Genomic Space of MGMT in Human Glioma Revisited: Novel Motifs, Regulatory RNAs, NRF1, 2, and CTCF Involvement in Gene Expression
Abstract
1. Introduction
2. Results and Discussion
2.1. Recapitulation of MGMT Genomic Space and the mRNA
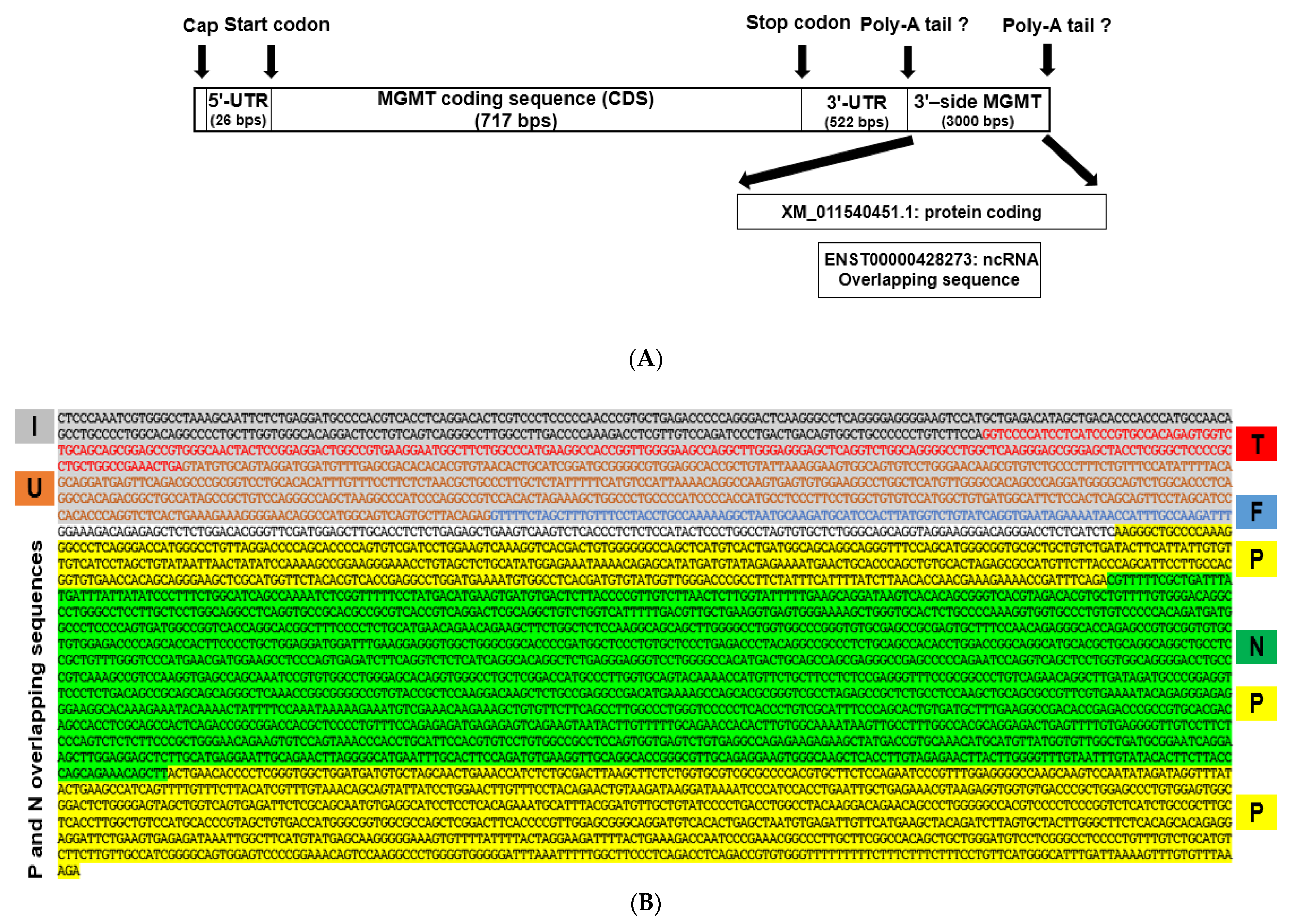
2.2. New Features of 3′ Untranslated Region (UTR) of MGMT Transcript
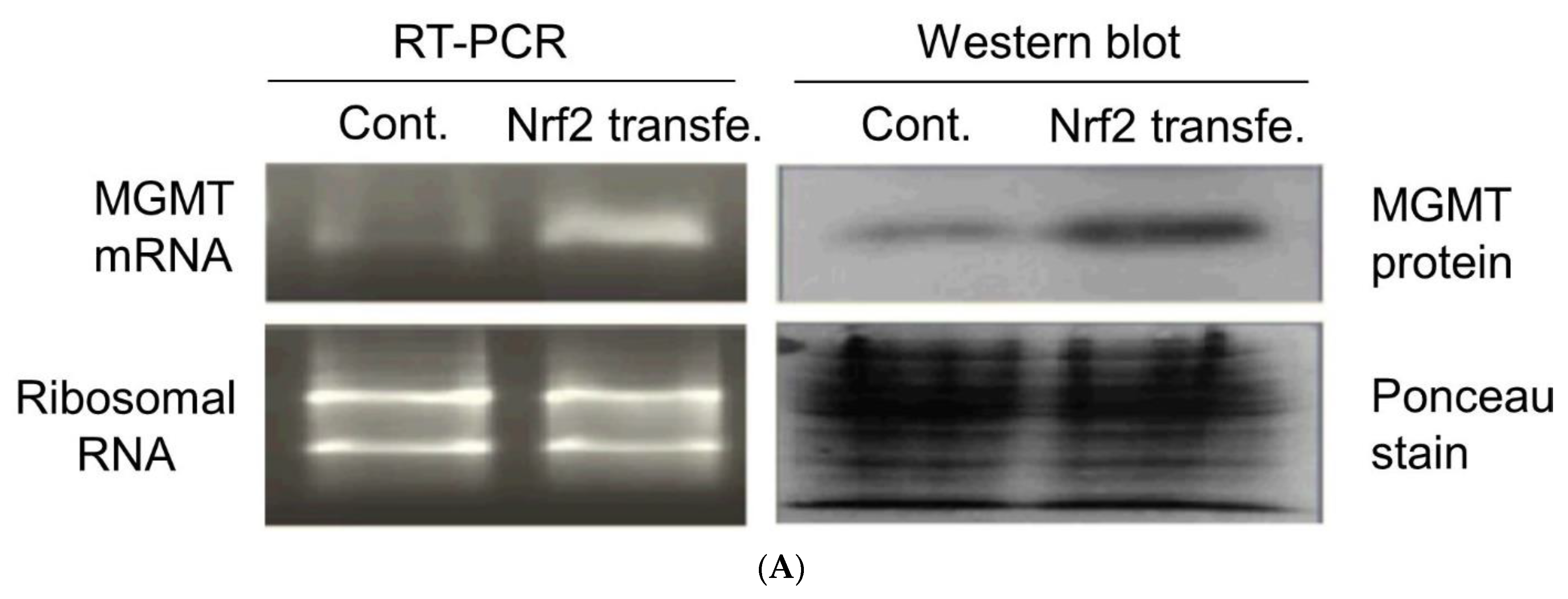
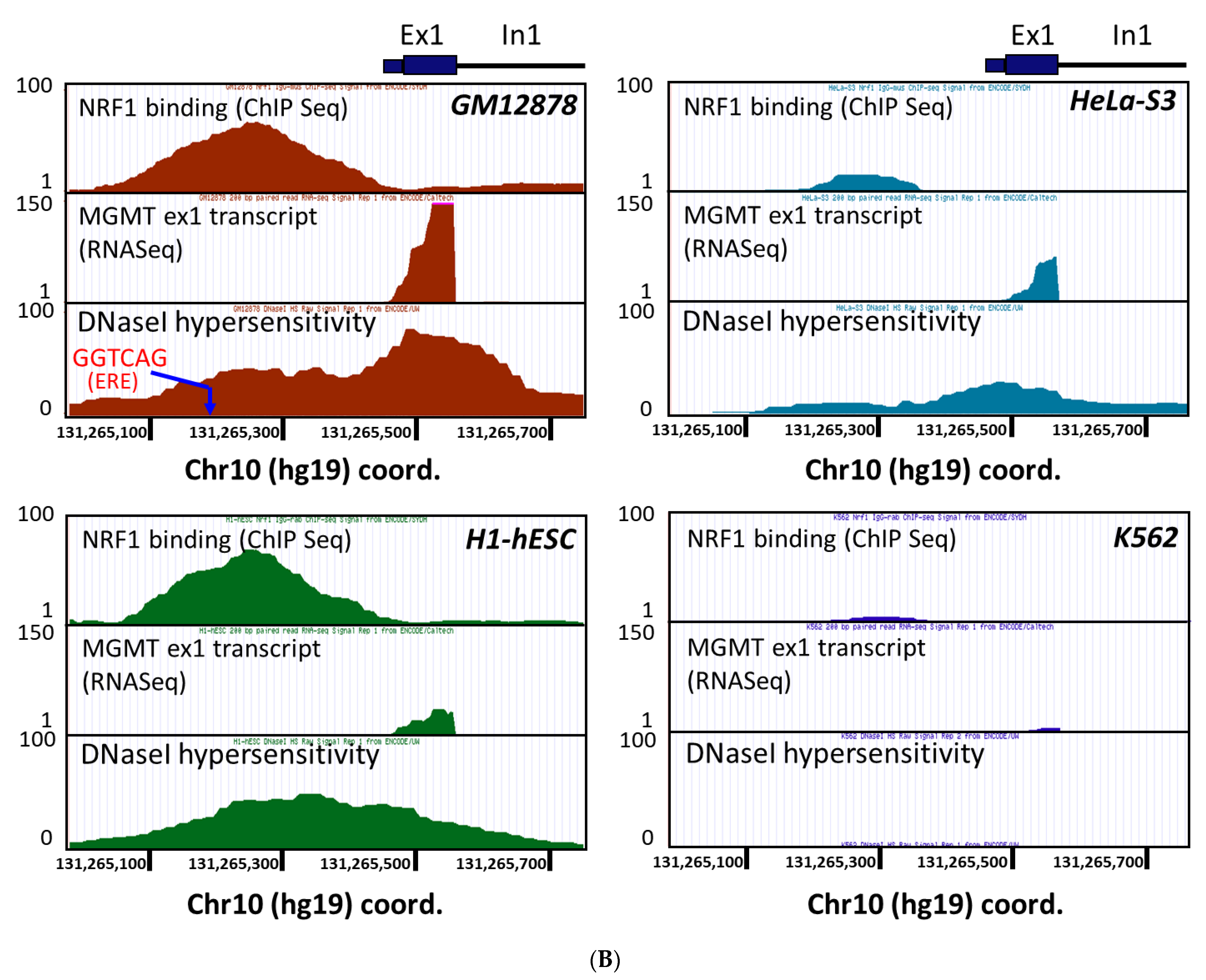
2.3. New Regulatory Motifs in MGMT Promoter; ARE, ERE, and NRF1/NRF2 -Mediated Upregulation of MGMT Expression
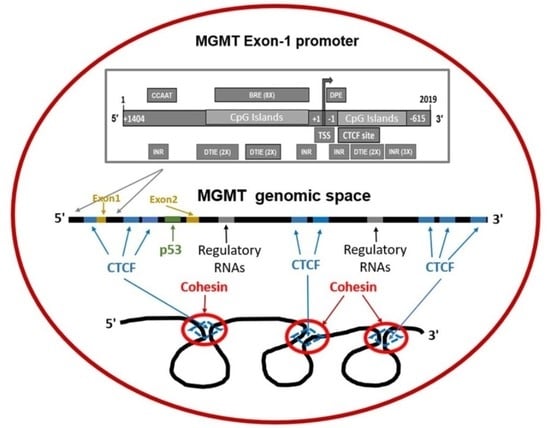
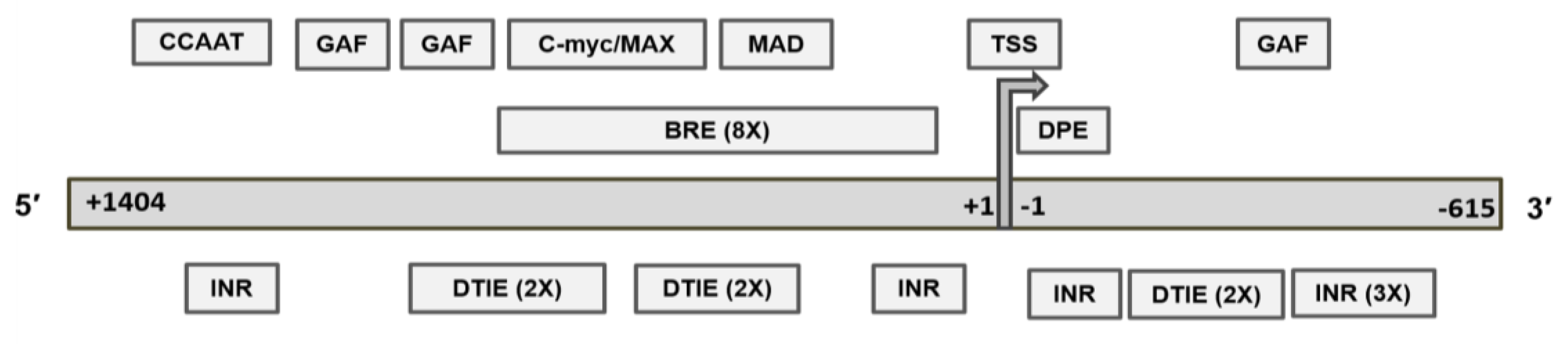
2.4. Revised MGMT Promoter That Includes Alternate Promoters, CGI Status, and Other Promoter-Like Sequences in MGMT Genome
2.5. Recognition Motifs for GAF and MYC/MAD/MAX Switch within the MGMT Promoter
2.6. Identification of p53 Response Elements (PREs) in Intron 1
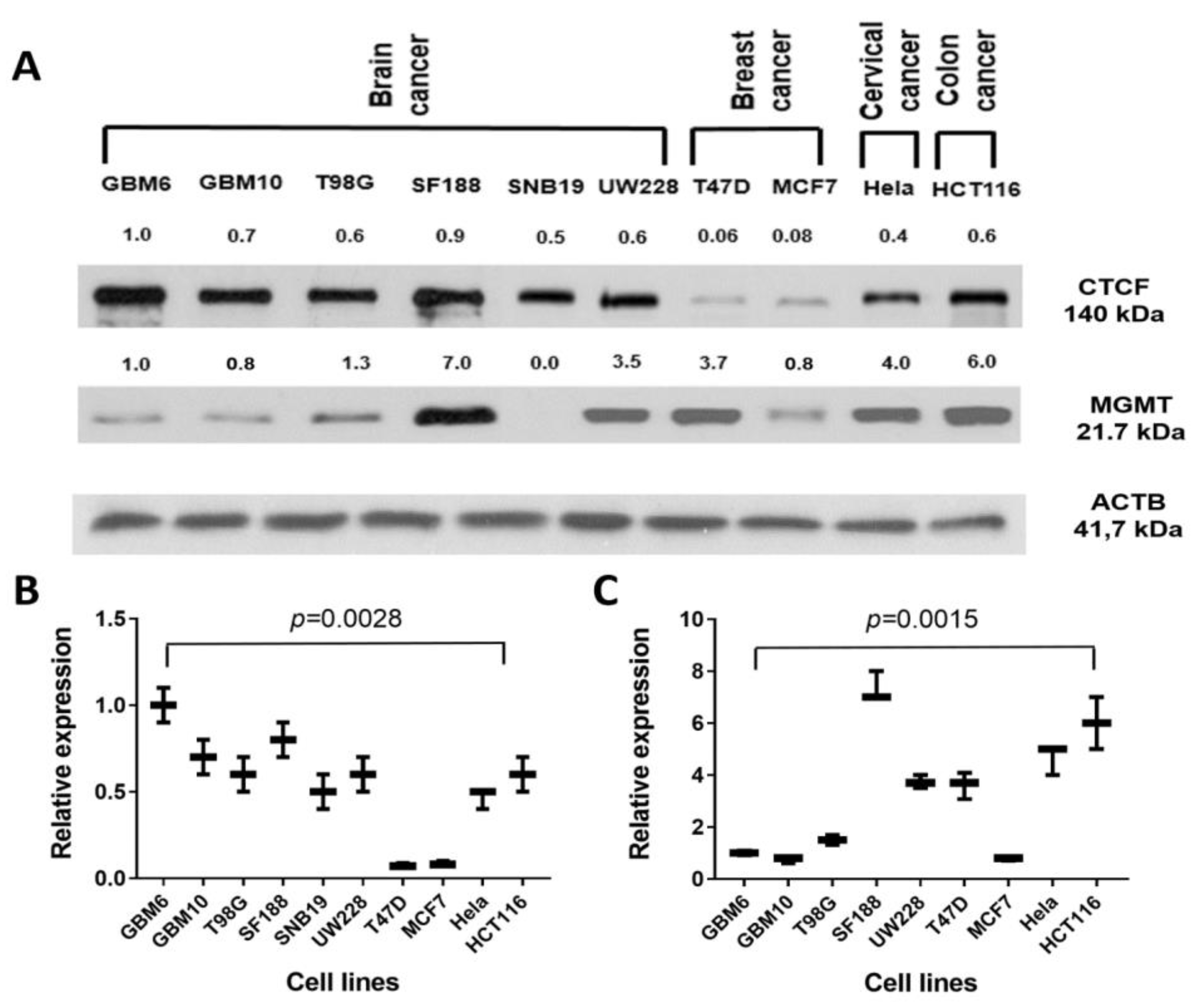
2.7. Presence of CTCF Recognition Motifs in the MGMT Promoter and Rest of the Gene: Evidence for CTCF as a Positive Regulator of MGMT Expression
2.8. Potential Involvement of RNA Regulatory Elements (Long Non-Coding RNA, Antisense RNA, Micro RNA) in the MGMT- EBF3 Region in the Regulation of MGMT Expression
3. Conclusions
4. Materials and Methods
4.1. Cell Culture
4.2. Western Blotting
4.3. siRNA and shRNA Transient Transfections and NRF2 Stable Transfection
4.4. Chromatin Immunoprecipitation (ChIP) Assay
4.5. Database Search
4.6. Regulatory Sequences and MGMT Alternative Promoters
4.7. Sequencing-Based Data from ENCODE and Bioinformatic Analysis of Non-Coding RNA Transcripts
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mitra, S.; Kaina, B. Regulation of repair of alkylation damage in mammalian genomes. Prog. Nucleic Acid Res. Mol. Biol. 1993, 44, 109–142. [Google Scholar]
- Pegg, A.E. Repair of O6-alkylguanine by alkyltransferases. Mutat. Res. 2000, 462, 83–100. [Google Scholar] [CrossRef]
- Mishina, Y.; Duguid, E.M.; He, C. Direct Reversal of DNA Alkylation Damage. Chem. Rev. 2006, 106, 215–232. [Google Scholar] [CrossRef]
- Silber, J.R.; Blank, A.; Bobola, M.S.; Mueller, B.A. Lack of DNA repair protein O6-methylguanine-DNA methyltransferase in histologically normal brain adjacent to primary human brain tumors. Proc. Natl. Acad. Sci. USA 1996, 93, 6941–6946. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Guo, J.; Wang, W.; Wang, D. Relationship between MGMT gene expression and treatment effectiveness and prognosis in glioma. Oncol. Lett. 2017, 14, 229–233. [Google Scholar] [CrossRef] [PubMed]
- Middlemas, D.S.; Stewart, C.F.; Kirstein, M.N.; Poquette, C.; Friedman, H.S.; Houghton, P.J.; Brent, T.P. Biochemical correlates of temozolomide sensitivity in pediatric solid tumor xenograft models. Clin. Cancer Res. 2000, 6, 998–1007. [Google Scholar]
- Gerson, S.L. Clinical Relevance of MGMT in the Treatment of Cancer. J. Clin. Oncol. 2002, 20, 2388–2399. [Google Scholar] [CrossRef]
- Weller, M.; Stupp, R.; Reifenberger, G.; Brandes, A.A.; van den Bent, M.J.; Wick, W.; Hegi, M.E. MGMT promoter methylation in malignant gliomas: Ready for personalized medicine? Nat. Rev. Neurol. 2010, 6, 39–51. [Google Scholar] [CrossRef]
- Mansouri, A.; Hachem, L.D.; Mansouri, S.; Nassiri, F.; Laperriere, N.J.; Xia, D.; Lindeman, N.I.; Wen, P.Y.; Chakravarti, A.; Mehta, M.P.; et al. MGMT promoter methylation status testing to guide therapy for glioblastoma: Refining the approach based on emerging evidence and current challenges. Neuro Oncol. 2019, 21, 167–178. [Google Scholar] [CrossRef]
- Kaina, B.; Margison, G.P.; Christmann, M. Targeting O⁶-methylguanine-DNA methyltransferase with specific inhibitors as a strategy in cancer therapy. Cell Mol. Life Sci. 2010, 67, 3663–3681. [Google Scholar] [CrossRef]
- Dolan, M.E.; Pegg, A.E. O6-benzylguanine and its role in chemotherapy. Clin. Cancer Res. 1997, 3, 837–847. [Google Scholar]
- Turriziani, M.; Caporaso, P.; Bonmassar, L.; Buccisano, F.; Amadori, S.; Venditti, A.; Cantonetti, M.; D’Atri, S.; Bonmassar, E. O6-(4-bromothenyl)guanine (PaTrin-2), a novel inhibitor of O6-alkylguanine DNA alkyl-transferase, increases the inhibitory activity of temozolomide against human acute leukemia cells in vitro. Pharmacol Res. 2006, 53, 317–323. [Google Scholar] [CrossRef]
- Paranjpe, A.; Zhang, R.; Ali-Osman, F.; Bobustuc, G.C.; Srivenugopal, K.S. Disulfiram is a direct and potent inhibitor of human O6-methylguanine-DNA methyltransferase (MGMT) in brain tumor cells and mouse brain and markedly increases the alkyl-ating DNA damage. Carcinogenesis 2014, 35, 692–702. [Google Scholar] [CrossRef]
- Kast, R.E.; Boockvar, J.A.; Brüning, A.; Cappello, F.; Chang, W.W.; Cvek, B.; Dou, Q.P.; Duenas-Gonzalez, A.; Efferth, T.; Focosi, D.; et al. A conceptually new treatment approach for relapsed glioblastoma: Coordinated undermining of survival paths with nine re-purposed drugs (CUSP9) by the International Initiative for Accelerated Improvement of Glioblastoma. Care Oncotarget. 2013, 4, 502–530. [Google Scholar] [CrossRef] [PubMed]
- Hegi, M.E.; Diserens, A.C.; Gorlia, T.; Hamou, M.F.; de Tribolet, N.; Weller, M.; Kros, J.M.; Hainfellner, J.A.; Mason, W.; Mariani, L.; et al. MGMT Gene silencing and benefit from temozolomide in glioblastoma. N. Engl. J. Med. 2005, 352, 997–1003. [Google Scholar] [CrossRef]
- Esteller, M.; Garcia-Foncillas, J.; Andion, E.; Goodman, S.N.; Hidalgo, O.F.; Vanaclocha, V.; Baylin, S.B.; Herman, J.G. Inactivation of the DNA-Repair Gene MGMT and the Clinical Response of Gliomas to Alkylating Agents. N. Engl. J. Med. 2000, 343, 1350–1354. [Google Scholar] [CrossRef] [PubMed]
- Gerson, S.L. MGMT: Its role in cancer etiology and cancer therapeutics. Nat. Rev. Cancer 2004, 4, 296–307. [Google Scholar] [CrossRef] [PubMed]
- Butler, M.; Lorinc, P.; Su, Y.-T.; Raffeld, M.; Quezado, M.; Trepel, J.; Aldape, K.; Pommier, Y.; Wu, J. MGMT status as a clinical biomarker in glioblastoma. Trends in Cancer 2020, 6, 380–391. [Google Scholar] [CrossRef]
- Bacolod, M.D.; Barany, F. MGMT epigenetics: The influence of gene body methylation and other insights derived from integrated methylomic, transcriptomic, and chromatin analyses for various cancer types. Curr. Cancer Drug Targets, 2020; in press. [Google Scholar]
- Costello, J.F.; Futscher, B.W.; Tano, K.; Graunke, D.M.; Pieper, R.O. Graded methylation in the promoter and body of the O6-methylguanine DNA methyltransferase (MGMT) gene correlates with MGMT expression in human glioma cells. J. Biol. Chem. 1994, 269, 17228–17237. [Google Scholar] [CrossRef]
- Moen, E.L.; Stark, A.L.; Zhang, W.; Dolan, M.E.; Godley, L.A. The role of gene body cytosine modifications in MGMT expression and sensitivity to temozolomide. Mol. Cancer Ther. 2014, 13, 1334–1344. [Google Scholar] [CrossRef][Green Version]
- Cabrini, G.; Fabbri, E.; Lo Nigro, C.; Dechecchi, M.C.; Gambari, R. Regulation of expression of O6-methylguanine-DNA methyltransferase and the treatment of glioblastoma. Int. J. Oncol. 2015, 47, 417–428. [Google Scholar] [CrossRef]
- Harris, L.C.; Potter, P.M.; Tano, K.; Shiota, S.; Mitra, S.; Brent, T.P. Characterization of the promoter region of the human O6-methylguanine-DNA methyltransferase gene. Nucleic Acids Res. 1991, 19, 6163–6167. [Google Scholar] [CrossRef]
- Harris, L.C.; Remack, J.S.; Brent, T.P. Identification of a 59 bp enhancer located at the first exon/intron boundary of the human O6-methylguanine DNA methyltransferase gene. Nucleic Acids Res. 1994, 22, 4614–4619. [Google Scholar] [CrossRef]
- Aasland, D.; Reich, T.R.; Tomicic, M.T.; Switzeny, O.J.; Kaina, B.; Christmann, M. Repair gene O6 -methylguanine-DNA methyltransferase is controlled by SP1 and up-regulated by glucocorticoids, but not by temozolomide and radiation. J. Neurochem. 2018, 144, 139–151. [Google Scholar] [CrossRef]
- Hughes, L.A.E.; Melotte, V.; Schrijver, J.D.; Maat, M.D.; Smit, V.T.H.B.M.; Bovee, J.V.M.G.; French, P.J.; Brandt, P.A.V.D.; Schouten, L.J.; Meyer, T.D.; et al. The CpG island methylator phenotype: What is in a name? Cancer Res. 2013, 73, 5858–5868. [Google Scholar] [CrossRef]
- Malta, T.M.; De Souza, C.F.; Sabedot, T.S.; Silva, T.C.; Mosella, M.S.; Kalkanis, S.N.; Snyder, J.; Castro, A.V.B.; Noushmehr, H. Glioma CpG island methylator phenotype (G-CIMP): Biological and clinical implications. Neuro Oncol. 2018, 20, 608–620. [Google Scholar] [CrossRef]
- Madala, H.R.; Punganuru, S.R.; Arutla, V.; Misra, S.; Thomas, T.J.; Srivenugopal, K.S. Beyond brooding on oncometabolic havoc in IDH-mutant gliomas and AML: Current and future therapeutic strategies. Cancers 2018, 10, 49. [Google Scholar] [CrossRef] [PubMed]
- Reitman, Z.J.; Yan, H. Isocitrate dehydrogenase 1 and 2 mutations in cancer: Alterations at a rossroads of cellular metabolism. J. Natl. Cancer Inst. 2010, 102, 932–941. [Google Scholar] [CrossRef]
- Flanagan, S.; Lee, M.; Li, C.C.Y.; Suter, C.M.; Buckland, M.E. Promoter Methylation Analysis of IDH Genes in Human Gliomas. Front. Oncol. 2012, 2, 193. [Google Scholar] [CrossRef] [PubMed]
- Krex, D.; Klink, B.; Hartmann, C.; von Deimling, A.; Pietsch, T.; Simon, M.; Sabel, M.; Steinbach, J.P.; Heese, O.; Reifenberger, G.; et al. German Glioma Network. Long-term survival with glioblastoma multiforme. Brain 2007, 130, 2596–2606.32. [Google Scholar] [CrossRef] [PubMed]
- von Deimling, A.; Korshunov, A.; Hartmann, C. The next generation of glioma biomarkers: MGMT methylation, BRAF fusions and IDH1 mutations. Brain Pathol. 2011, 21, 74–87. [Google Scholar] [CrossRef]
- Braccioli, L.; de Wit, E. CTCF: A Swiss-army knife for genome organization and transcription regulation. Essays Biochem. 2019, 63, 157–165. [Google Scholar]
- Ghirlando, R.; Felsenfeld, G. CTCF: Making the right connections. Genes Dev. 2016, 30, 881–891. [Google Scholar] [CrossRef]
- Bulger, M.; Groudine, M. Looping versus linking: Toward a model for long-distance gene activation. Genes Dev. 1999, 13, 2465–2477. [Google Scholar] [CrossRef]
- Liao, D. Emerging Roles of the EBF Family of Transcription Factors in Tumor Suppression. Mol. Cancer Res. 2009, 7, 1893–1901. [Google Scholar] [CrossRef]
- Zardo, G.; Tiirikainen, M.I.; Hong, C.; Misra, A.; Feuerstein, B.G.; Volik, S.; Collins, C.C.; Lamborn, K.R.; Bollen, A.; Pinkel, D.; et al. Integrated genomic and epigenomic analyses pinpoint biallelic gene inactivation in tumors. Nat. Genet. 2002, 32, 453–458. [Google Scholar] [CrossRef]
- Parsons, D.W.; Jones, S.; Zhang, X.; Lin, J.C.-H.; Leary, R.J.; Angenendt, P.; Mankoo, P.; Carter, H.; Siu, I.-M.; Gallia, G.L.; et al. An Integrated Genomic Analysis of Human Glioblastoma Multiforme. Science 2008, 321, 1807–1812. [Google Scholar] [CrossRef]
- Rodger, E.J.; Chatterjee, A.; Stockwell, P.A.; Eccles, M.R. Characterization of DNA methylation changes in EBF3 and TBC1D16 associated with tumor progression and metastasis in multiple cancer types. Clin. Epigenet. 2019, 11, 114. [Google Scholar] [CrossRef] [PubMed]
- Bennett, K.L.; Karpenko, M.; Lin, M.T.; Claus, R.; Arab, K.; Dyckhoff, G.; Plinkert, P.; Herpel, E.; Smiraglia, D.; Plass, C. Frequently methylated tumor suppressor genes in head and neck squamous cell carcinoma. Cancer Res. 2008, 68, 4494–4499. [Google Scholar] [CrossRef] [PubMed]
- Hawkins, N.J.; Lee, J.H.-F.; Wong, J.J.-L.; Kwok, C.-T.; Ward, R.L.; Hitchins, M.P. MGMT methylation is associated primarily with the germline C > T SNP (rs16906252) in colorectal cancer and normal colonic mucosa. Mod. Pathol. 2009, 22, 1588–1599. [Google Scholar] [CrossRef]
- Kim, J.; Min, S.Y.; Lee, H.E.; Kim, W.H. Aberrant DNA methylation and tumor suppressive activity of the EBF3 gene in gastric carcinoma. Int. J. Cancer 2012, 130, 817–826. [Google Scholar] [CrossRef]
- Pesole, G.; Liuni, S.; Grillo, G.; Ippedico, M.; Larizza, A.; Makalowski, W.; Saccone, C. UTRdb: A specialized database of 5’ and 3’ untranslated regions of eukaryotic mRNAs. Nucleic Acids Res. 1999, 27, 188–191. [Google Scholar] [CrossRef]
- Biswas, T.; Ramana, C.V.; Srinivasan, G.; Boldogh, I.; Hazra, T.K.; Chen, Z.; Tano, K.; Thompson, E.B.; Mitra, S. Activation of human O6-methylguanine-DNA methyltransferase gene by glucocorticoid hormone. Oncogene 1999, 18, 525–532. [Google Scholar] [CrossRef] [PubMed]
- Raghunath, A.; Sundarraj, K.; Nagarajan, R.; Arfuso, F.; Bian, J.; Kumar, A.P.; Sethi, G.; Perumal, E. Antioxidant response elements: Discovery, classes, regulation and potential applications. Redox Biol. 2018, 17, 297–314. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q. Role of Nrf2 in Oxidative Stress and Toxicity. Annu. Rev. Pharmacol. Toxicol. 2013, 53, 401–426. [Google Scholar] [CrossRef]
- Niture, S.K.; Velu, C.S.; Smith, Q.R.; Bhat, G.; Srivenugopal, K.S. Increased expression of the MGMT repair protein mediated by cysteine prodrugs and chemopreventative natural products in human lymphocytes and tumor cell lines. Carcinogenesis 2006, 28, 378–389. [Google Scholar] [CrossRef] [PubMed]
- Niture, S.K.; Rao, U.S.; Srivenugopal, K.S. Chemopreventative strategies targeting the MGMT repair protein: Augmented expression in human lymphocytes and tumor cells by ethanolic and aqueous extracts of several Indian medicinal plants. Int. J. Oncol. 2006, 29, 1269–1278. [Google Scholar] [CrossRef] [PubMed]
- Paranjpe, A.; Bailey, N.I.; Konduri, S.; Bobustuc, G.C.; Ali-Osman, F.; Yusuf, M.A.; Punganuru, S.R.; Madala, H.R.; Basak, D.; Mostofa, A.; et al. New insights into estrogenic regulation of O6-methylguanine DNA-methyltransferase (MGMT) in human breast cancer cells: Co-degradation of ER-α and MGMT proteins by fulvestrant or O6-benzylguanine indicates fresh avenues for therapy. J. Biomed. Res. 2016, 30, 393–410. [Google Scholar] [CrossRef] [PubMed]
- Ohtsuji, M.; Katsuoka, F.; Kobayashi, A.; Aburatani, H.; Hayes, J.D.; Yamamoto, M. Nrf1 and Nrf2 play distinct roles inactivation of antioxidant response element-dependent genes. J. Biol. Chem. 2008, 283, 33554–33562. [Google Scholar] [CrossRef]
- Kiyama, T.; Chen, C.-K.; Wang, S.W.; Pan, P.; Ju, Z.; Wang, J.; Takada, S.; Klein, W.H.; Mao, C.-A. Essential roles of mitochondrial biogenesis regulator Nrf1 in retinal development and homeostasis. Mol. Neurodegener. 2018, 13, 1–23. [Google Scholar] [CrossRef]
- Mattingly, K.A.; Ivanova, M.M.; Riggs, K.A.; Wickramasinghe, N.S.; Barch, M.J.; Klinge, C.M. Estradiol Stimulates Transcription of Nuclear Respiratory Factor-1 and Increases Mitochondrial Biogenesis. Mol. Endocrinol. 2008, 22, 609–622. [Google Scholar] [CrossRef] [PubMed]
- Batut, P.; Dobin, A.; Plessy, C.; Carninci, P.; Gingeras, T.R. High-fidelity promoter profiling reveals widespread alternative promoter usage and transposon-driven developmental gene expression. Genome Res. 2013, 23, 169–180. [Google Scholar] [CrossRef] [PubMed]
- Jacox, E.; Gotea, V.; Ovcharenko, I.; Elnitski, L. Tissue-Specific and Ubiquitous Expression Patterns from Alternative Promoters of Human Genes. PLoS ONE 2010, 5, e12274. [Google Scholar] [CrossRef]
- Ko, L.J.; Engel, J.D. DNA-binding specificities of the GATA transcription factor family. Mol. Cell. Biol. 1993, 13, 4011–4022. [Google Scholar] [CrossRef] [PubMed]
- Granok, H.; Leibovitch, B.A.; Shaffer, C.D.; Elgin, S.C. Chromatin. Gaga over GAGA factor. Curr. Biol. 1995, 5, 238–241. [Google Scholar] [CrossRef]
- Tsai, S.-Y.; Chang, Y.-L.; Swamy, K.B.S.; Chiang, R.-L.; Huang, D.-H. GAGA factor, a positive regulator of global gene expression, modulates transcriptional pausing and organization of upstream nucleosomes. Epigenetics Chromatin 2016, 9, 32. [Google Scholar] [CrossRef]
- Grandori, C.; Cowley, S.M.; James, L.P.; Eisenman, R.N. The Myc/Max/Mad Network and the Transcriptional Control of Cell Behavior. Annu. Rev. Cell Dev. Biol. 2000, 16, 653–699. [Google Scholar] [CrossRef]
- Giardino Torchia, M.L.; Ashwell, J.D. Getting MAD at MYC. Proc. Natl. Acad. Sci. USA 2018, 115, 9821–9823. [Google Scholar] [CrossRef]
- Grombacher, T.; Eichhorn, U.; Kaina, B. p53 is involved in regulation of the DNA repair gene O6-methylguanine-DNA methyltransferase (MGMT) by DNA damaging agents. Oncogene 1998, 17, 845–851. [Google Scholar] [CrossRef]
- Bocangel, D.; Sengupta, S.; Mitra, S.; Bhakat, K.K. p53-Mediated down-regulation of the human DNA repair gene O6-methylguanine-DNA methyltransferase (MGMT) via interaction with Sp1 transcription factor. AntiCancer. Res. 2009, 29, 3741–3750. [Google Scholar] [PubMed]
- Harris, L.C.; Remack, J.S.; Houghton, P.J.; Brent, T.P. Wild-type p53 suppresses transcription of the human O6-methylguanine-DNA methyltransferase gene. Cancer Res. 1996, 56, 2029–2032. [Google Scholar] [PubMed]
- Srivenugopal, K.S.; Shou, J.; Mullapudi, S.R.; Lang, F.F., Jr.; Rao, J.S.; Ali-Osman, F. Enforced expression of wild-type p53 curtails the transcription of the O6-methylguanine-DNA methyltransferase gene in human tumor cells and enhances their sensitivity to alkylating agents. Clin. Cancer Res. 2001, 7, 1398–1409. [Google Scholar] [PubMed]
- Ma, B.; Pan, Y.; Zheng, J.; Levine, A.J.; Nussinov, R. Sequence analysis of p53 response-elements suggests multiple binding modes of the p53 tetramer to DNA targets. Nucleic Acids Res. 2007, 35, 2986–3001. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Yu, N.-K.; Kaang, B.-K. CTCF as a multifunctional protein in genome regulation and gene expression. Exp. Mol. Med. 2015, 47, e166. [Google Scholar] [CrossRef]
- Song, S.H.; Kim, T.Y. CTCF, Cohesin, and Chromatin in Human Cancer. Genom. Inform. 2017, 15, 114–122. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.C.; Wang, W.; Zhang, L.; Wang, X. A tour of 3D genome with a focus on CTCF. Semin. Cell Dev. Biol. 2019, 90, 4–11. [Google Scholar] [CrossRef]
- Kim, T.H.; Abdullaev, Z.K.; Smith, A.D.; Ching, K.A.; Loukinov, D.I.; Green, R.D.; Zhang, M.Q.; Lobanenkov, V.V.; Ren, B. Analysis of the Vertebrate Insulator Protein CTCF-Binding Sites in the Human Genome. Cell 2007, 128, 1231–1245. [Google Scholar] [CrossRef]
- Wang, H.; Maurano, M.T.; Qu, H.; Varley, K.E.; Gertz, J.; Pauli, F.; Lee, K.; Canfield, T.; Weaver, M.; Sandstrom, R.; et al. Widespread plasticity in CTCF occupancy linked to DNA methylation. Genome Res. 2012, 22, 1680–1688. [Google Scholar] [CrossRef]
- Holwerda, S.J.; de Laat, W. CTCF: The protein, the binding partners, the binding sites and their chromatin loops. Philos. Trans. R Soc. Lond. B Biol. Sci. 2013, 368, 20120369. [Google Scholar] [CrossRef] [PubMed]
- Renda, M.; Baglivo, I.; Burgess-Beusse, B.; Esposito, S.; Fattorusso, R.; Felsenfeld, G.; Pedone, P.V. Critical DNA binding interactions of the insulator protein CTCF: A small number of zinc fingers mediate strong binding, and a single finger-DNA interaction controls binding at imprinted loci. J. Biol. Chem. 2007, 282, 33336–33345. [Google Scholar] [CrossRef]
- Dávalos-Salas, M.; Furlan-Magaril, M.; González-Buendía, E.; Valdes-Quezada, C.; Ayala-Ortega, E.; Recillas-Targa, F. Gain of DNA methylation is enhanced in the absence of CTCF at the human retinoblastoma gene promoter. BMC Cancer 2011, 11, 232. [Google Scholar] [CrossRef]
- Rodriguez, C.; Borgel, J.; Court, F.; Cathala, G.; Forné, T.; Piette, J. CTCF is a DNA methylation-sensitive positive regulator of the INK/ARF locus. Biochem. Biophys. Res. Commun. 2010, 392, 129–134. [Google Scholar] [CrossRef]
- Chen, F.Y.; Harris, L.C.; Remack, J.S.; Brent, T.P. Cytoplasmic sequestration of an O6-methylguanine-DNA methyltransferase en-hancer binding protein in DNA repair-deficient human cells. Proc. Natl. Acad. Sci. USA 1997, 94, 4348–4353. [Google Scholar] [CrossRef] [PubMed]
- Flavahan, W.A.; Drier, Y.; Liau, B.B.; Gillespie, S.M.; Venteicher, A.S.; Stemmer-Rachamimov, A.O.; Suvà, M.L.; Bernstein, B.E. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature 2016, 529, 110–114. [Google Scholar] [CrossRef] [PubMed]
- Kirstein, A.; Schmid, T.E.; Combs, S.E. The Role of miRNA for the Treatment of MGMT Unmethylated Glioblastoma Multiforme. Cancers 2020, 12, 1099. [Google Scholar] [CrossRef] [PubMed]
- Slaby, O.; Lakomy, R.; Fadrus, P.; Hrstka, R.; Kren, L.; Lzicarova, E.; Smrcka, M.; Svoboda, M.; Dolezalova, H.; Novakova, J.; et al. Mi-croRNA-181 family predicts response to concomitant chemoradiotherapy with temozolomide in glioblastoma patients. Neoplasma 2010, 57, 264–269. [Google Scholar] [CrossRef] [PubMed]
- Kushwaha, D.; Ramakrishnan, V.; Ng, K.; Steed, T.; Nguyen, T.; Futalan, D.; Akers, J.C.; Sarkaria, J.; Jiang, T.; Chowdhury, D.; et al. A genome-wide miRNA screen revealed miR-603 as a MGMT-regulating miRNA in glioblastomas. Oncotarget 2014, 5, 4026–4039. [Google Scholar] [CrossRef] [PubMed]
- Khalil, S.; Fabbri, E.; Santangelo, A.; Bezzerri, V.; Cantù, C.; Di Gennaro, G.; Finotti, A.; Ghimenton, C.; Eccher, A.; Dechecchi, M.; et al. miRNA array screening reveals cooperative MGMT-regulation between miR-181d-5p and miR-409-3p in glioblastoma. Oncotarget 2016, 7, 28195–28206. [Google Scholar] [CrossRef]
- Gao, Y.T.; Chen, X.B.; Liu, H.L. Up-regulation of miR-370-3p restores glioblastoma multiforme sensitivity to temozolomide by in-fluencing MGMT expression. Sci. Rep. 2016, 6, 32972. [Google Scholar] [CrossRef]
- Slack, F.J.; Chinnaiyan, A.M. The Role of Non-coding RNAs in Oncology. Cell 2019, 179, 1033–1055. [Google Scholar] [CrossRef] [PubMed]
- Saini, H.K.; Griffiths-Jones, S.; Enright, A.J. Genomic analysis of human microRNA transcripts. Proc. Natl. Acad. Sci. USA 2007, 104, 17719–17724. [Google Scholar] [CrossRef] [PubMed]
- Tafrihi, M.; Hasheminasab, E. MiRNAs: Biology, Biogenesis, their Web-based Tools, and Databases. MicroRNA 2019, 8, 4–27. [Google Scholar] [CrossRef] [PubMed]
- Rehmsmeier, M.; Steffen, P.; Höchsmann, M.; Giegerich, R. Fast and effective prediction of microRNA/target duplexes. RNA 2004, 10, 1507–1517. [Google Scholar] [CrossRef]
- Nadaradjane, A.; Briand, J.; Bougras-Cartron, G.; Disdero, V.; Vallette, F.M.; Frenel, J.S.; Cartron, P.F. miR-370-3p is a therapeutic tool in anti-glioblastoma therapy but is not an intratumoral or cell-free circulating biomarker. Mol. Ther. Nucleic Acids 2018, 13, 642–650. [Google Scholar] [CrossRef] [PubMed]
- Hermisson, M.; Klumpp, A.; Wick, W.; Wischhusen, J.; Nagel, G.; Roos, W.; Kaina, B.; Weller, M. O6-methylguanine DNA methyltransferase and p53 status predict temozolomide sensitivity in human malignant glioma cells. J. Neurochem. 2006, 96, 766–776. [Google Scholar] [CrossRef]
- Chen, H.; Li, X.; Li, W.; Zheng, H. miR-130a can predict response to temozolomide in patients with glioblastoma multiforme, independently of O6-methylguanine-DNA methyltransferase. J. Transl. Med. 2015, 13, 69. [Google Scholar] [CrossRef]
- Qiu, S.; Lin, S.; Hu, D.; Feng, Y.; Tan, Y.; Peng, Y. Interactions of miR-323/miR-326/miR-329 and miR-130a/miR-155/miR-210 as prognostic indicators for clinical outcome of glioblastoma patients. J. Transl. Med. 2013, 11, 10. [Google Scholar] [CrossRef]
- Ziebarth, J.D.; Bhattacharya, A.; Cui, Y. CTCFBSDB 2.0: A database for CTCF-binding sites and genome organization. Nucleic Acids Res. 2013, 41, D188–D194. [Google Scholar] [CrossRef]
- Bao, L.; Zhou, M.; Cui, Y. CTCFBSDB: A CTCF-binding site database for characterization of vertebrate genomic insulators. Nucleic Acids Res. 2008, 36, D83–D87. [Google Scholar] [CrossRef]
- Gardiner-Garden, M.; Frommer, M. CpG islands in vertebrate genomes. J. Mol. Biol. 1987, 196, 261–282. [Google Scholar] [CrossRef]
- Hackenberg, M.; Barturen, G.; Carpena, P.; Luque-Escamilla, P.L.; Previti, C.; Oliver, J.L. Prediction of CpG-island function: CpG clustering vs. sliding-window methods. BMC Genom. 2010, 11, 327. [Google Scholar] [CrossRef]
- Marbach-Bar, N.; Bahat, A.; Ashkenazi, S.; Golan-Mashiach, M.; Haimov, O.; Wu, S.-Y.; Chiang, C.-M.; Puzio-Kuter, A.; Hirshfield, K.M.; Levine, A.J.; et al. DTIE, a novel core promoter element that directs start site selection in TATA-less genes. Nucleic Acids Res. 2016, 44, 1080–1094. [Google Scholar] [CrossRef]
- ENCODE_project_consortium. The ENCODE (ENCyclopedia of DNA Elements). Project Sci. 2004, 306, 636–640. [Google Scholar]
- Euskirchen, G.M.; Rozowsky, J.S.; Wei, C.-L.; Lee, W.H.; Zhang, Z.D.; Hartman, S.; Emanuelsson, O.; Stolc, V.; Weissman, S.; Gerstein, M.B.; et al. Mapping of transcription factor binding regions in mammalian cells by ChIP: Comparison of array- and sequencing-based technologies. Genome Res. 2007, 17, 898–909. [Google Scholar] [CrossRef]
- Sabo, P.J.; Kuehn, M.S.; Thurman, R.; E Johnson, B.; Johnson, E.M.; Cao, H.; Yu, M.; Rosenzweig, E.; Goldy, J.; Haydock, A.; et al. Genome-scale mapping of DNase I sensitivity in vivo using tiling DNA microarrays. Nat. Methods 2006, 3, 511–518. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; Van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Muller, R.Y.; Hammond, M.C.; Rio, D.C.; Lee, Y.J. An Efficient Method for Electroporation of Small Interfering RNAs into ENCODE Project Tier 1 GM12878 and K562 Cell Lines. J. Biomol. Tech. 2015, 26, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Preuss, I.; Thust, R.; Kaina, B. Protective effect of O6-methylguanine-DNA methyltransferase (MGMT) on the cytotoxic and re-combinogenic activity of different antineoplastic drugs. Int. J. Cancer 1996, 65, 506–512. [Google Scholar] [CrossRef]
- Cai, S.; Xu, Y.; Cooper, R.J.; Ferkowicz, M.J.; Hartwell, J.R.; Pollok, K.E.; Kelley, M.R. Mitochondrial Targeting of Human O6-Methylguanine DNA Methyltransferase Protects against Cell Killing by Chemotherapeutic Alkylating Agents. Cancer Res. 2005, 65, 3319–3327. [Google Scholar] [CrossRef]
- Griffiths-Jones, S.; Grocock, R.J.; van Dongen, S.; Bateman, A.; Enright, A.J. miRBase: microRNA sequences, targets, and gene no-menclature. Nucleic Acids Res. 2006, 34, D140–D144. [Google Scholar] [CrossRef]
- Miranda, K.C.; Huynh, T.; Tay, Y.; Ang, Y.S.; Tam, W.L.; Thomson, A.M.; Lim, B.; Rigoutsos, I. A pattern-based method for the identifi-cation of MicroRNA binding sites and their corresponding heteroduplexes. Cell 2006, 126, 1203–1217. [Google Scholar] [CrossRef] [PubMed]






| Source | Promoter ID | Map Locations at Chromosome 10/+ Strand | Span (bp) |
|---|---|---|---|
| PrESSTo/FANTOM | P1@MGMT | 129,466,944–129,467,344 | 401 |
| P2@MGMT | 129,466,905–129,467,305 | 401 | |
| Eukaryotic Promoter Database (EPD) | MGMT_1 | 129,466,745–129,467,344 | 600 |
| NCBI/Nucleotide | X61657.1 | 129,466,183–129,467,339 | 1157 |
| Ensembl 84 | ENSR00001428452 | 129,466,558–129,468,201 | 1644 |
| Revised MGMT exon-1 promoter (this study) | MGMT-P1 | 129,466,183–129,468,201 | 2019 |
| Transcriptional Regulatory Element Database (TRED) | TRED-5071 (MGMT-P2) | 129,535,540–129,536,539 | 1000 |
| CGI Annotation | Nucleotide Position 5’–3’ | CGI Length (bp) | Map Location | %GC | Obs/Exp CpG |
|---|---|---|---|---|---|
| P1-CGI | 1–2019 | 2019 | chr10:129,466,183–129,468,201 | 63 | 0.63 |
| US-CGI | 241–720 | 480 | chr10: 129,466,423–129,466,902 | 65.2 | 1.1 |
| DS-CGI | 901–1440 | 540 | chr10:129,467,083–129,467,622 | 73.1 | 0.79 |
| ncRNA | ncRNA Locus ID | ncRNA Transcript ID | The Similarity of Predicted miR with miRBase Mature miRNA Sequences | MRE Locations in MGMT and EBF3 mRNAs | No. of Identified MRE in MGMT and EBF3 mRNAs | Leftmost Position (5’→3’) of Predicted MGMT and EBF3 mRNAs Target Sites. |
|---|---|---|---|---|---|---|
| miRNA | ENSG00000266061 | ENST00000585165.1 | hsa-miR-574-5p | MGMT-3’-UTR | 1 | 908 |
| EBF3-E8, E13, 3’-UTR | 5 | 759, 1309, 3500, 3821, 4101 | ||||
| MIR4297 | ENST00000579857.1 | has-miR-4297 | MGMT-E4 | 1 | 406 | |
| EBF3 | 0 | - | ||||
| Antisense RNA | ENSG00000275005 | ENST00000614150.1 | hsa-miR-4645-3p | MGMT | 0 | - |
| EBF3-E2, E4 | 2 | 353, 423 | ||||
| ENSG00000275327 | ENST00000617939.1 | hsa-miR-942-5p | MGMT-3’-UTR | 1 | 1093 | |
| EBF3-E5 & 3’-UTR | 2 | 499, 3989 | ||||
| IncRNA | ENSG00000227374 | ENST00000428273.1 | hsa-miR-339-5p | MGMT | 0 | - |
| EBF3-E1, E5, 3’-UTR | 3 | 186, 505, 3290 | ||||
| ENSG00000283141 | ENST00000635764.1 | hsa-miR-6862-5p | MGMT-3’-UTR | 1 | 946 | |
| EBF3 | 0 | - |
| ncRNA Locus ID | Predicted miR by miRBase Search Tool | Predicted MRE by RNA22 v2 Tool | ||||
|---|---|---|---|---|---|---|
| Sequence | hsa-miR | Score | E-value | Heteroduplex | p Value | |
| ENSG00000266061 | UGUGUGUGUGUGUGUGUGUGUGU | hsa-miR-574-5p | 100 | 0.009 | 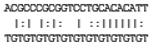 | 0.36 |
| miRNA4297 | UGCCUUCCUGUCUGUG | hsa-miR-4297 | 88 | 0.22 |  | 0.14 |
| ENSG00000275005 | UAGUUCUUGCCUGG | hsa-miR-4645-3p | 70 | 4.2 | - | - |
| ENSG00000275327 | ACAUGGCCAAAACAGAG | hsa-miR-942-5p | 67 | 3.6 |  | 0.072 |
| ENSG00000227374 | AGGUUCCCUCUGGCCGC | hsa-miR-4726-3p | 67 | 6.4 | - | - |
| ENSG00000283141 | GCAUGCUGGGAGAGACU | hsa-miR-6862-5p | 67 | 2.4 |  | 0.225 |
| MIR127 | UCGGAUCCGUCUGAGCUUGGCU | hsa-miR-127-3p | 110 | 6 × 10−4 | 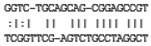 | 0.107 |
| MIR370 | GCCUGCUGGGGUGGAACCUGGU | hsa-miR-370-3p | 110 | 7 × 10−4 |  | 0.225 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ibrahim Al-Obaide, M.A.; Arutla, V.; Bacolod, M.D.; Wang, W.; Zhang, R.; Srivenugopal, K.S. Genomic Space of MGMT in Human Glioma Revisited: Novel Motifs, Regulatory RNAs, NRF1, 2, and CTCF Involvement in Gene Expression. Int. J. Mol. Sci. 2021, 22, 2492. https://doi.org/10.3390/ijms22052492
Ibrahim Al-Obaide MA, Arutla V, Bacolod MD, Wang W, Zhang R, Srivenugopal KS. Genomic Space of MGMT in Human Glioma Revisited: Novel Motifs, Regulatory RNAs, NRF1, 2, and CTCF Involvement in Gene Expression. International Journal of Molecular Sciences. 2021; 22(5):2492. https://doi.org/10.3390/ijms22052492
Chicago/Turabian StyleIbrahim Al-Obaide, Mohammed A., Viswanath Arutla, Manny D. Bacolod, Wei Wang, Ruiwen Zhang, and Kalkunte S. Srivenugopal. 2021. "Genomic Space of MGMT in Human Glioma Revisited: Novel Motifs, Regulatory RNAs, NRF1, 2, and CTCF Involvement in Gene Expression" International Journal of Molecular Sciences 22, no. 5: 2492. https://doi.org/10.3390/ijms22052492
APA StyleIbrahim Al-Obaide, M. A., Arutla, V., Bacolod, M. D., Wang, W., Zhang, R., & Srivenugopal, K. S. (2021). Genomic Space of MGMT in Human Glioma Revisited: Novel Motifs, Regulatory RNAs, NRF1, 2, and CTCF Involvement in Gene Expression. International Journal of Molecular Sciences, 22(5), 2492. https://doi.org/10.3390/ijms22052492