Changes in the Flower and Leaf Proteome of Common Buckwheat (Fagopyrum esculentum Moench) under High Temperature
Abstract
1. Introduction
2. Results
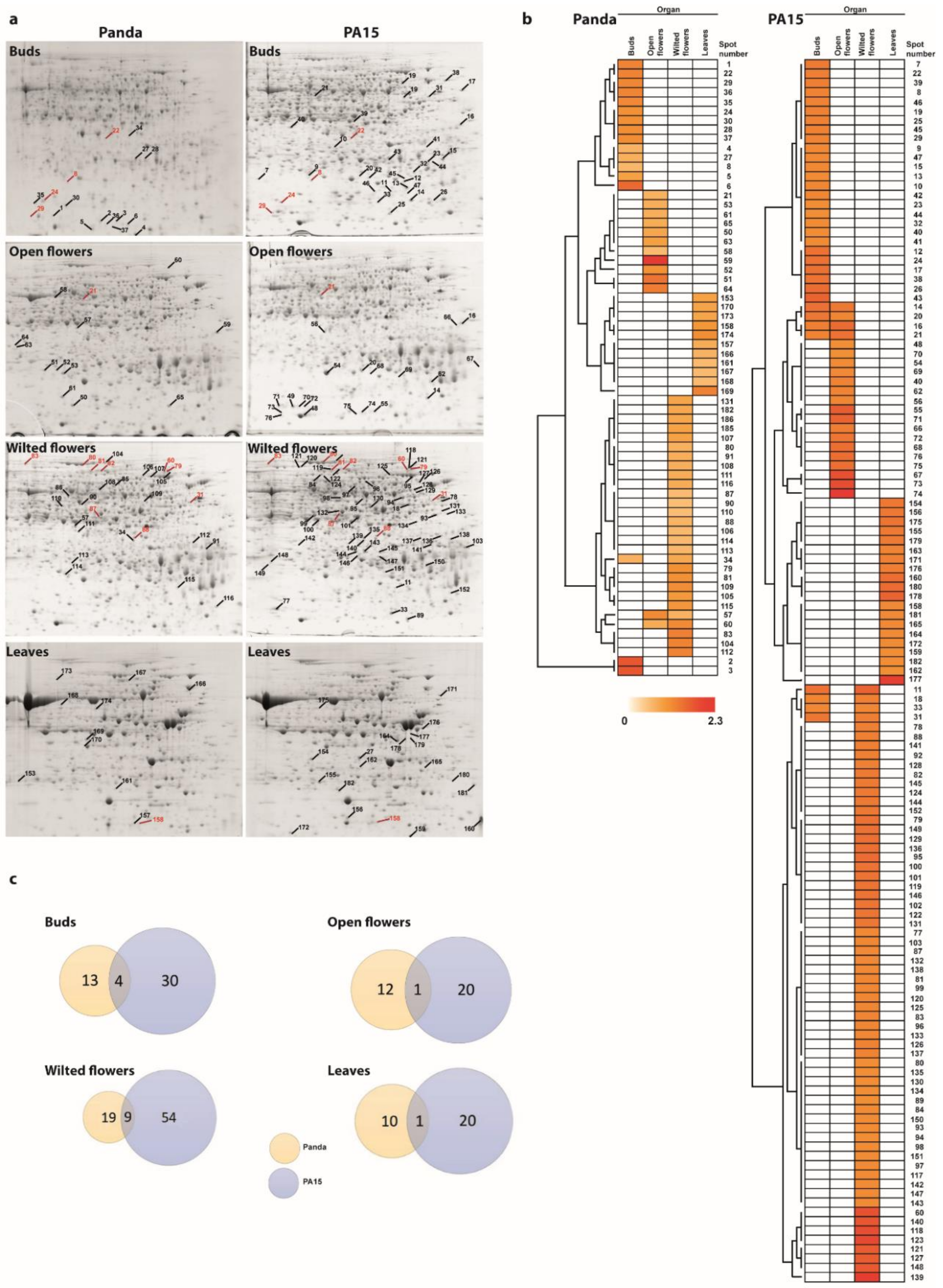
2.1. Protein Profiles in Flowers and Leaves
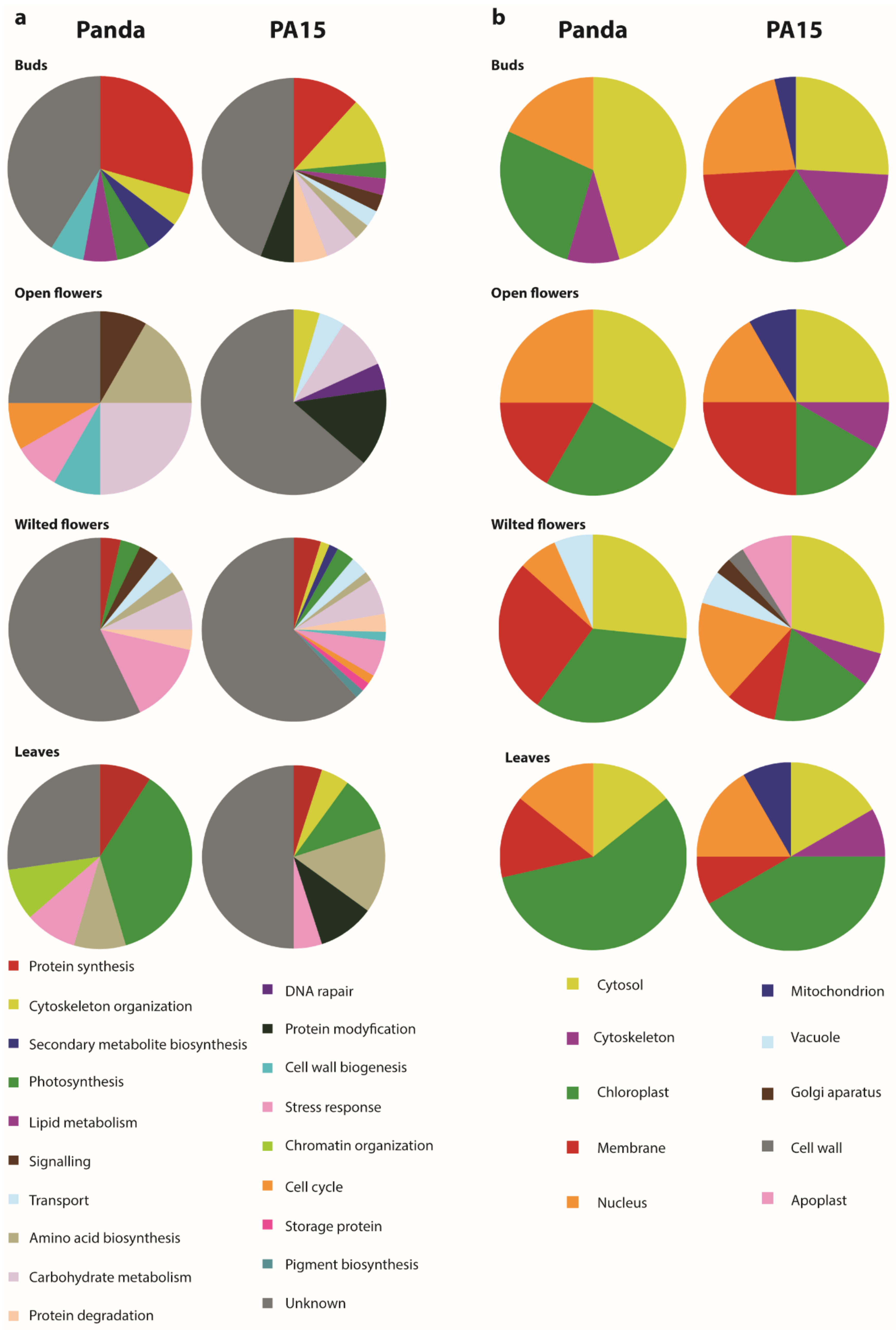
2.2. Identification of Differentially Accumulated Proteins
3. Discussion
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Protein Extraction
4.3. Two-Dimensional Gel Electrophoresis
4.4. Gel Image Analysis
4.5. Protein Identification
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 2-DE | Two-dimensional gel electrophoresis |
| 6PGDH | 6-phosphogluconate dehydrogenase |
| ABA | Abscisic acid |
| ACP | Acyl-[acyl-carrier-protein] |
| CHAPS | 3-[(3-cholamidopropyl)dimethyl-ammonio]-1-propane sulfonate |
| DREB2A | Dehydration-responsive element binding protein 2A |
| DTT | Dithiothreitol |
| FBA | Fructose-bisphosphate aldolase |
| FBP | Fructose-1,6-bisphosphate |
| GLR | Glutamate receptor-like channel |
| HSF | Heat-shock transcription factor |
| HsfA1 | Heat-shock transcription factor A1 |
| HSP | Heat-shock proteins |
| IEF | Isoelectricfocusing |
| IGP | Indole-3-glycerol phosphate |
| IGPS | Indole-3-glycerol phosphate synthase |
| JA | Jasmonic acid |
| JA-Met | Methyl ester of jasmonic acid |
| OPPP | Oxidative pentose phosphate pathway |
| PGK | Phosphoglycerate kinase |
| PMF | Peptide mass fingerprinting |
| ROS | Reactive oxygen species |
| SA | Salicylic acid |
| SAM | S-adenosylmethionine |
| SAMS | S-adenosylmethionine synthetase |
| SDS | Sodium dodecyl sulfate |
| SFBA | Sedoheptulose/fructose-bisphosphate aldolase |
| SuBP | Sedoheptulose-1,7-bisphosphate |
| TF | Transcription factor |
| UFA | Unsaturated fatty acids |
| V-ATPase | V-type proton ATPase |
References
- Christa, K.; Soral-Śmietana, M. Buckwheat grains and buckwheat products—Nutritional and prophylactic value of their components—A review. Czech J. Food Sci. 2008, 26, 153–162. [Google Scholar] [CrossRef]
- Halbrecq, B.; Romedenne, P.; Ledent, J.F. Evolution of flowering, ripening and seed set in buckwheat (Fagopyrum esculentum Moench): Quantitative analysis. Eur. J. Agron. 2005, 23, 209–224. [Google Scholar] [CrossRef]
- Farooq, S.; Rehman, R.U.; Pirzadah, T.B.; Malik, B.; Dar, F.A.; Tahir, I. Cultivation, agronomic practices, and growth performance of buckwheat. In Molecular Breeding and Nutritional Aspects of Buckwheat; Zhou, M., Kreft, I., Woo, S.H., Chrungoo, N., Wieslander, G., Eds.; Academic Press: Oxford, UK, 2016; pp. 299–320. [Google Scholar] [CrossRef]
- Cawoy, V.; Ledent, J.-F.; Kinet, J.-M.; Jacquemart, A.-L. Floral biology of common buckwheat (Fagopyrum esculentum Moench). Eur. J. Plant Sci. Biotechnol. 2009, 3, 1–9. [Google Scholar]
- Adachi, T. How to combine the reproductive system with biotechnology in order to overcome the breeding barrier in buckwheat. Fagopyrum 1990, 10, 1. [Google Scholar]
- Slawinska, J.; Obendorf, R.L. Buckwheat seed set in planta and during in vitro inflorescence culture: Evaluation of temperature and water deficit stress. Seed Sci. Res. 2001, 11, 223–233. [Google Scholar] [CrossRef]
- Słomka, A.; Michno, K.; Dubert, F.; Dziurka, M.; Kopeć, P.; Plażek, A. Embryological background of low seed set in distylous common buckwheat (Fagopyrum esculentum Moench) with biased morph ratios, and biostimulant-induced improvement of it. Crop Pasture Sci. 2017, 68, 680–690. [Google Scholar] [CrossRef]
- Hornyák, M.; Płazek, A.; Kopeć, P.; Dziurka, M.; Pastuszak, J.; Szczerba, A.; Hura, T. Photosynthetic activity of common buckwheat (Fagopyrum esculentum Moench) exposed to thermal stress. Photosynthetica 2020, 58, 45–53. [Google Scholar] [CrossRef]
- Płażek, A.; Słomka, A.; Kopeć, P.; Dziurka, M.; Hornyák, M.; Sychta, K.; Pastuszak, J.; Dubert, F. Effects of high temperature on embryological development and hormone profile in flowers and leaves of common buckwheat (Fagopyrum esculentum Moench). Int. J. Mol. Sci. 2019, 20, 5. [Google Scholar] [CrossRef]
- Timperio, A.M.; Egidi, M.G.; Zolla, L. Proteomics applied on plant abiotic stresses: Role of heat shock proteins (HSP). J. Proteom. 2008, 71, 391–411. [Google Scholar] [CrossRef]
- Kosová, K.; Vítámvás, P.; Prášil, I.T.; Renaut, J. Plant proteome changes under abiotic stress: Contribution of proteomics studies to understanding plant stress response. J. Proteom. 2011, 74, 1301–1322. [Google Scholar] [CrossRef]
- Lee, D.-G.; Ahsan, N.; Lee, S.-H.; Kang, K.Y.; Bahk, J.D.; Lee, I.-J.; Lee, B.-H. A proteomic approach in analyzing heat-responsive proteins in rice leaves. Proteomics 2007, 7, 3369–3383. [Google Scholar] [CrossRef]
- Mittler, R.; Zilinskas, B.A. Molecular cloning and characterization of a gene encoding pea cytosolic ascorbate peroxidase. J. Biol. Chem. 1992, 267, 21802–21807. [Google Scholar] [CrossRef]
- Miernyk, J.A. The 70 kDa stress-related proteins as molecular chaperones. Trends Plant Sci. 1997, 2, 180–187. [Google Scholar] [CrossRef]
- Renaut, J.; Hausman, J.F.; Wisniewski, M.E. Proteomics and low-temperature studies: Bridging the gap between gene expression and metabolism. Physiol. Plant 2006, 126, 97–109. [Google Scholar] [CrossRef]
- Miernyk, J.A.; Pret’ová, A.; Olmedilla, A.; Klubicová, K.; Obert, B.; Hajduch, M. Using proteomics to study sexual reproduction in angiosperms. Sex. Plant Reprod. 2011, 24, 9–22. [Google Scholar] [CrossRef] [PubMed]
- Feng, B.; Li, L.; Zhou, X.; Stanley, B.; Ma, H. Analysis of the Arabidopsis floral proteome: Detection of over 2000 proteins and evidence for postrranslational modifications. J. Integr. Plant Biol. 2009, 51, 207–223. [Google Scholar] [CrossRef] [PubMed]
- Kerim, T.; Imin, N.; Weinman, J.J.; Rolfe, B.G. Proteome analysis of male gametophyte development in rice anthers. Proteomics 2003, 3, 738–751. [Google Scholar] [CrossRef]
- Uchiumi, T.; Shinkawa, T.; Isobe, T.; Okamoto, T. Identification of the major protein components of rice egg cells. Int. J. Plant Res. 2007, 120, 575–579. [Google Scholar] [CrossRef]
- Liu, H.; Liu, Y.-Z.; Zheng, S.-Q.; Jiang, J.-m.; Wang, P.; Chen, W. Comparative proteomic analysis of longan (Dimocarpus longan Lour.) seed abortion. Planta 2010, 231, 847–860. [Google Scholar] [CrossRef]
- Das, A.; Eldakak, M.; Paudel, B.; Kim, D.-W.; Hemmati, H.; Basu, C.; Rohila, J.S. Leaf proteome analysis reveals prospective drought and heat stress response mechanisms in soybean. Biomed. Res. Int. 2016, 2016. [Google Scholar] [CrossRef]
- Rodziewicz, P.; Chmielewska, K.; Sawikowska, A.; Marczak, Ł.; Łuczak, M.; Bednarek, P.; Mikołajczak, K.; Ogrodowicz, P.; Kuczyńska, A.; Krajewski, P.; et al. Identification of drought responsive proteins and related proteomic QTLs in barley. J. Exp. Bot. 2019, 70, 2823–2837. [Google Scholar] [CrossRef]
- Singh, P.; Song, Q.-Q.; Singh, R.K.; Li, H.-B.; Solanki, M.K.; Malviya, M.K.; Verma, K.K.; Yang, L.-T.; Li, Y.-R. Proteomic analysis of the resistance mechanism in sugercane during Sporisorium scitamineum infection. Int. J. Mol. Sci. 2019, 20, 569. [Google Scholar] [CrossRef] [PubMed]
- Awana, M.; Jain, N.; Samota, M.K.; Rani, K.; Kumar, A.; Ray, M.; Gaikwad, K.; Praveen, S.; Singh, N.K.; Singh, A. Protein and gene integration analysis through proteome and transcriptome brings new insight into salt stress tolerance in pigeonpea (Cajanus cajan L.). Int. J. Biol. Macromol. 2020, 164, 3589–3602. [Google Scholar] [CrossRef]
- Li, H.; Li, Y.; Ke, Q.; Kwak, S.-S.; Zhang, S.; Deng, X. Physiological and differential proteomic analysis of imitation drought stress response in Sorghum bicolor root at the seedling stage. Int. J. Mol. Sci. 2020, 21, 9174. [Google Scholar] [CrossRef] [PubMed]
- Ohama, N.; Sato, H.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Transcriptional regulatory network of plant heat stress response. Trends Plant Sci. 2017, 22, 53–65. [Google Scholar] [CrossRef]
- Janni, M.; Gulli, M.; Maestri, E.; Marmiroli, M.; Valliyodan, B.; Nguyen, H.T.; Marmiroli, N. Molecular and genetic bases of heat stress responses in crop plants and breeding for increased resilience and productivity. J. Exp. Bot. 2020, 71, 3780–3802. [Google Scholar] [CrossRef]
- Płażek, A.; Hura, K.; Hura, T.; Słomka, A.; Hornyák, M.; Sychta, K. Synthesis of heat-shock proteins HSP-70 and HSP-90 in flowers of common buckwheat (Fagopyrum esculentum) under thermal stress. Crop Pasture Sci. 2020, 70, 760–767. [Google Scholar] [CrossRef]
- Vadivel, A.-K.-A. Gel-based proteomics in plants: Time to move on from the tradition. Front. Plant Sci. 2015, 6, 369. [Google Scholar] [CrossRef]
- Thiede, B.; Höhenwarter, W.; Krah, A.; Mattow, J.; Schmid, M.; Schmidt, F.; Jungblut, P.R. Peptide mass fingerprinting. Methods 2005, 35, 237–247. [Google Scholar] [CrossRef] [PubMed]
- Bocian, A.; Kosmala, A.; Rapacz, M.; Jurczyk, B.; Marczak, L.; Zwierzykowski, Z. Differences in leaf proteome response to cold acclimation between Lolium perenne plants with distinct levels of frost tolerance. J. Plant Physiol. 2011, 168, 1271–1279. [Google Scholar] [CrossRef]
- Yasui, Y.; Hirakawa, H.; Ueno, M.; Matsui, K.; Katsube-Tanaka, T.; Yang, S.J.; Aii, J.; Sato, S.; Mori, M. Assembly pf the draft genome of buckwheat and its applications in identifying agronomically useful genes. DNA Res. 2016, 23, 215–224. [Google Scholar] [CrossRef]
- Hernandez, M.L.; Sicardo, M.D.; Alfonso, M.; Martinez-Rivas, J.M. Transcriptional regulation of stearoyl-acyl carrier protein desaturase genes in response abiotic stresses leads to changes in the unsaturated fatty acids composition of olive mesocarp. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef]
- Kachroo, A.; Shanklin, J.; Whittle, E.; Lapchyk, L.; Hildebrand, D.; Kachroo, P. The Arabidopsis stearoyl-acyl carrier protein-desaturase family and the contribution of leaf isoforms to oleic acid synthesis. Plant Mol. Biol. 2007, 63, 257–271. [Google Scholar] [CrossRef]
- Magwanga, R.O.; Kirungu, J.N.; Lu, P.; Yang, X.; Dong, Q.; Cai, X.; Xu, Y.; Wang, X.; Zhou, Z.; Hou, Y.; et al. Genome wide identification of the trihelix transcription factors and overexpression of Gh_A05G2067 (GT-2), a novel gene contributing to increased drought and salt stresses tolerance in cotton. Physiol. Plant 2019, 167, 447–464. [Google Scholar] [CrossRef]
- Hou, F.-Y.; Huang, J.; Yu, S.-L.; Zhang, H.-S. The 6-phosphogluconate dehydrogenase genes are responsive to abiotic stresses in rice. J. Integr. Plant Biol. 2007, 49, 655–663. [Google Scholar] [CrossRef]
- Can’ani, A.; Seifan, M.; Tzin, V. Indole is an essential molecule for plant interactions with herbivores and pollinators. J. Plant Biol. Crop Res. 2018, 1, 1003. [Google Scholar] [CrossRef]
- Ouyang, J.; Shao, X.; Li, J. Indole-3-glycerol phosphate, a branchpoint of indole-3-acetic acid biosynthesis from tryptohan biosynthetic pathway in Arabidopsis thaliana. Plant J. 2000, 24, 327–333. [Google Scholar] [CrossRef] [PubMed]
- Sakata, T.; Oshino, T.; Miura, S.; Tomabechi, M.; Tsunaga, Y.; Higashitani, N.; Miyazawa, Y.; Takahashi, H.; Watanabe, M.; Higashitani, A. Auxins reverse plant male sterility caused by high temperatures. Proc. Natl. Acad. Sci. USA 2010, 107, 8569–8574. [Google Scholar] [CrossRef]
- Cao, X.; Yang, H.; Shang, C.; Ma, S.; Liu, L.; Cheng, J. The roles of auxin biosynthesis YUCCA gene family in plants. Int. J. Mol. Sci. 2019, 20, 6343. [Google Scholar] [CrossRef] [PubMed]
- Nahar, K.; Hasanuzzaman, M.; Ahamed, K.U.; Hakeem, K.R.; Ozturk, M.; Fujita, M. Plant responses and tolerance to high temperature stress: Role of exogenous phytoprotectants. In Crop Production and Global Environmental Issues; Hakeem, K., Ed.; Springer: Cham, Switzerland, 2015; pp. 385–435. [Google Scholar] [CrossRef]
- Landi, S.; Capasso, G.; Ben Azaiez, F.E.; Jallouli, S.; Ayadi, S.; Trifa, Y.; Esposito, S. Different roles of heat shock proteins (70 kDa) during abiotic stresses in barley (Hordeum vulgare) genotypes. Plants 2019, 8, 248. [Google Scholar] [CrossRef] [PubMed]
- Sjögren, L.L.; MacDonald, T.M.; Sutinen, S.; Clarke, A.K. Inactivation of the clpC1 gene encoding a chloroplast Hsp100 molecular chaperone causes growth retardation, leaf chlorosis, lower photosynthetic activity, and a specific reduction in photosystem content. Plant Physiol. 2004, 136, 4114–4126. [Google Scholar] [CrossRef]
- Ratajczak, R. Structure, function and regulation of the plant vacuolar H+-translocating ATPase. Biochim. Biophys. Acta Biomembr. 2000, 1465, 17–36. [Google Scholar] [CrossRef]
- Zhao, S.; Liang, Z.; Demko, V.; Wilson, R.; Johansen, W.; Olsen, O.-A.; Shalchian-Tabrizi, K. Massive expansion of the calpain gene family in unicellular eukaryotes. BMC Evol. Biol. 2012, 12. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.-H.; Lee, C.-E.; Lin, Y.-S.; Lee, M.-H.; Chen, P.-Y.; Chang, H.-C.; Chang, I.-F. The glutamate receptor-like protein GLR3. 7 interacts with 14-3-3ω and participates in salt stress response in Arabidopsis thaliana. Front. Plant Sci. 2019, 10, 1169. [Google Scholar] [CrossRef] [PubMed]
- Rosa-Tellez, S.; Djoro Anoman, A.; Flores-Tornero, M.; Toujani, W.; Alseek, S.; Fernie, A.R.; Nebauer, S.G.; Munoz-Bertomeu, J.; Segura, J.; Ros, R. Phosphoglycerate kinases are co-regulated to adjust metabolism and to optimize growth. Plant Physiol. 2018, 176, 1182–1198. [Google Scholar] [CrossRef] [PubMed]
- Lv, G.-Y.; Guo, X.-G.; Xie, L.-P.; Xie, C.-G.; Zhang, X.-H.; Yang, Y.; Xiao, L.; Tang, Y.-Y.; Pan, X.-L.; Guo, A.-G.; et al. Molecular characterization, gene evolution, and expression analysis of the fructose-1, 6-bisphosphate aldolase (FBA) gene family in wheat (Triticum aestivum L.). Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Heidari, P.; Mazloomi, F.; Nussbaumer, T.; Barcaccia, G. Insights into the SAM synthetase gene family and its roles in tomato seedlings under abiotic stresses and hormone treatments. Plants 2020, 9, 586. [Google Scholar] [CrossRef]
- Hurkman, W.J.; Tanaka, C.K. Solubilization of plant membrane proteins for analysis by two-dimensional gel electrophoresis. Plant Physiol. 1986, 81, 802–806. [Google Scholar] [CrossRef]
- Hajduch, M.; Ganapathy, A.; Stein, J.W.; Thelen, J.J. A systematic proteomic study of seed filling in soybean. Establishment of high-resolution two-dimensional reference maps, expression profiles, and an interactive proteome database. Plant Physiol. 2005, 137, 1397–1419. [Google Scholar] [CrossRef]
- Neuhoff, V.; Arold, N.; Taube, D.; Ehrhardt, W. Improved staining of proteins in polyacrylamide gels including isoelectric focusing gels with clear background at nanogram sensitivity using Coomassie Brilliant Blue G-250 and R-250. Electrophoresis 1988, 9, 255–262. [Google Scholar] [CrossRef]
- Hartman, K.; Mielczarek, P.; Silberring, J. Synthesis of the novel covalent cysteine proteases inhibitor with iodoacetic functional group. Molecules 2020, 25, 813. [Google Scholar] [CrossRef] [PubMed]
- Drabik, A.; Ner-Kluza, J.; Mielczarek, P.; Civit, L.; Mayer, G.; Silberring, J. Advances in the study of aptamer-protein target identification using the chromatographic approach. J. Proteome Res. 2018, 17, 2174–2181. [Google Scholar] [CrossRef] [PubMed]
| Spot No. a | Organ b | Technique c | UniProt No. d | Protein Name e | Reference Organism f | Mt [kDa] g | pIt h | Protein Score i | Peptide Count j | Coverage (%) k |
|---|---|---|---|---|---|---|---|---|---|---|
| 2 | B | LC-MS/MS | RS122_ARATH | 40S ribosomal protein S12-2 | Arabidopsis thaliana | 15.3 | 5.55 | 169.24 | 2 | 12.5 |
| 4 | B | PMF | RL26_BRACM | 60S ribosomal protein L26 | Brassica campestris | 16.9 | 11.60 | 135.90 | 4 | 8.9 |
| 5 | B | PMF | EIF3A_MAIZE | Eukaryotic translation initiation factor 3 subunit A | Zea mays | 111.5 | 9.80 | 134.80 | 4 | 5.8 |
| 6 | B | PMF | CLDS_TOBAC | Copal-8-ol diphosphatehydratase, chloroplastic | Nicotiana tabacum | 93.2 | 5.50 | 128.00 | 2 | 3.6 |
| 7 | B | LC-MS/MS | PDRP2_ARATH | Pyruvate, phosphatedikinase regulatory protein 2 | Arabidopsis thaliana | 41.4 | 9.64 | 97.92 | 1 | 2.1 |
| 8 | B | PMF | STAD6_ORYSI | Acyl-[acyl-carrier-protein] desaturase 6, chloroplastic | Oryza sativa subsp. indica | 46.5 | 7.2 | 136.80 | 2 | 9.0 |
| 9 | B | LC-MS/MS | PS4_PINST | Putative LRR diseaseresistance protein/transmembrane receptor kinase PS4 (fragment) | Pinus strobus | 0.90 | 11.10 | 95.06 | 1 | 100.0 |
| 10 | B | LC-MS/MS | AB5F_ARATH | ABC transporter F family member 5 | Arabidopsis thaliana | 78 | 6.49 | 94.56 | 1 | 1.7 |
| 13 | B | PMF | RHM2_ARATH | Trifunctional UDP-glucose 4,6-dehydratase/UDP-4-keto-6-deoxy-D-glucose 3,5-epimerase/UDP-4-keto-L-rhamnose reductase RHM2 | Arabidopsis thaliana | 75.2 | 6.00 | 116.70 | 1 | 2.4 |
| 15 | B | LC-MS/MS | PSA5_ORYSJ | Proteasome subunit alpha type-5 | Oryza sativa subsp. japonica | 26 | 4.60 | 557.20 | 11 | 37.1 |
| 16 | B; OF | PMF | UPL1_ARATH | E3 ubiquitin-protein ligase UPL1 | Arabidopsis thaliana | 404.7 | 4.80 | 126.20 | 3 | 1.3 |
| 17 | B | LC-MS/MS | PSMD4_ARATH | 26S proteasome non-ATPase regulatory subunit 4 homolog | Arabidopsis thaliana | 40.7 | 4.30 | 183.00 | 2 | 4.4 |
| 18 | B; WF | PMF | Y1765_ARATH | Probable LRR receptor-likeserine/threonine-protein kinase At1g07650 | Arabidopsis thaliana | 112.8 | 9.50 | 118.90 | 3 | 4.0 |
| 20 | B; OF | PMF | KN12D_ARATH | Kinesin-like protein KIN-12D | Arabidopsis thaliana | 314.9 | 5.10 | 118.00 | 3 | 1.3 |
| 21 | B; OF | LC-MS/MS | 6PGD1_SPIOL | 6-phosphogluconate dehydrogenase, decarboxylating 1 | Spinacia oleracea | 53.2 | 6.00 | 786.00 | 13 | 20.1 |
| 22 | B | PMF | RL51_ARATH | 60S ribosomal protein L5-1 | Arabidopsis thaliana | 34.3 | 9.70 | 130.40 | 3 | 11.6 |
| 23 | B | PMF | QWRF4_ARATH | QWRF motif-containing protein 4 | Arabidopsis thaliana | 66.9 | 10.20 | 123.00 | 3 | 4.2 |
| 24 | B | PMF | GTL2_ARATH | Trihelix transcription factor GTL2 | Arabidopsis thaliana | 71.2 | 6.70 | 113.50 | 2 | 2.7 |
| 25 | B | PMF | KN12F_ORYSJ | Kinesin-like protein KIN-12F | Oryza sativa subsp. japonica | 317.1 | 5.00 | 137.70 | 5 | 2.0 |
| 26 | B | PMF | MYB98_ARATH | Transcription factor MYB98 | Arabidopsis thaliana | 50.1 | 6.10 | 129.70 | 4 | 6.8 |
| 27 | B | LC-MS/MS | TPIC_ARATH | Triosephosphate isomerase, chloroplastic | Arabidopsis thaliana | 33.3 | 8.90 | 355.70 | 5 | 16.5 |
| 28 | B | LC-MS/MS | IPYR2_ARATH | Soluble inorganic pyrophosphatase 2 | Arabidopsis thaliana | 24.7 | 5.70 | 125.80 | 1 | 5.5 |
| 33 | B; WF | LC-MS/MS | ADF2_ORYSJ | Actin-depolymerizing factor 2 | Oryza sativa subsp. japonica | 16.8 | 5.60 | 102.50 | 1 | 8.3 |
| 48 | OF | LC-MS/MS | AB2C_ARATH | ABC transporter C family member 2 | Arabidopsis thaliana | 182 | 6.00 | 103.88 | 0 | 0.0 |
| 49 | OF | PMF | HXK4_ORYSJ | Hexokinase-4, chloroplastic | Oryza sativa subsp. japonica | 54.7 | 6.50 | 122.80 | 3 | 6.5 |
| 51 | OF | PMF | SWTIE_ARATH | Protein SWEETIE | Arabidopsis thaliana | 244.2 | 5.10 | 123.00 | 3 | 1.8 |
| 52 | OF | PMF | CALS2_ARATH | Callosesynthase 2 | Arabidopsis thaliana | 225.9 | 9.20 | 132.50 | 4 | 1.9 |
| 53 | OF | LC-MS/MS | PS4_PINST | Putative LRR diseaseresistance protein/transmembrane receptor kinase PS4 (fragment) | Pinus strobus | 0.90 | 11.10 | 90.53 | 1 | 100.0 |
| 54 | OF | LC-MS/MS | ZDHC8_ARATH | Probable protein S-acyltransferase 20 | Arabidopsis thaliana | 76.8 | 9.60 | 123.05 | 2 | 2.4 |
| 57 | OF; WF | PMF | MDHC2_ARATH | Malate dehydrogenase 2, cytoplasmic | Arabidopsis thaliana | 35.7 | 6.30 | 466.10 | 8 | 26.8 |
| 58 | OF | PMF | UGDH2_ARATH | UDP-glucose 6-dehydrogenase 2 | Arabidopsis thaliana | 53.1 | 5.60 | 162.80 | 3 | 7.7 |
| 60 | OF; WF | LC-MS/MS | HSP70_MAIZE | Heatshock 70 kDa protein | Zea mays | 70.5 | 5.10 | 470.40 | 8 | 14.7 |
| 61 | OF | PMF | AUG8_ARATH | AUGMIN subunit 8 | Arabidopsis thaliana | 69.8 | 10.70 | 117.50 | 4 | 5.7 |
| 77 | WF | PMF | RLT2_ARATH | Homeobox-DDT domain protein RLT2 | Arabidopsis thaliana | 190.3 | 5.30 | 116.30 | 4 | 2.2 |
| 78 | WF | PMF | RCA_MALDO | Ribulose bisphosphate carboxylase/oxygenase activase, chloroplastic | Malus domestica | 48 | 8.20 | 277.70 | 5 | 13.5 |
| 79 | WF | PMF | HSP7N_ARATH | Heatshock 70 kDa protein 18 | Arabidopsis thaliana | 68.3 | 5.20 | 539.30 | 7 | 16.4 |
| 80 | WF | PMF | CLPC_PEA | Chaperone protein ClpC, chloroplastic | Pisum sativum | 102.6 | 6.50 | 853.70 | 16 | 16.3 |
| 81 | WF | PMF | VATA_BRANA | V-type proton ATPase catalytic subunit A | Brassica napus | 68.7 | 5.10 | 356.60 | 6 | 9.0 |
| 82 | WF | PMF | DEK1_ARATH | Calpain-typecysteineprotease DEK1 | Arabidopsis thaliana | 238.1 | 6.10 | 131.20 | 4 | 2.4 |
| 83 | WF | PMF | GLR34_ARATH | Glutamate receptor 3.4 | Arabidopsis thaliana | 107.1 | 9.10 | 120.00 | 4 | 5.0 |
| 84 | WF | PMF | C76AD_BETVU | Cytochrome P450 76AD1 | Beta vulgaris | 56.2 | 8.10 | 133.30 | 3 | 4.6 |
| 85 | WF | PMF | ILV5_ARATH | Ketol-acid reductoisomerase, chloroplastic | Arabidopsis thaliana | 63.8 | 6.40 | 181.00 | 2 | 4.9 |
| 86 | WF | PMF | Y5537_ARATH | G-typelectin S-receptor-likeserine/threonine-protein kinase At5g35370 | Arabidopsis thaliana | 96 | 6.60 | 127.20 | 3 | 4.5 |
| 87 | WF | LC-MS/MS | PGKY_TOBAC | Phosphoglycerate kinase, cytosolic | Nicotiana tabacum | 42.3 | 5.60 | 557.90 | 10 | 28.9 |
| 88 | WF | PMF | ALFP2_ARATH | Fructose-bisphosphatealdolase 2, chloroplastic | Arabidopsis thaliana | 43 | 6.80 | 451.50 | 8 | 15.8 |
| 89 | WF | PMF | SMC3_ARATH | Structural maintenance of chromosomes protein 3 | Arabidopsis thaliana | 139.3 | 6.10 | 124.50 | 4 | 2.9 |
| 91 | WF | LC-MS/MS | 1433_HELAN | 14-3-3-like protein | Helianthus annuus | 28.9 | 4.50 | 356.30 | 6 | 19.7 |
| 92 | WF | LC-MS/MS | HSP7C_PETHY | Heatshock cognate 70 kDa protein | Petunia hybrida | 71.2 | 5.00 | 1145.10 | 20 | 30.7 |
| 93 | WF | LC-MS/MS | SPD1_DATST | Spermidine synthase 1 | Datura stramonium | 34 | 5.10 | 246.70 | 3 | 8.8 |
| 95 | WF | LC-MS/MS | GDI_ARATH | Guanosine nucleotide diphosphate dissociation inhibitor | Arabidopsis thaliana | 49.5 | 5.00 | 266.90 | 5 | 10.1 |
| 96 | WF | LC-MS/MS | PMG2_ARATH | Probable 2,3-bisphosphoglycerate-independent phosphoglyceratemutase 2 | Arabidopsis thaliana | 60.7 | 5.50 | 267.00 | 3 | 6.8 |
| 97 | WF | LC-MS/MS | PMGI_MESCR | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase | Mesembryanthemum crystallinum | 61.1 | 5.30 | 340.60 | 6 | 12.7 |
| 98 | WF | LC-MS/MS | RH15_ARATH | DEAD-box ATP-dependent RNA helicase 15 | Arabidopsis thaliana | 48.3 | 5.30 | 369.40 | 6 | 12.6 |
| 99 | WF | LC-MS/MS | ALF_ORYSJ | Fructose-bisphosphate aldolase cytoplasmic isozyme | Oryza sativa subsp. japonica | 38.8 | 7.70 | 254.80 | 4 | 7.8 |
| 100 | WF | LC-MS/MS | GLYG1_SOYBN | Glycinin G1 | Glycine max | 55.7 | 5.80 | 394.60 | 8 | 21.0 |
| 101 | WF | LC-MS/MS | UPTG_MAIZE | Alpha-1,4-glucan-protein synthase [UDP-forming] | Zea mays | 41.2 | 5.70 | 423.00 | 9 | 27.7 |
| 102 | WF | LC-MS/MS | PSA1_ORYSJ | Proteasome subunit alpha type-1 | Oryza sativa subsp. japonica | 29.6 | 5.60 | 151.70 | 2 | 10.0 |
| 153 | L | LC-MS/MS | CAHC_TOBAC | Carbonic anhydrase, chloroplastic | Nicotiana tabacum | 34.5 | 6.46 | 139.33 | 1 | 3.1 |
| 154 | L | LC-MS/MS | CAHC_TOBAC | Carbonic anhydrase, chloroplastic | Nicotiana tabacum | 34.5 | 6.46 | 110.55 | 1 | 5.6 |
| 158 | L | PMF | METK_CAMSI | S-adenosylmethionine synthase | Camellia sinensis | 42.8 | 5.20 | 150.70 | 3 | 6.9 |
| 160 | L | PMF | PP207_ARATH | Pentatricopeptide repeat-containing protein At3g02330, mitochondrial | Arabidopsis thaliana | 101.6 | 6.00 | 128.60 | 3 | 4.9 |
| 161 | L | PMF | CHR4_ARATH | Protein CHROMATIN REMODELING 4 | Arabidopsis thaliana | 247.8 | 5.90 | 128.70 | 4 | 3.2 |
| 162 | L | LC-MS/MS | CAHC_PEA | Carbonic anhydrase, chloroplastic | Pisum sativum | 35.4 | 7.74 | 263.63 | 1 | 5.5 |
| 163 | L | LC-MS/MS | CDSP_ARATH | Thioredoxin-like protein CDSP32, chloroplastic | Arabidopsis thaliana | 33.7 | 9.40 | 214.50 | 3 | 7.9 |
| 164 | L | LC-MS/MS | CYSKP_SPIOL | Cysteine synthase, chloroplastic/chromoplastic | Spinacia oleracea | 40.6 | 7.60 | 211.70 | 4 | 12.5 |
| 165 | L | PMF | CRK20_ARATH | Putativecysteine-rich receptor-like protein kinase 20 | Arabidopsis thaliana | 74 | 6.60 | 118.50 | 3 | 7.1 |
| 166 | L | PMF | RUB2_BRANA | RuBisCO large subunit-binding protein subunit alpha, chloroplastic | Brassica napus | 61.6 | 5.00 | 569.20 | 9 | 15.8 |
| 167 | L | PMF | TKTC_SPIOL | Transketolase, chloroplastic | Spinacia oleracea | 80.2 | 6.20 | 262.90 | 5 | 6.5 |
| 168 | L | PMF | HUAL1_ARATH | Protein HUA2-LIKE 1 | Arabidopsis thaliana | 156.5 | 8.90 | 113.20 | 3 | 2.4 |
| 169 | L | PMF | GRDP1_ARATH | Glycine-richdomain-containing protein 1 | Arabidopsis thaliana | 89.4 | 6.60 | 129.60 | 5 | 6.7 |
| 170 | L | PMF | FENR1_ORYSI | Ferredoxin--NADP reductase, leaf isozyme 1, chloroplastic | Oryza sativa subsp. indica | 40 | 8.70 | 192.90 | 3 | 7.7 |
| 171 | L | PMF | KN12F_ORYSJ | Kinesin-like protein KIN-12F | Oryza sativa subsp. japonica | 317.1 | 5.00 | 123.70 | 4 | 1.9 |
| 172 | L | PMF | GLTB2_ARATH | Ferredoxin-dependent glutamatesynthase 2, chloroplastic | Arabidopsis thaliana | 177.6 | 6.60 | 128.00 | 4 | 2.8 |
| Spot No. a | Organ b | UniProt No. | Protein Name c | Reference Organism d | Mt e [kDa] | pIt f | Protein Score g | Peptide Count h | Coverage [%] i | Homologous Protein Name j | NCBI No. | Reference Organism f | I k | P l |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 3 | B | CAPZA_KLULA | F-actin-capping protein subunit alpha | Kluyveromyces lactis | 29.9 | 4.54 | 99.31 | 1 | 4.2 | F-actin-capping protein subunit alpha-like | XP_023907009.1 | Quercus suber | 33% | 53% |
| 11 | B; WF | MTNC_GLUDA | Enolase-phosphatase E1 | Gluconacetobacter diazotrophicus | 24.9 | 4.97 | 97.57 | 1 | 3.0 | Probable bifunctional methylthioribulose-1-phosphate dehydratase/enolase-phosphatase E1 1 | XP_028794642.1 | Prosopis alba | 39% | 53% |
| 19 | B | IF2_THEFY | Translation initiation factor IF-2 | Thermobifida fusca | 100.5 | 9.82 | 101.74 | 1 | 1.2 | Translation initiation factor IF-2, chloroplastic | GEZ89434.1 | Tanacetum cinerariifolium | 52% | 71% |
| 50 | OF | MURA_PSEU5 | UDP-N-acetylglucosamine 1-carboxy-vinyltransferase | Pseudomonas stutzer | 44.6 | 5.62 | 94.32 | 1 | 2.1 | Glutamate synthase 1 [NADH], chloroplastic isoform X1 | GEU28314.1 | Tanacetum cinerariifolium | 59% | 74% |
| 56 | OF | LEXA_MYXXD | LexA repressor | Myxococcus xanthus | 24.7 | 9.29 | 99.27 | 1 | 5.4 | DNA-3-methyladenine glycosylase 1 | GEX09398.1 | Tanacetum cinerariifolium | 37% | 58% |
| 59 | OF | TRPC_ACIF2 | Indole-3-glycerol phosphate synthase | Acidithiobacillus ferrooxidans | 28.7 | 5.02 | 101.36 | 1 | 3.4 | Indole-3-glycerol phosphate synthase, chloroplastic-like isoform X2 | XP_026448585.1 | Physcomitrella patens | 48% | 63% |
| 159 | L | SCP_CHIOP | Sarcoplasmic calcium-binding protein (fragment) | Chionoecetes opilio | 0.8 | 11.1 | 114.44 | 1 | 100.0 | F-box protein At3g58530 isoform X1 | XP_021283280.1 | Herrania umbratica | 87% | 100% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kopeć, P.; Hornyák, M.; Pastuszak, J.; Szczerba, A.; Rapacz, M.; Waga, J.; Płażek, A. Changes in the Flower and Leaf Proteome of Common Buckwheat (Fagopyrum esculentum Moench) under High Temperature. Int. J. Mol. Sci. 2021, 22, 2678. https://doi.org/10.3390/ijms22052678
Kopeć P, Hornyák M, Pastuszak J, Szczerba A, Rapacz M, Waga J, Płażek A. Changes in the Flower and Leaf Proteome of Common Buckwheat (Fagopyrum esculentum Moench) under High Temperature. International Journal of Molecular Sciences. 2021; 22(5):2678. https://doi.org/10.3390/ijms22052678
Chicago/Turabian StyleKopeć, Przemysław, Marta Hornyák, Jakub Pastuszak, Anna Szczerba, Marcin Rapacz, Jacek Waga, and Agnieszka Płażek. 2021. "Changes in the Flower and Leaf Proteome of Common Buckwheat (Fagopyrum esculentum Moench) under High Temperature" International Journal of Molecular Sciences 22, no. 5: 2678. https://doi.org/10.3390/ijms22052678
APA StyleKopeć, P., Hornyák, M., Pastuszak, J., Szczerba, A., Rapacz, M., Waga, J., & Płażek, A. (2021). Changes in the Flower and Leaf Proteome of Common Buckwheat (Fagopyrum esculentum Moench) under High Temperature. International Journal of Molecular Sciences, 22(5), 2678. https://doi.org/10.3390/ijms22052678








