Development of Novel Inhibitors Targeting the D-Box of the DNA Binding Domain of Androgen Receptor
Abstract
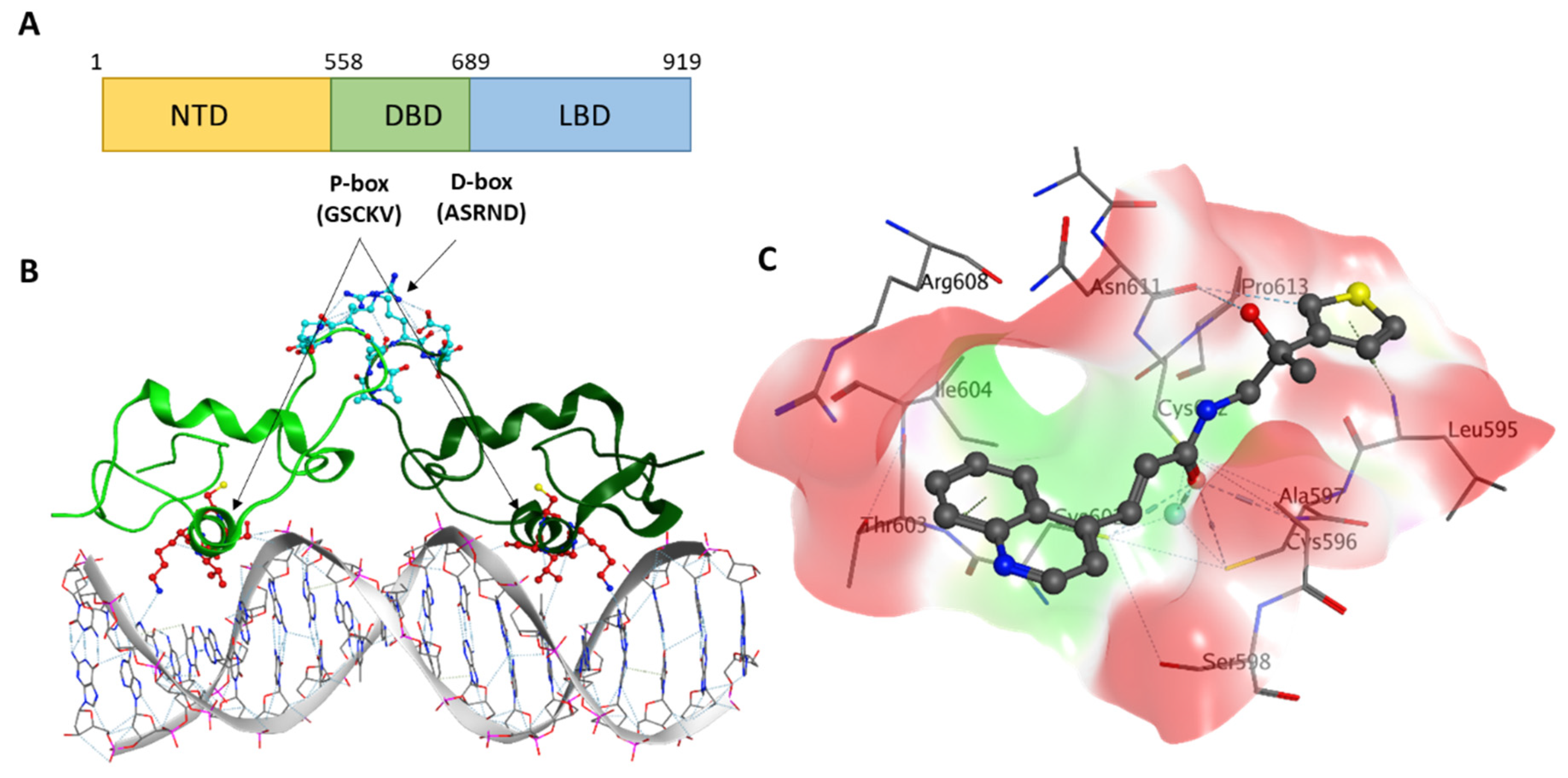
1. Introduction
2. Results
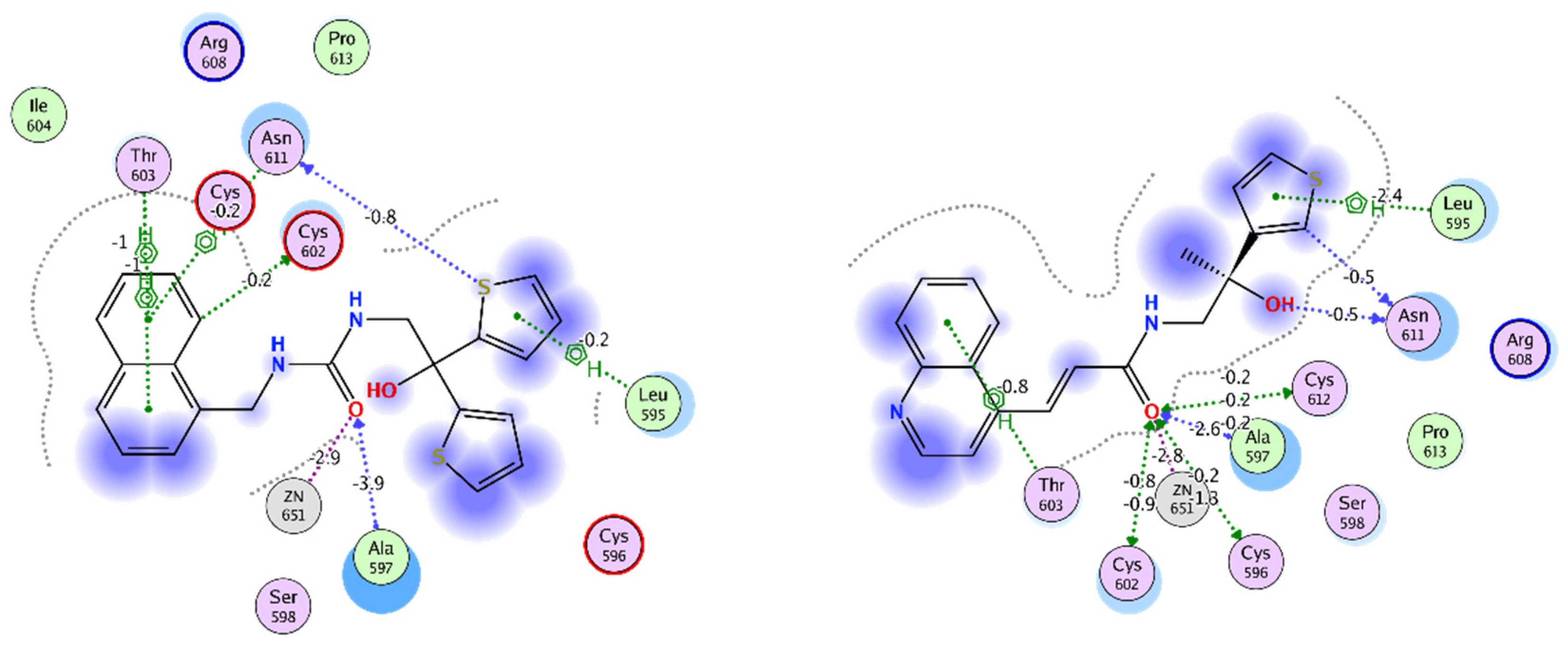
2.1. In Silico Identification of VPC-17160 and VPC-17281
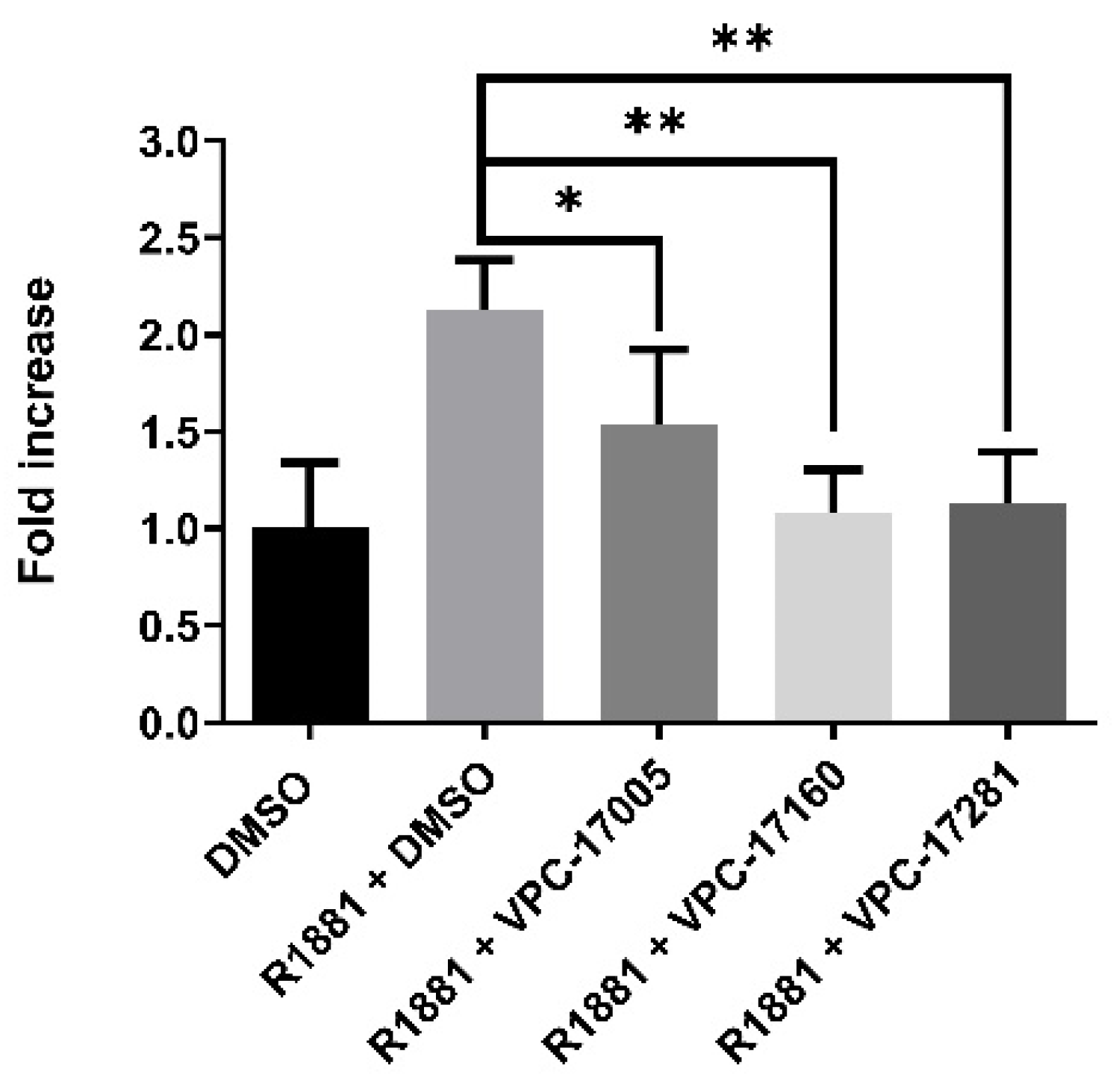
2.2. VPC-17160 and VPC-17281 Reduces AR-FL and AR-V7 Transcriptional Activities
2.3. Evidence for an Anti-Dimer Mechanism
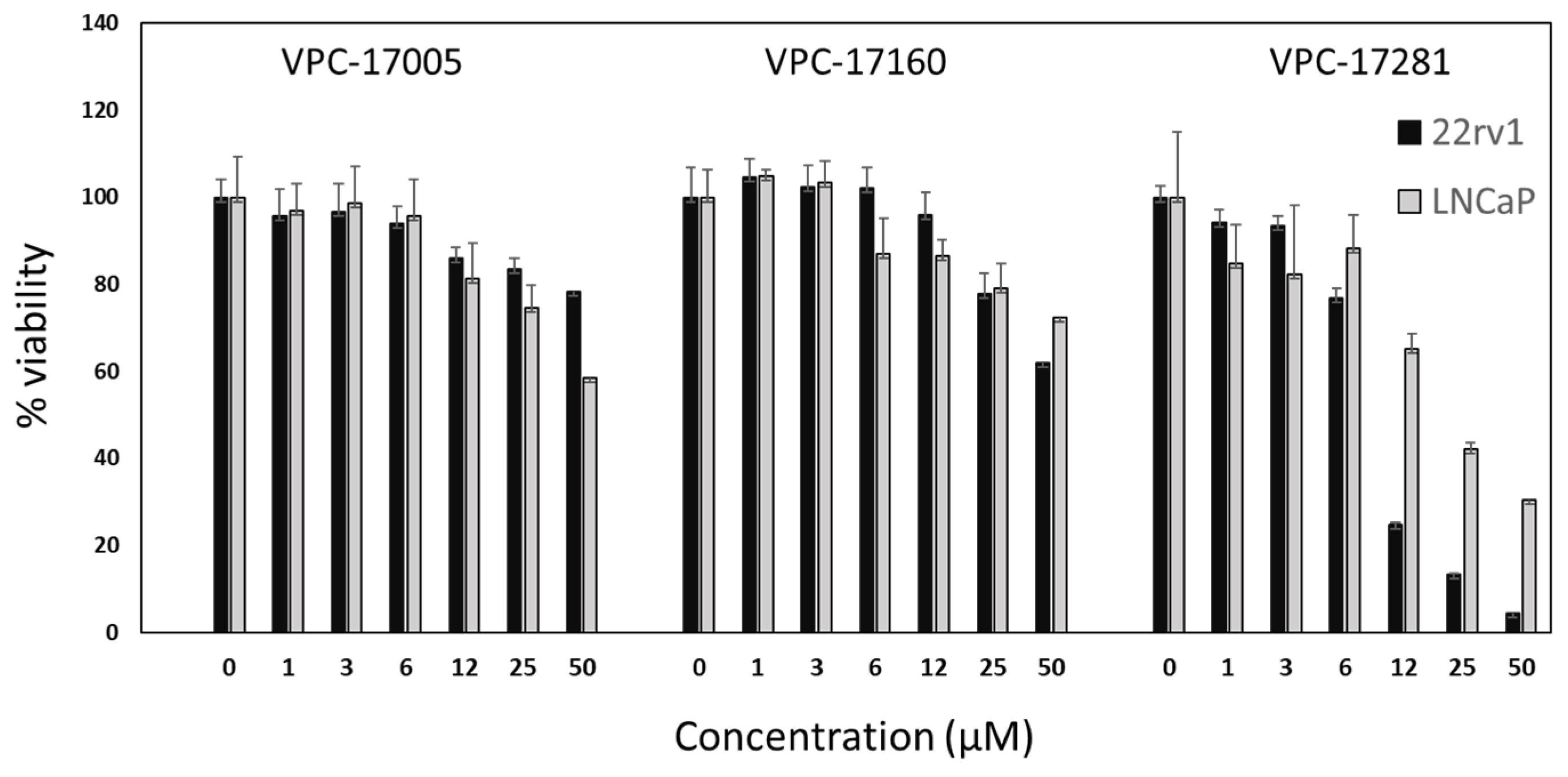
2.4. Effect on the Growth of Prostate Cancer Cell Lines
3. Discussion
4. Methods
4.1. In Silico Identification Experiments
4.2. Compounds
4.3. Plasmids and Constructructs
4.4. PCa Cell Lines
4.5. Androgen Displacement Assay
4.6. Reporter Assays and Cell Viability Experiments
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA A Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef]
- Huggins, C.; Hodges, C.V. Studies on prostatic cancer: I. The effect of castration, of estrogen and of androgen injection on serum phosphatases in metastatic carcinoma of the prostate. CA A Cancer J. Clin. 1972, 22, 232–240. [Google Scholar] [CrossRef] [PubMed]
- La Vignera, S.; Condorelli, R.; Russo, G.; Morgia, G.; Calogero, A. Endocrine control of benign prostatic hyperplasia. Andrology 2016, 4, 404–411. [Google Scholar] [CrossRef] [PubMed]
- Decker, K.F.; Zheng, D.; He, Y.; Bowman, T.; Edwards, J.R.; Jia, L. Persistent androgen receptor-mediated transcription in castration-resistant prostate cancer under androgen-deprived conditions. Nucleic Acids Res. 2012, 40, 10765–10779. [Google Scholar] [CrossRef]
- Green, S.M.; Mostaghel, E.A.; Nelson, P.S. Androgen action and metabolism in prostate cancer. Mol. Cell. Endocrinol. 2012, 360, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Eder, I.E.; Culig, Z.; Putz, T.; Nessler-Menardi, C.; Bartsch, G.; Klocker, H. Molecular biology of the androgen receptor: From molecular understanding to the clinic. Eur. Urol. 2001, 40, 241–251. [Google Scholar] [CrossRef]
- Kirby, M.; Hirst, C.; Crawford, E. Characterising the castration-resistant prostate cancer population: A systematic review. Int. J. Clin. Pract. 2011, 65, 1180–1192. [Google Scholar] [CrossRef] [PubMed]
- Lorente, D.; Mateo, J.; Zafeiriou, Z.; Smith, A.D.; Sandhu, S.; Ferraldeschi, R.; De Bono, J.S. Switching and withdrawing hormonal agents for castration-resistant prostate cancer. Nat. Rev. Urol. 2015, 12, 37. [Google Scholar] [CrossRef] [PubMed]
- Snow, O.; Lallous, N.; Singh, K.; Lack, N.; Rennie, P.; Cherkasov, A. Androgen receptor plasticity and its implications for prostate cancer therapy. Cancer Treat. Rev. 2019, 81, 101871. [Google Scholar] [CrossRef] [PubMed]
- Bohl, C.E.; Miller, D.D.; Chen, J.; Bell, C.E.; Dalton, J.T. Structural basis for accommodation of nonsteroidal ligands in the androgen receptor. J. Biol. Chem. 2005, 280, 37747–37754. [Google Scholar] [CrossRef]
- Davey, R.A.; Grossmann, M. Androgen receptor structure, function and biology: From bench to bedside. Clin. Biochem. Rev. 2016, 37, 3. [Google Scholar]
- Beer, T.M.; Armstrong, A.J.; Rathkopf, D.E.; Loriot, Y.; Sternberg, C.N.; Higano, C.S.; Iversen, P.; Bhattacharya, S.; Carles, J.; Chowdhury, S. Enzalutamide in metastatic prostate cancer before chemotherapy. N. Engl. J. Med. 2014, 371, 424–433. [Google Scholar] [CrossRef]
- Hussain, M.; Fizazi, K.; Saad, F.; Rathenborg, P.; Shore, N.; Ferreira, U.; Ivashchenko, P.; Demirhan, E.; Modelska, K.; Phung, D. Enzalutamide in men with nonmetastatic, castration-resistant prostate cancer. N. Engl. J. Med. 2018, 378, 2465–2474. [Google Scholar] [CrossRef]
- Rice, M.A.; Malhotra, S.V.; Stoyanova, T.I. Second-generation antiandrogens in castration resistant prostate cancer. Front. Oncol. 2019, 9, 801. [Google Scholar] [CrossRef]
- Korpal, M.; Korn, J.M.; Gao, X.; Rakiec, D.P.; Ruddy, D.A.; Doshi, S.; Yuan, J.; Kovats, S.G.; Kim, S.; Cooke, V.G. An F876L mutation in androgen receptor confers genetic and phenotypic resistance to MDV3100 (enzalutamide). Cancer Discov. 2013, 3, 1030–1043. [Google Scholar] [CrossRef] [PubMed]
- Antonarakis, E.S.; Lu, C.; Wang, H.; Luber, B.; Nakazawa, M.; Roeser, J.C.; Chen, Y.; Mohammad, T.A.; Chen, Y.; Fedor, H.L. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N. Engl. J. Med. 2014, 371, 1028–1038. [Google Scholar] [CrossRef] [PubMed]
- Antonarakis, E.S.; Lu, C.; Luber, B.; Wang, H.; Chen, Y.; Zhu, Y.; Silberstein, J.L.; Taylor, M.N.; Maughan, B.L.; Denmeade, S.R. Clinical significance of androgen receptor splice variant-7 mRNA detection in circulating tumor cells of men with metastatic castration-resistant prostate cancer treated with first-and second-line abiraterone and enzalutamide. J. Clin. Oncol. 2017, 35, 2149. [Google Scholar] [CrossRef]
- Ban, F.; Dalal, K.; Li, H.; LeBlanc, E.; Rennie, P.S.; Cherkasov, A. Best Practices of Computer-Aided Drug Discovery: Lessons Learned from the Development of a Preclinical Candidate for Prostate Cancer with a New Mechanism of Action. J. Chem. Inf. Modeling 2017, 57, 1018–1028. [Google Scholar] [CrossRef]
- Dalal, K.; Roshan-Moniri, M.; Sharma, A.; Li, H.; Ban, F.; Hassona, M.D.; Hsing, M.; Singh, K.; LeBlanc, E.; Dehm, S.; et al. Selectively targeting the DNA-binding domain of the androgen receptor as a prospective therapy for prostate cancer. J. Biol. Chem. 2014, 289, 26417–26429. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Ban, F.; Dalal, K.; Leblanc, E.; Frewin, K.; Ma, D.; Adomat, H.; Rennie, P.S.; Cherkasov, A. Discovery of small-molecule inhibitors selectively targeting the DNA-binding domain of the human androgen receptor. J. Med. Chem. 2014, 57, 6458–6467. [Google Scholar] [CrossRef]
- Shaffer, P.L.; Jivan, A.; Dollins, D.E.; Claessens, F.; Gewirth, D.T. Structural basis of androgen receptor binding to selective androgen response elements. Proc. Natl. Acad. Sci. USA 2004, 101, 4758–4763. [Google Scholar] [CrossRef]
- van Royen, M.E.; van Cappellen, W.A.; de Vos, C.; Houtsmuller, A.B.; Trapman, J. Stepwise androgen receptor dimerization. J. Cell Sci. 2012, 125, 1970–1979. [Google Scholar] [CrossRef]
- Chan, S.C.; Selth, L.A.; Li, Y.; Nyquist, M.D.; Miao, L.; Bradner, J.E.; Raj, G.V.; Tilley, W.D.; Dehm, S.M. Targeting chromatin binding regulation of constitutively active AR variants to overcome prostate cancer resistance to endocrine-based therapies. Nucleic Acids Res. 2015, 43, 5880–5897. [Google Scholar] [CrossRef]
- Azoitei, A.; Merseburger, A.S.; Godau, B.; Hoda, M.R.; Schmid, E.; Cronauer, M.V. C-terminally truncated constitutively active androgen receptor variants and their biologic and clinical significance in castration-resistant prostate cancer. J. Steroid Biochem. Mol. Biol. 2017, 166, 38–44. [Google Scholar] [CrossRef] [PubMed]
- Zhan, Y.; Zhang, G.; Wang, X.; Qi, Y.; Bai, S.; Li, D.; Ma, T.; Sartor, O.; Flemington, E.K.; Zhang, H. Interplay between cytoplasmic and nuclear androgen receptor splice variants mediates castration resistance. Mol. Cancer Res. 2017, 15, 59–68. [Google Scholar] [CrossRef]
- Dalal, K.; Ban, F.; Li, H.; Morin, H.; Roshan-Moniri, M.; Tam, K.J.; Shepherd, A.; Sharma, A.; Peacock, J.; Carlson, M.L. Selectively targeting the dimerization interface of human androgen receptor with small-molecules to treat castration-resistant prostate cancer. Cancer Lett. 2018, 437, 35–43. [Google Scholar] [CrossRef]
- Sterling, T.; Irwin, J.J. ZINC 15–ligand discovery for everyone. J. Chem. Inf. Modeling 2015, 55, 2324–2337. [Google Scholar] [CrossRef] [PubMed]
- Tavassoli, P.; Snoek, R.; Ray, M.; Rao, L.G.; Rennie, P.S. Rapid, non-destructive, cell-based screening assays for agents that modulate growth, death, and androgen receptor activation in prostate cancer cells. Prostate 2007, 67, 416–426. [Google Scholar] [CrossRef] [PubMed]
- Dhanasekaran, S.M.; Barrette, T.R.; Ghosh, D.; Shah, R.; Varambally, S.; Kurachi, K.; Pienta, K.J.; Rubin, M.A.; Chinnaiyan, A.M. Delineation of prognostic biomarkers in prostate cancer. Nature 2001, 412, 822–826. [Google Scholar] [CrossRef]
- Dang, C.V.; Barrett, J.; Villa-Garcia, M.; Resar, L.; Kato, G.J.; Fearon, E.R. Intracellular leucine zipper interactions suggest c-Myc hetero-oligomerization. Mol. Cell. Biol. 1991, 11, 954–962. [Google Scholar] [CrossRef]
- Sastry, G.M.; Adzhigirey, M.; Day, T.; Annabhimoju, R.; Sherman, W. Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided Mol. Des. 2013, 27, 221–234. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The protein data bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Molecular Operating Environment (MOE); Chemical Computing Group (ULC): Montreal, QC, Canada, 2019.
- Schrödinger Release 2020-4: Glide; Schrödinger, LLC: New York, NY, USA, 2020.
- Peacock, J.W.; Palmer, J.; Fink, D.; Ip, S.; Pietras, E.M.; Mui, A.L.-F.; Chung, S.W.; Gleave, M.E.; Cox, M.E.; Parsons, R. PTEN loss promotes mitochondrially dependent type II Fas-induced apoptosis via PEA-15. Mol. Cell. Biol. 2009, 29, 1222–1234. [Google Scholar] [CrossRef] [PubMed]
- Klock, H.E.; Koesema, E.J.; Knuth, M.W.; Lesley, S.A. Combining the polymerase incomplete primer extension method for cloning and mutagenesis with microscreening to accelerate structural genomics efforts. Proteins Struct. Funct. Bioinform. 2008, 71, 982–994. [Google Scholar] [CrossRef] [PubMed]

| Compound | % Displacement | Half-Life (min) | |
|---|---|---|---|
| (1 μM) | (10 μM) | ||
| Enzalutamide | 63 | 85 | >1000 |
| VPC-17005 | 26 | 56 | 14 |
| VPC-17160 | 10 | 29 | 3 |
| VPC-17281 | 0 | 0 | 90 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Radaeva, M.; Ban, F.; Zhang, F.; LeBlanc, E.; Lallous, N.; Rennie, P.S.; Gleave, M.E.; Cherkasov, A. Development of Novel Inhibitors Targeting the D-Box of the DNA Binding Domain of Androgen Receptor. Int. J. Mol. Sci. 2021, 22, 2493. https://doi.org/10.3390/ijms22052493
Radaeva M, Ban F, Zhang F, LeBlanc E, Lallous N, Rennie PS, Gleave ME, Cherkasov A. Development of Novel Inhibitors Targeting the D-Box of the DNA Binding Domain of Androgen Receptor. International Journal of Molecular Sciences. 2021; 22(5):2493. https://doi.org/10.3390/ijms22052493
Chicago/Turabian StyleRadaeva, Mariia, Fuqiang Ban, Fan Zhang, Eric LeBlanc, Nada Lallous, Paul S. Rennie, Martin E. Gleave, and Artem Cherkasov. 2021. "Development of Novel Inhibitors Targeting the D-Box of the DNA Binding Domain of Androgen Receptor" International Journal of Molecular Sciences 22, no. 5: 2493. https://doi.org/10.3390/ijms22052493
APA StyleRadaeva, M., Ban, F., Zhang, F., LeBlanc, E., Lallous, N., Rennie, P. S., Gleave, M. E., & Cherkasov, A. (2021). Development of Novel Inhibitors Targeting the D-Box of the DNA Binding Domain of Androgen Receptor. International Journal of Molecular Sciences, 22(5), 2493. https://doi.org/10.3390/ijms22052493






