1. Introduction
Industrial and consumer interest in lupins grew recently due to their wide range of agricultural and health benefits, as well as their significant contribution toward achieving sustainable farming [
1,
2]. Lupin seeds are increasingly considered a valuable protein source in terms of both the human diet and as animal fodder, due to their high protein content [
1]. Like other legumes, lupins fix atmospheric nitrogen, thus supplying the soil with bioavailable nitrogen necessary for the cultivation of other crops, and allowing for reduction of mineral fertilizer use [
3,
4]. The main restraint obstructing the wider adoption of lupin as a crop results from the presence of alkaloids, which although considered to provide substantial chemical defense against herbivores and pathogens, are also considered to be antinutritional factors in terms of both food and feed, due to their bitter taste, simultaneous toxicity, and tendency to accumulate in lupin seeds [
5,
6]. The alkaloids occurring in lupins mostly belong to the group of quinolizidine alkaloids (QAs), which are lysine-derived molecules distributed mainly in the Leguminosae family [
7,
8].
Narrow-leafed lupin (NLL,
Lupinus angustifolius L.) is an important legume crop with a relatively short breeding history [
9]. During the process of NLL domestication, a natural, recessive mutant allele,
iucundus, underpinning a reduced alkaloid content was discovered, which was since incorporated into most
L. angustifolius cultivars, along with other domestication genes [
9,
10]. It is evident that breeding efforts might reduce the alkaloid content below the established safety threshold (0.02% of seed dry weight) [
11]. However, success in the effective and sustained reduction of seed alkaloid content relies on the detailed clarification of the molecular mechanisms involved in the QA synthesis pathway, as well as its underlying regulation.
Whereas the molecular machinery underlying alkaloid accumulation is quite well-resolved in other plant species (e.g.,
Catharanthus roseus and
Nicotiana tabacum), researchers working with lupins are still struggling to paint a general picture of QA synthesis and pathway regulation [
8,
12,
13,
14,
15,
16,
17]. Various theories about the sites of QA synthesis in lupins emerged over the years [
12,
18,
19,
20], coalescing into a consensus that the vast majority of QA synthesis takes place in green aerial tissues [
21], with a small contribution from the roots [
14]. The existing knowledge about leaf chloroplasts, as a site of QA biosynthesis [
22], is enriched by recognition of the input of other lupin organs, especially pods [
14,
23] and stems [
24]. The very first steps of QA synthesis seem to be relatively well understood. At the beginning, inside plant chloroplasts, lysine is decarboxylated into cadaverine polyamine [
15,
22,
25], which is further converted into 1-piperideine [
16]. These two reactions are carried out by lysine decarboxylase (LDC) [
15] and copper amine oxidase (LaCAO) [
16] enzymes, respectively. Another enzyme, namely the mitochondria (−)-13α-hydroxymultiflorine/(+)-13α-hydroxylupanine O-tigloyltransferase (HMT/HLT), is involved in the formation of ester-type alkaloids, which are assumed to be a form of alkaloid transport and storage [
13,
20,
26]. Additionally, acyltransferase enzyme (LaAT) is described as possibly being involved in the formation of polyamine conjugates. However, its specific function needs further investigation [
8]. We recently demonstrated that the APETALA2/ethylene response RAP2-7 transcription factor (TF) likely plays a crucial role in the regulation of alkaloid accumulation in NLL [
27]. The
RAP2-7 gene is found to co-segregate with the
iucundus locus, conferring low seed alkaloid content, and is located within a region with major QTLs that affect the QA composition. The contribution of this genomic region to QA biosynthesis is further supported by the 4-hydroxy-tetrahydrodipicolinate synthase (
DHDPS) gene that is possibly involved in L-lysine biosynthesis, which is found to be closely linked with the
iucundus locus [
27]. Furthermore, a gene-targeted marker developed based on the
RAP2-7 point mutation showed 100% marker–trait association (199
iucundus/
Iucundus accessions comprising domesticated and wild germplasm) [
28]. Last, but not least, we confirmed that the
RAP2-7 expression was much higher in the leaves of high-alkaloid
Iucundus vs. low-alkaloid
iucundus accessions, in a previous transcriptome-derived investigation [
27]. Nonetheless, until now,
RAP2-7 expression profiles were not yet investigated in other NLL organs.
Lupin anthracnose, caused by the fungal pathogen
Colletotrichum lupini, is one of the most devastating lupin diseases and leads to major agricultural losses [
29]. Plants exposed to various stress-inducing agents produce a wide range of different secondary metabolites, many of which play an important role in the response to biotic stresses [
5,
30]. However, the detailed involvement of QAs in response to anthracnose infection remains obscure. It was shown [
31] that a high alkaloid content in
Lupinus arboreus correlated with lesser lesions caused by
Colletotrichum sp. pathogens. In contrast, an evaluation of secondary metabolite profiles of NLL cultivars exposed to
Colletotrichum lupini revealed that alkaloid accumulation was not induced by the infection; however, the correlation between plant resistance and QA accumulation is not yet investigated [
32]. At present, only limited sources of resistance against anthracnose are available in breeding programs [
29]. Therefore, an improved understanding of the relationship between plant exposure to
Colletotrichum lupini and QA synthesis is crucial in considering them as another line of defense against this pathogen in lupins. Moreover, determination of whether the infection of resistant accessions is accompanied by an increase in seed QA content is also pivotal from the perspective of food security.
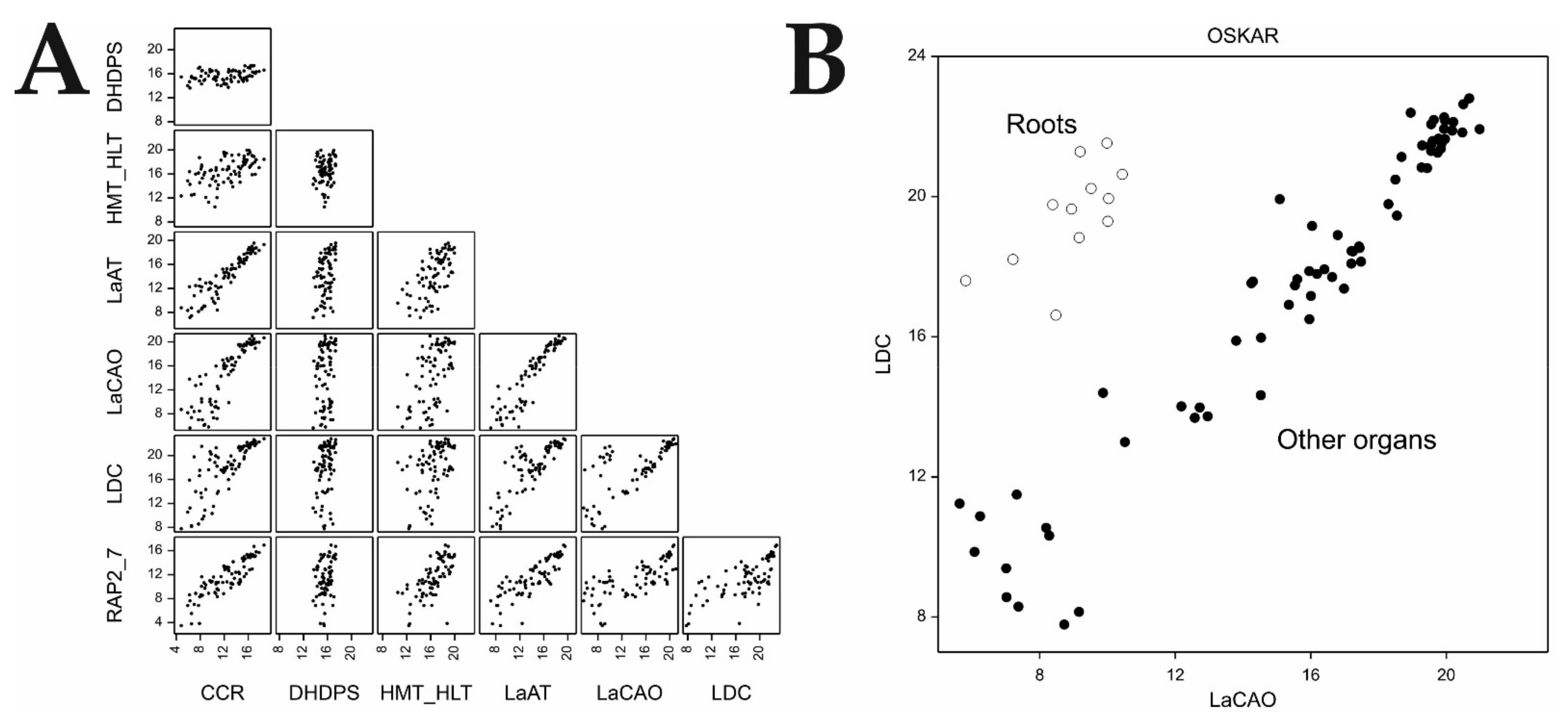
In the current study, we explored the association between the expression profiles of QA genes and alkaloid content/composition, as estimated in selected organs of NLL, to further discern the significance of the tested organs in QA biosynthesis. We analyzed organ-specific expression patterns of QA structural and candidate genes, with special attention paid to the iucundus candidate, in the form of RAP2-7, to further explain its role in the regulation of QA biosynthesis. Moreover, we also examined the transcriptional changes of QA genes in NLL plants infected with Colletotrichum lupini, to investigate the possible role of lysine-based alkaloids as a secondary line of plant defense against infection by this fungus.
4. Materials and Methods
4.1. Plant Material Grown in a Field Experiment to Study the Association Between QA Gene Expression Profiles and Alkaloid Content/Composition
The analyses of gene expression, in conjunction with measurements of alkaloid content, were carried out using two Polish NLL cultivars characterized by their contrasting content of quinolizidine alkaloids (QAs) in dry seeds. These were the sweet (low-alkaloid) cultivar, Regent (Catalogue No. 96245), and the bitter (high-alkaloid) cultivar, Oskar (Catalogue No. 96247), the seeds of which were retrieved from the Polish Lupinus Genebank (Poznan Plant Breeders Ltd., Wiatrowo Branch, Poland). Ten to fifteen seeds of each cultivar were sown each year in a field experiment in Wiatrowo, Poland. A natural vernalization process was followed by standard agricultural treatments—mechanical weed control treatment, and fertilizers P2O5 (60 kg/h) and K2O (90 kg/h), during plant growth and development. The field experiment was conducted over four vegetative seasons—2015, 2017, 2018, and 2019.
Plant material was collected yearly for all three phenological phases (i.e., flowering, pod setting, and pod maturing). A total of 13 different organs were sampled during the experiments (
Table 2). For a detailed list of plant organs collected and analyzed each year see
Tables S1 and S2. Three plants per cultivar, each representing a single biological replicate, were chosen for qPCR and GC analyses. Samples for expression analyses were kept frozen upon collection, until further analysis. The corresponding samples harvested for GC analysis were left to dry.
4.2. Plant Material Grown in a Greenhouse Experiment to Study Expression Changes of QA Genes in Relation to Colletotrichum lupini Infection
Eight NLL cultivars characterized with different susceptibilities to Colletotrichum lupini infection and contrasting seed alkaloid content were grown in a greenhouse experiment. The cultivars were contrasted by disease resistance into four cultivars mildly affected by the disease, referred to as ‘resistant’ (Regent, Roland, Sonate, and Tanjil), vs. the four cultivars severely affected by the disease, referred to as ‘susceptible’ (Ernani, Graf, Oskar, and Wersal). The cultivars were then categorized according to alkaloid content, as high seed alkaloid content cultivars (Ernani and Oskar) and low seed alkaloid content cultivars (Graf, Regent, Roland, Sonate, Tanjil, and Wersal). The resistant and susceptible cultivars were recommended by breeders based on their field observations over the years. The seeds were retrieved from the Polish Lupinus Genebank (Poznan Plant Breeders Ltd., Wiatrowo Branch, Poland) and Plant Breeding Smolice Ltd., Co. Three seeds were sown per pot (soil and sand mixture; 1:1), serving as one biological replicate. Three biological replicates (pots) were grown and further used for samples collection.
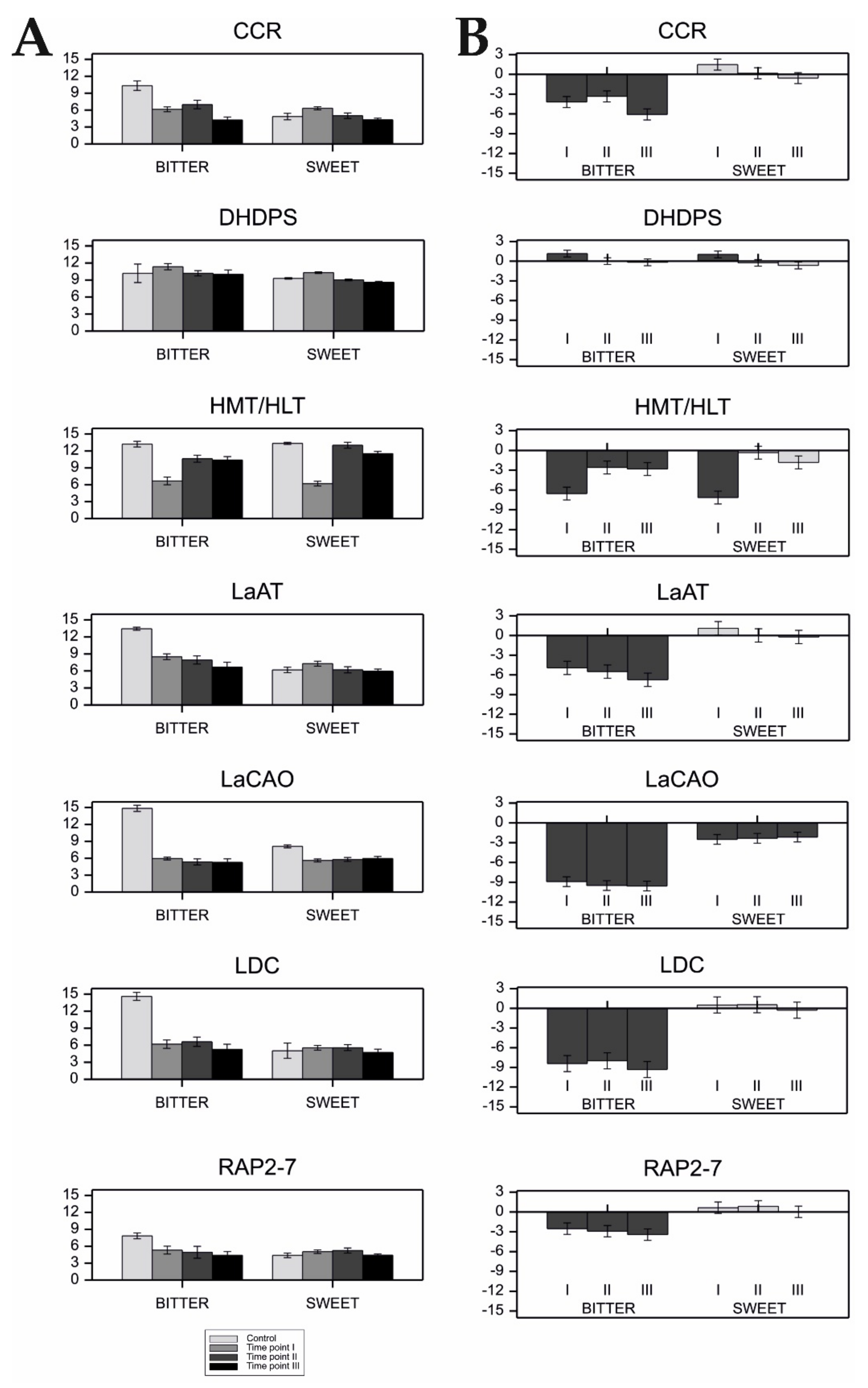
Seedling were grown for 14 days in controlled environment greenhouse (22 °C day/18 °C night, 16 h photoperiod, air humidity ~60–65%), and were manually watered once a week (200 mL). After two weeks, the seedlings were sprayed with a suspension of conidial spores of Colletotrichum lupini (2 × 106 conidia per mL) and incubated in darkness for 48 h (25 °C, air humidity ~100%). In the subsequent stage, the growing conditions were set as follows—temperature 22 °C/day and 19–20 °C/night, air humidity ~60–65%. Plant leaves for qPCR analyses were collected, in triplicate, 48 h, 72 h, and 120 h after inoculation with the pathogen (designated as time-point I, II, and III, respectively); leaves collected before inoculation served as controls.
4.3. RNA Extraction and Quality Control
RNA was extracted using an SV Total RNA Isolation System Kit (Promega, Madison, WI, USA) from 15–50 mg (leaves and green seeds) or 100–120 mg (remaining organs) of frozen and ground material. RNA purity and concentration were measured with a NanoDrop® Spectrophotometer ND-1000 (Thermo Fisher Scientific, Waltham, MA, USA). The integrity of RNA samples was visualized on non-denaturing 1% agarose gel (90 mV voltage, 30 min); samples with clear 28S and 18S rRNA bands were used for reverse-transcribing into cDNA.
4.4. Reverse Transcription
Reverse transcription was performed with 1000 ng RNA, using a Transcriptor First Strand cDNA Synthesis Kit (Roche, Mannheim, Germany), according to the manufacturer’s protocol, with anchored oligo(dT)18 primer. Two technical replicates were prepared for each sample and subsequently tested for the presence of genomic DNA (gDNA) contamination. For this purpose, conventional PCR (GoTaq® G2 Hot Start DNA polymerase; Promega, Madison, WI, USA) was carried out, with a primer pair designed against two different exons of an ATP synthase (ATPsyn; LOC109361517) gene sequence (primer sequences: F: AGTATGCTGTTCCTGTTCGTCA; R: ATGGTGATCTTCTCCTTCTTTAG). Two non-contaminated technical replicates of cDNA were mixed and 10-fold or 2-fold samples dilutions were used as template for the evaluation of gene expression. A mixture of non-diluted cDNA (5 µL of each sample) was prepared and used as a template for optimizing qPCR thermal conditions/efficiency, performed separately for the field and greenhouse experiment samples.
4.5. Target Genes in Gene Expression Studies and Experimental Design of qPCR Assay
A total of seven genes with known or putative involvement in alkaloid biosynthesis/accumulation selected on the basis of our previous transcriptome-derived investigation [
27] were examined within the current study (
Table 1). qPCR analyses were conducted using a LightCycler
® 480 II instrument and dedicated software (Roche, Mannheim, Germany). Assays were performed with aid of LightCycler
® 480 Probes Master (Roche, Mannheim, Germany) and TaqMan probes (Genomed S.A., Poland) labeled with 5′6-FAM and 3′BHQ-1, following the manufacturer’s protocols (reaction volume 10 µL). Primer pairs and probes of target and reference genes were mostly taken from [
27] and were designed on the basis of gDNA of 83A:476 (low-alkaloid) and P27255 (high-alkaloid) accessions (
Table S8). Three biological and two technical replicates of each sample, as well as a negative control, were included in each assay.
Reaction efficiency was determined for each gene, using a standard curve derived from a pooled cDNA mixture (5–7 serial, 2-fold dilution; three technical replicates) and the 2nd Derivative Max method. PCR amplification efficiencies of the reference genes ranged from 0.94 to 0.97, while those of the tested target genes ranged from 0.94 to 1.00 (
Table S8).
To calculate changes in target gene expression, the expression values of individual samples were normalized to the reference genes and subjected to efficiency corrections (these values are referred to as CP_NORM observations) [
66].
4.6. Determination of Reference Genes Expression Stability
Nine candidate reference genes [
27] were evaluated for their expression stability in a current experimental design (
Table S8). Expression stability of these genes was analyzed using the NormFinder and geNorm algorithms, incorporated into the GenEx6 ver. 6 package (Multid Analyses AB, Göteborg, Sweden) as well as the BestKeeper software [
67]. Assessment of the most stable reference genes was conducted separately for two subsets of samples: (1) for expression analyses across different plant organs; and (2) for expression analyses after
Colletotrichum infection, taking into account different time-points of infection. Eventually, the three most stable reference genes for each subset of samples were used to normalize the expression of QA-related genes. For the first subset of samples these were—alcohol dehydrogenase class-3 (
ADH3) gene, elongation factor 1-beta 2-like (
ELFB) gene, and tubulin alpha-5 chain (
TUBA5) gene. For the second subset selected, the reference genes were—
ADH3,
ELF1B, and
TUB5 (Control); actin-2-like (
ACT2/7),
ADH3, and
TUB5 (Time-point I); glucose-6-phosphate 1-dehydrogenase (
G6PD),
ACT2/7 and
TUB5 (Time-point II);
ACT2/7,
ADH3, and
TUB5 (Time-point III).
4.7. Analysis of QA Qualitative and Quantitative Content in Different Lupin Organs by the GC Method
The analysis of alkaloid profiles was carried out using the gas chromatography (GC) assay (GC-2014, Shimadzu, Kyoto, Japan). A detailed description of the alkaloid extraction method, including the technical details of instrument settings, were previously described [
68]. Due to the high amount of plant material required for extraction (ca. 500 mg of dry tissue per one technical replicate), each plant organ was analyzed as one representative sample pooled from three biological replicates. Consequently, various types of certain organs (i.e., lateral and main parts of roots and stems) were also combined. Each sample was analyzed using two technical replicates. Quantitative analysis was carried out with the aid of a linear calibration curve made for lupanine using caffeine as an internal standard. Total alkaloid content was evaluated as the percentage of the sum of alkaloids present in the analyzed sample (% of sample dry weight). The relative abundance of each alkaloid was assessed as the percentage of the total alkaloid content in a particular plant organ.
4.8. Statistical Analysis
The observations of gene expression were transformed using the formula log
2(CP_NORM × 10
6). Observations of alkaloid levels were transformed using the formula log
10((x + 0.01) × 10
2). Analysis of variance was performed in a mixed linear model containing the fixed effects of plant organs, cultivars, groups of cultivars, or time-points, and their interactions, as well as the random effects of years (according to the needs of analysis for various experiments and traits). Mean total alkaloid content in organs was obtained by Best Linear Unbiased Prediction in the mixed linear model. Post-hoc comparisons and groupings of mean values of observed traits were made using Fisher’s protected LSD test. The significance of Pearson’s correlation coefficients was assessed using the test, based on the
t-distribution. All above analyses and visualization of the results were carried out using the Genstat ver. 19 software [
69].
5. Conclusions
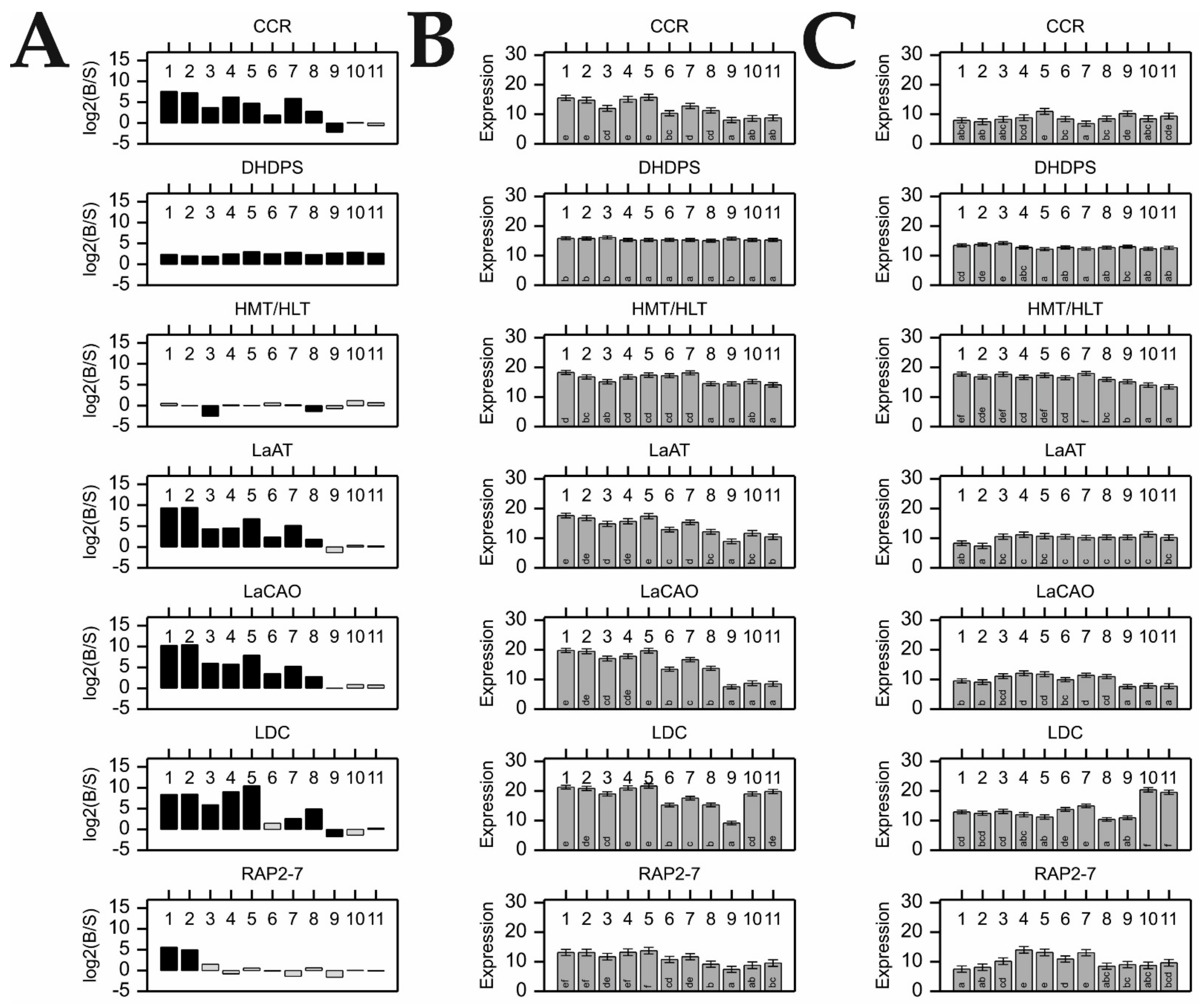
At present, there remain gaps in our knowledge about the precise regulation of QA synthesis in NLL. Within the framework of the current investigation, we identified the upregulation of RAP2-7 and other QA genes across the aerial organs of a bitter cultivar, in parallel with significant correlations of transcript expression, thus supporting the role of RAP2-7 as a vital regulatory gene of alkaloid biosynthesis/accumulation in narrow-leafed lupin. On the other hand, the unexpected expression patterns of RAP2-7 in the organs of a sweet cultivar implied that other aspects of QA biosynthesis regulation might be involved. Undoubtedly, in the aerial organs (i.e., stems and young pods) of the sweet cultivar, despite relatively high RAP2-7 expression, the expression of other genes of the QA synthesis pathway were maintained at a much lower level when compared with those of the corresponding organs of the bitter cultivar. Therefore, further research is required to resolve the question of how the RAP2-7 S196R substitution affects its function in the sweet cultivar.
We also demonstrated that at least several steps of alkaloid synthesis (converted by the products of investigated genes) might occur independently in organs other than leaves, thus supporting the role of RAP2-7 as a common regulator of the pathway. Follow-up studies on the mechanisms of its action and effect on regulating expression are necessary to obtain a clear understanding of this important metabolic pathway.
Moreover, in the limited context of the eight NLL cultivars characterized by their differing resistance/susceptibility to Colletotrichum lupini infection and contrasting seed alkaloid content, we also demonstrated that the alkaloid content in NLL seeds was seemingly unperturbed after anthracnose infection, even though other lysine-derived metabolites, such as polyamines, might play roles in the defense against this fungal disease. Direct investigation of the contribution of lupin polyamines to the plant defense response might cast new light on this important issue.