; Isserlin, R.; Bader, G.D. WordCloud: A Cytoscape plugin to create a visual semantic summary of networks. Source Code Biol. Med. 2011, 6. [Google Scholar] [CrossRef]
| tRFs ID | tRFs Seq | tRFs Type | tRFdb ID | MINTbase ID | Length | Fold Change | p-Value |
|---|---|---|---|---|---|---|---|
| 5′tiRNA-SeC-TCA-001 | GCCCGGATGATCCTCAGTGGTCTGGGGTGCAGGCTT | 5′tiRNA | - | - | 36 | 11.3 | 0.0011 |
| 5′tiRNA-Gly-CCC-004 | GCATTGGTGGTTCAATGGTAGAATTCTCGCC | 5′tiRNA | - | - | 31 | 10.3 | 0.0001 |
| 3′tiRNA-His-GTG-002 | TGAATCTGACAACAGAGGCTTACGACCCCTTATTTACCA | 3′tiRNA | - | - | 39 | 8.1 | 0.0072 |
| tRF-His-GTG-033 | GGTGGTTCTAACTTGCTGGGGTGGCGGTTTTT | tRF-1 | - | - | 32 | 7.8 | 0.0005 |
| 5′tiRNA-iMet-CAT-003 | AGCAGAGTGGCGCAGCGGAAGCGTGCTGGGCC | 5′tiRNA | - | tRF-32-FP18LPMBQ4NKJ | 32 | 7.2 | 0.0016 |
| 5′tiRNA-Gly-GCC-010 | GCATGGGTGGTTCAGTGGTAGAATTCTCGCC | 5′tiRNA | 5003c | tRF-31-P4R8YP9LON4VD | 31 | 7.1 | 0.0000 |
| tRF-His-GTG-034 | GGTGGTTCTAACTTGCTGGGGTGGCGGTTTTTT | tRF-1 | - | - | 33 | 6.9 | 0.0013 |
| 5′tiRNA-Val-AAC-009 | GTTTCCGTAGTGTAGTGGTCATCACGTTCGCC | 5′tiRNA | - | tRF-32-79MP9P9MH57SJ | 32 | 6.9 | 0.0001 |
| 5′tiRNA-Gly-CCC-013 | GCATTGGTGGTTCAGTGGTAGAATTCTCGCC | 5′tiRNA | 5004c | tRF-31-PNR8YP9LON4VD | 31 | 6.7 | 0.0001 |
| 5′tiRNA-Gly-TCC-010 | GCGTTGGTGGTATAGTGGTGAGCATAGCTGCC | 5′tiRNA | - | tRF-32-QNR8VP94FQFYJ | 32 | 6.7 | 0.0002 |
| tRF-Leu-CAG-013 | GTCAGGATGGCCGAGCGGTCTAA | tRF-5b | 5023b | tRF-23-SP5830MM0F | 23 | 6.3 | 0.0026 |
| 5′tiRNA-iMet-CAT-001 | AGCAGAGTGGCGCAGCGGAAGCGTGCTGGGCCC | 5′tiRNA | - | tRF-33-FP18LPMBQ4NKDJ | 33 | 6.3 | 0.0032 |
| 5′tiRNA-Gly-GCC-004 | GCATAGGTGGTTCAGTGGTAGAATTCTTGCC | 5′tiRNA | - | - | 31 | 6.2 | 0.0017 |
| 5′tiRNA-iMet-CAT-002 | AGCAGAGTGGCGCAGCGGAAGCGTGCTGGGC | 5′tiRNA | - | tRF-31-FP18LPMBQ4NKD | 31 | 6.2 | 0.0019 |
| tRF-Gly-GCC-012 | TCGATTCCCGGCCAATGCACCA | tRF-3b | 3027b | tRF-22-WE8SPOX52 | 22 | 6.1 | 0.0002 |
| tRF-Asn-GTT-015 | GCCCACCCAGGGACGCCA | tRF-3a | - | tRF-18-P6KP6HD2 | 18 | 6.0 | 0.0002 |
| 5′tiRNA-Val-CAC-045 | GTTTCCGTAGTGTAGCGGTTATCACATTCGC | 5′tiRNA | - | tRF-31-79MP9PMNH5ISD | 31 | 5.8 | 0.0148 |
| tRF-Gly-CCC-012 | GCATTGGTGGTTCAGTGGTAGAATTCTCGC | tRF-5c | - | tRF-30-PNR8YP9LON4V | 30 | 5.7 | 0.0002 |
| 5′tiRNA-Val-TAC-018 | GTTTCCGTGGTGTAGTGGTTATCACATTCGCC | 5′tiRNA | - | - | 32 | 5.7 | 0.0048 |
| tRF-His-GTG-044 | GGTGGTTCTAACTTGCTGGGGTGGCGGTTTT | tRFs-1 | - | - | 31 | 5.6 | 0.0022 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pereira, M.; Ribeiro, D.R.; Pinheiro, M.M.; Ferreira, M.; Kellner, S.; Soares, A.R. m5U54 tRNA Hypomodification by Lack of TRMT2A Drives the Generation of tRNA-Derived Small RNAs. Int. J. Mol. Sci. 2021, 22, 2941. https://doi.org/10.3390/ijms22062941
Pereira M, Ribeiro DR, Pinheiro MM, Ferreira M, Kellner S, Soares AR. m5U54 tRNA Hypomodification by Lack of TRMT2A Drives the Generation of tRNA-Derived Small RNAs. International Journal of Molecular Sciences. 2021; 22(6):2941. https://doi.org/10.3390/ijms22062941
Chicago/Turabian StylePereira, Marisa, Diana R. Ribeiro, Miguel M. Pinheiro, Margarida Ferreira, Stefanie Kellner, and Ana R. Soares. 2021. "m5U54 tRNA Hypomodification by Lack of TRMT2A Drives the Generation of tRNA-Derived Small RNAs" International Journal of Molecular Sciences 22, no. 6: 2941. https://doi.org/10.3390/ijms22062941
APA StylePereira, M., Ribeiro, D. R., Pinheiro, M. M., Ferreira, M., Kellner, S., & Soares, A. R. (2021). m5U54 tRNA Hypomodification by Lack of TRMT2A Drives the Generation of tRNA-Derived Small RNAs. International Journal of Molecular Sciences, 22(6), 2941. https://doi.org/10.3390/ijms22062941






