ANT1 Activation and Inhibition Patterns Support the Fatty Acid Cycling Mechanism for Proton Transport
Abstract
1. Introduction
2. Results
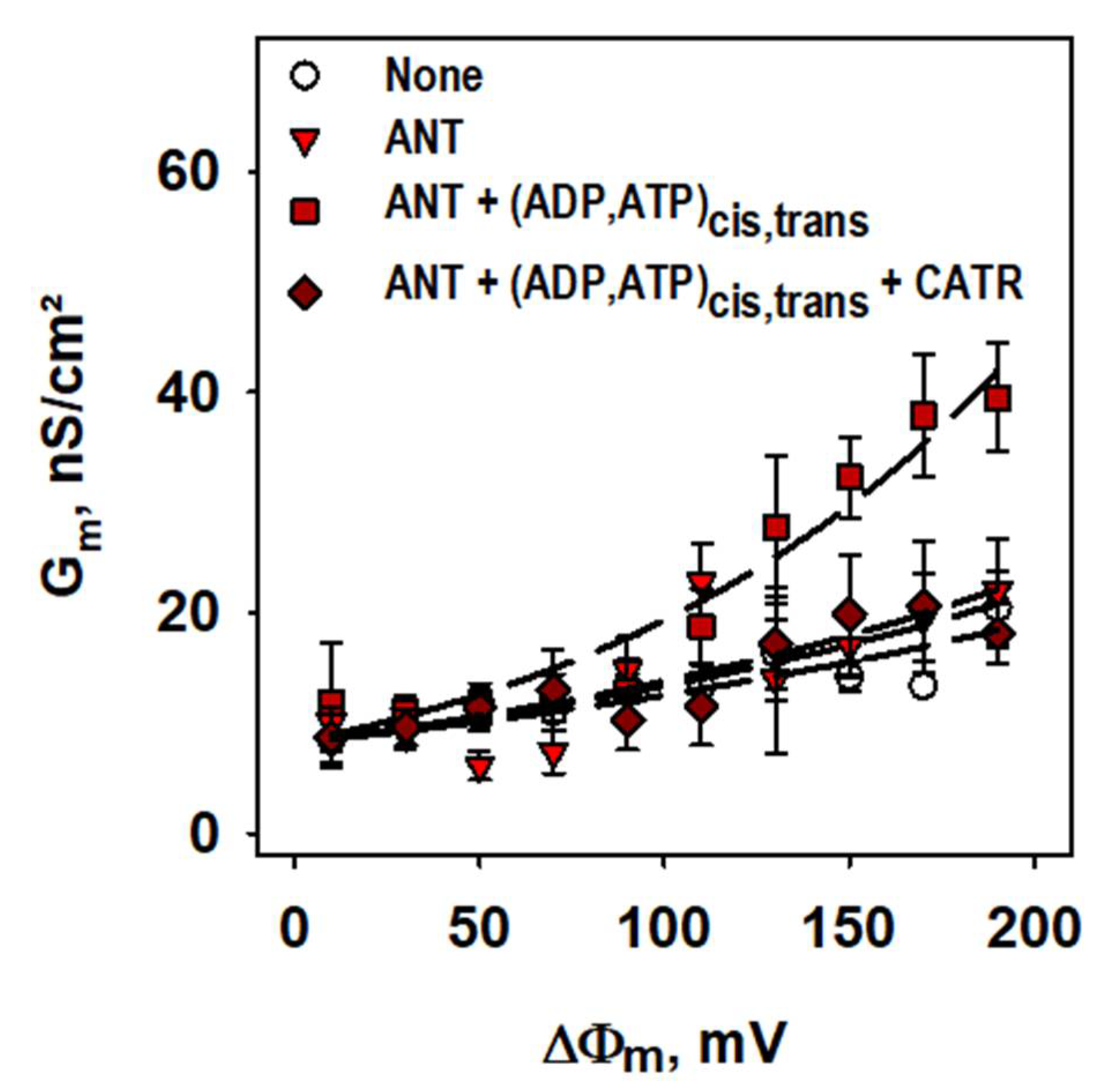
2.1. ANT1-Mediated Substrate Transport
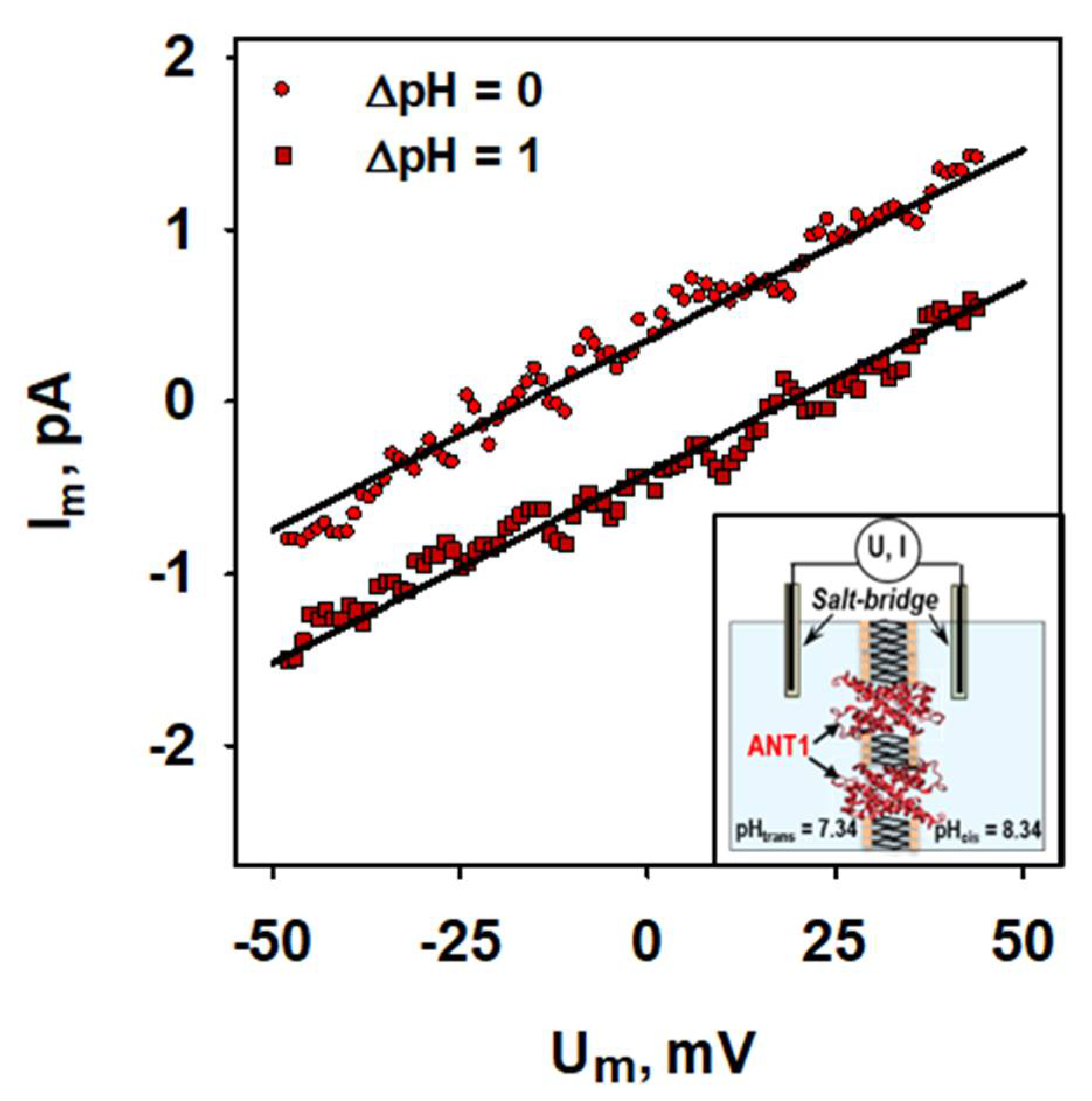
2.2. Basal Proton Leak
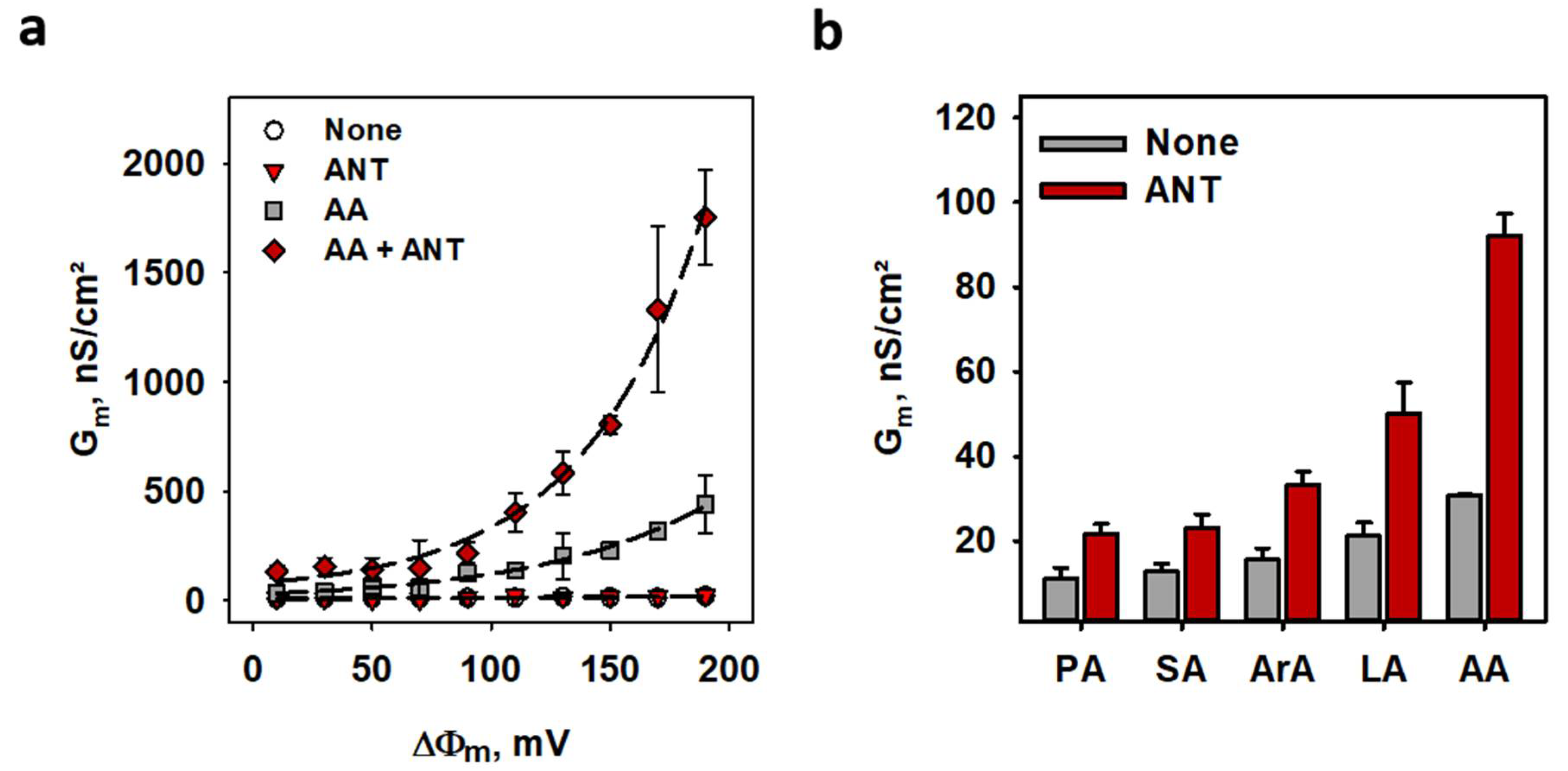
2.3. ANT1-Mediated Proton Transport in the Presence of FA
2.4. Proton Turnover Number of ANT1
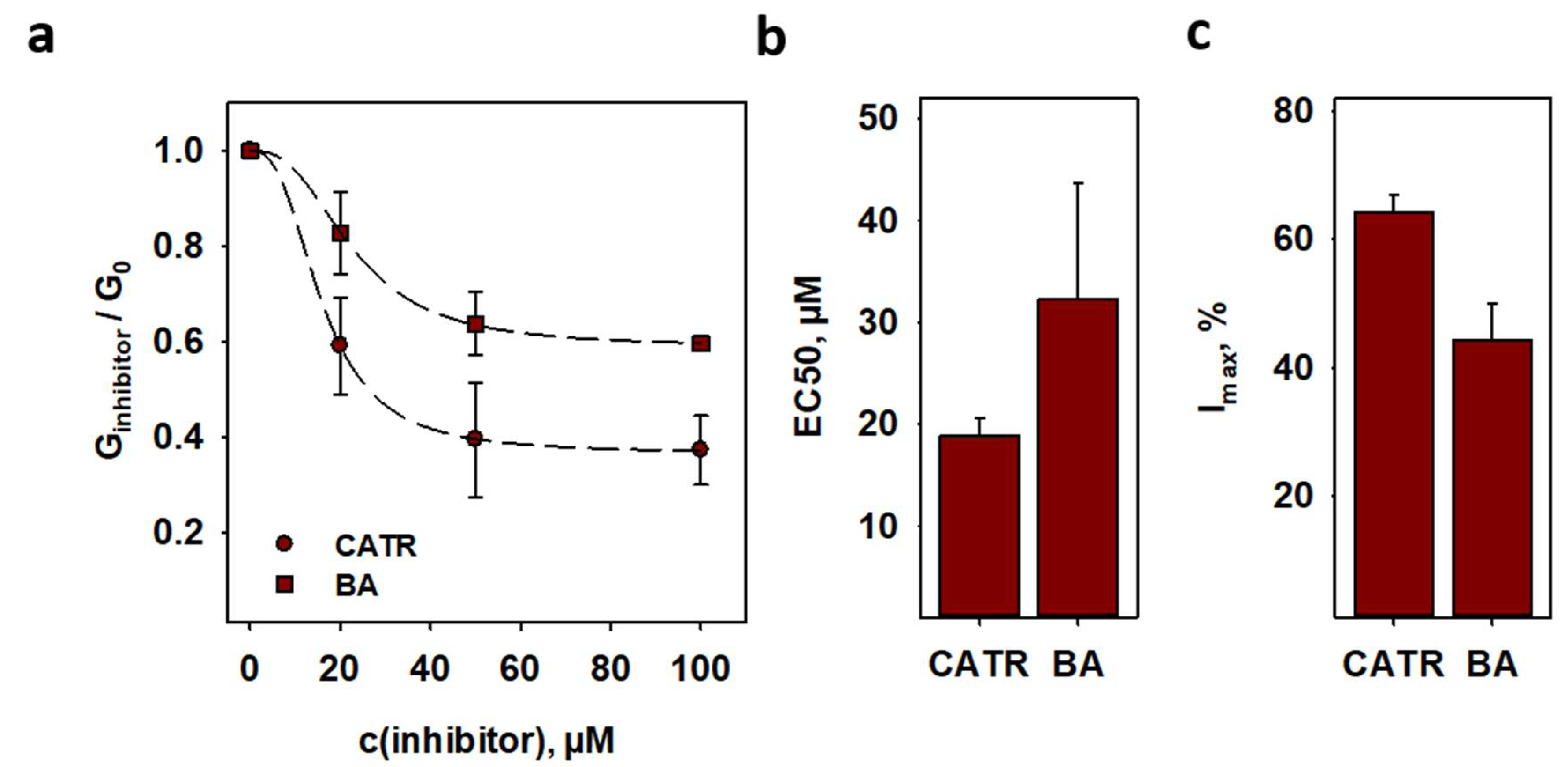
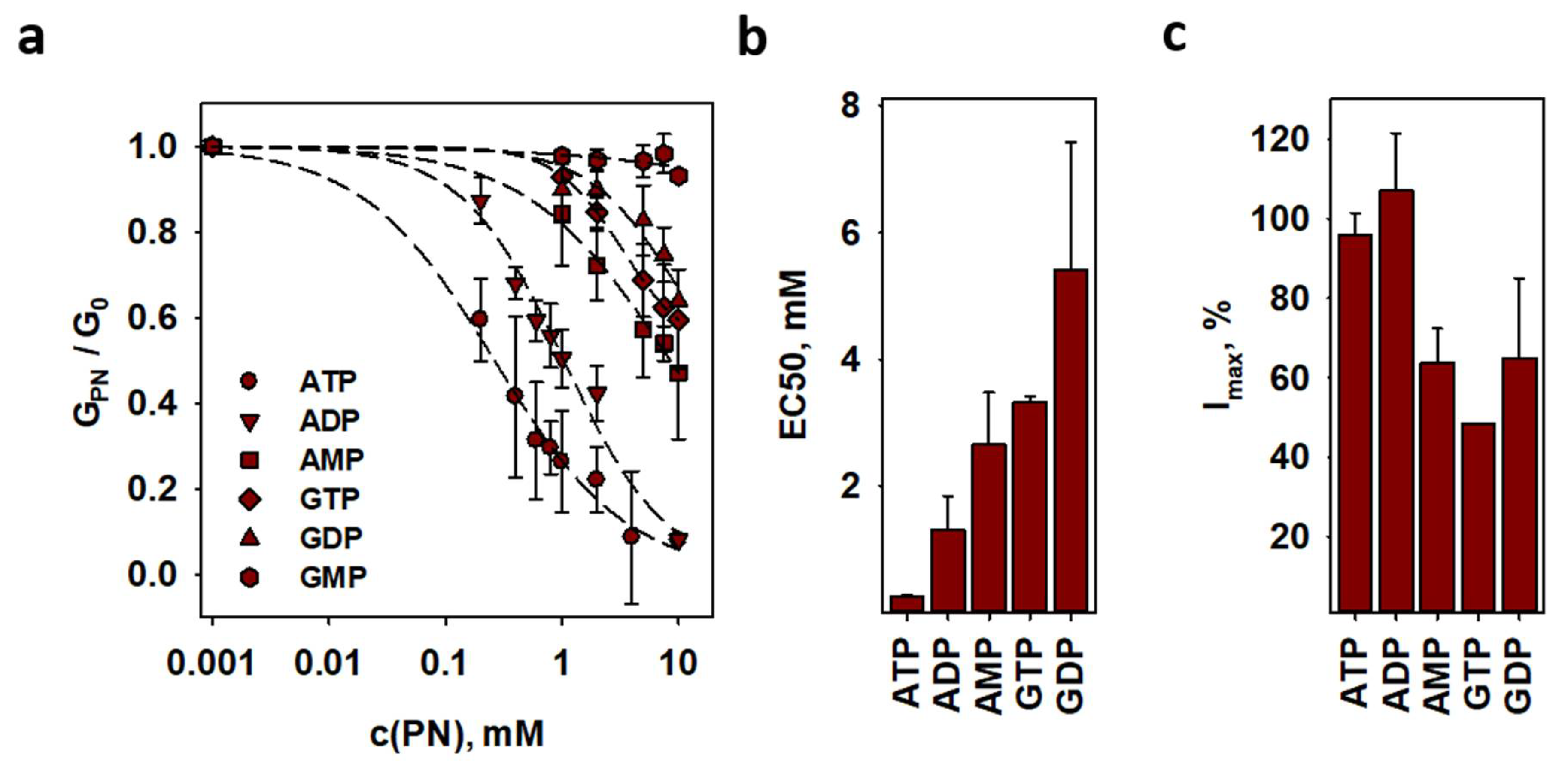
2.5. Inhibition of ANT1-Mediated Proton Transport
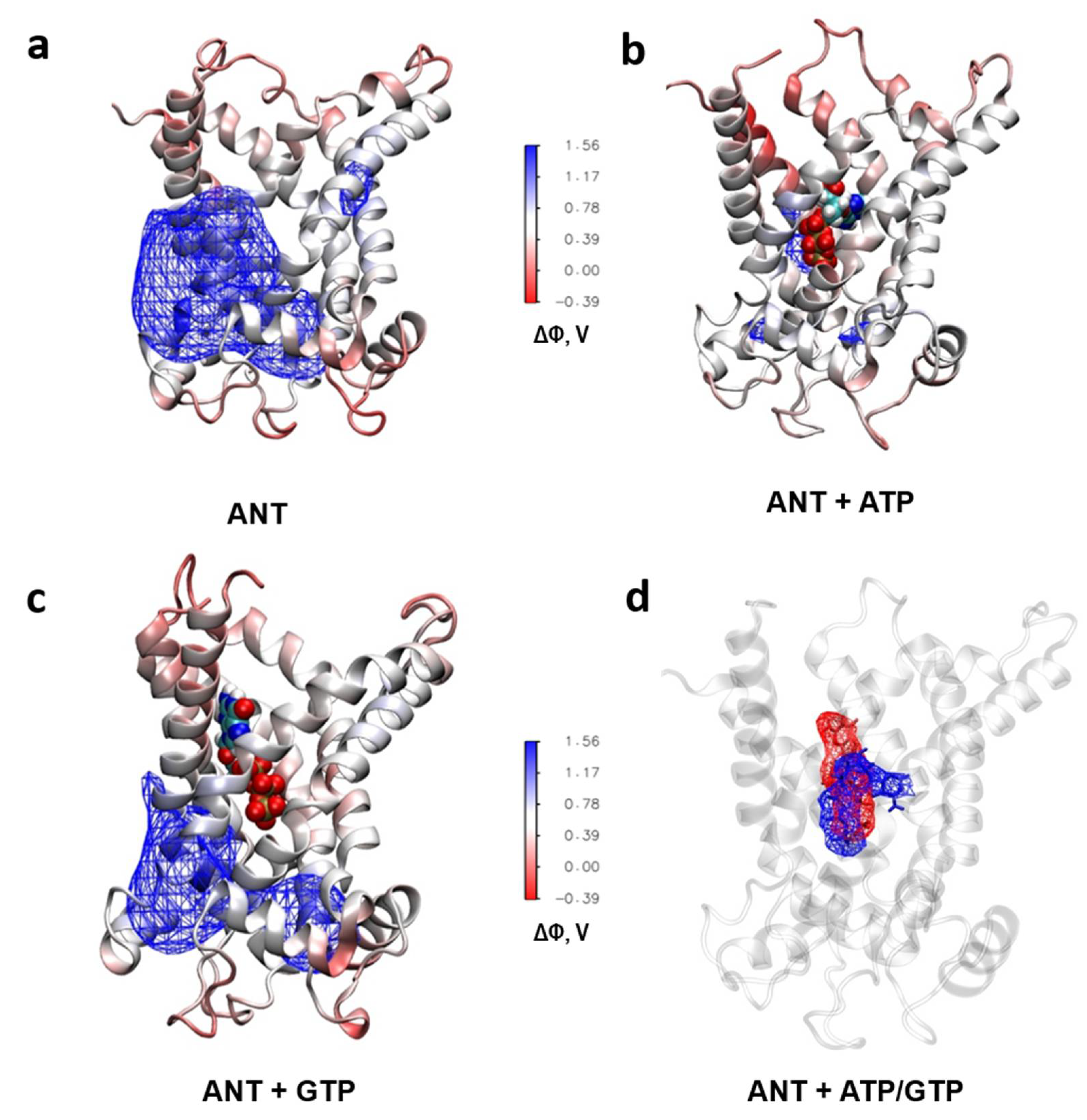
2.6. Analysis of the ANT´s Surface Electrostatic Potential using Molecular Dynamic Simulations
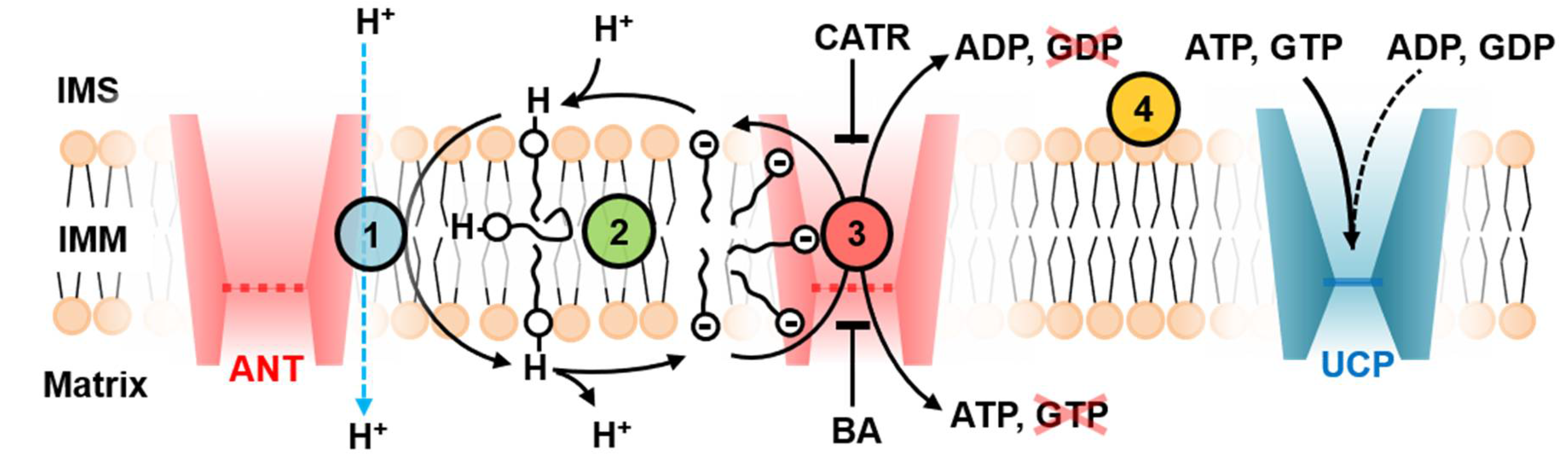
3. Discussion
4. Methods
4.1. Chemicals
4.2. Cloning, Purification and Reconstitution of Murine ANT1
4.3. Exchange Rate Measurements of mANT1
4.4. Electrophysiological Measurements of mANT1
4.5. Fluorescence Correlation Spectroscopy (FCS)
4.6. Molecular Dynamics Simulations
4.7. Statistics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brand, M.D. The proton leak across the mitochondrial inner membrane. Biochim. Biophys. Acta 1990, 1018, 128–133. [Google Scholar] [CrossRef]
- Rolfe, D.F.; Brand, M.D. Contribution of mitochondrial proton leak to skeletal muscle respiration and to standard metabolic rate. Am. J. Physiol. 1996, 271 Pt 1, C1380–C1389. [Google Scholar] [CrossRef]
- Rolfe, D.F.; Brand, M.D. The physiological significance of mitochondrial proton leak in animal cells and tissues. Biosci. Rep. 1997, 17, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Krauss, S.; Zhang, C.Y.; Lowell, B.B. The mitochondrial uncoupling-protein homologues. Nat. Rev. Mol. Cell Biol. 2005, 6, 248–261. [Google Scholar] [CrossRef] [PubMed]
- Skulachev, V.P. Uncoupling: New approaches to an old problem of bioenergetics. Biochim. Biophys. Acta 1998, 1363, 100–124. [Google Scholar] [CrossRef]
- Beck, V.; Jaburek, M.; Demina, T.; Rupprecht, A.; Porter, R.K.; Jezek, P.; Pohl, E.E. Polyunsaturated fatty acids activate human uncoupling proteins 1 and 2 in planar lipid bilayers. FASEB J. 2007, 21, 1137–1144. [Google Scholar] [CrossRef]
- Macher, G.; Koehler, M.; Rupprecht, A.; Kreiter, J.; Hinterdorfer, P.; Pohl, E.E. Inhibition of mitochondrial UCP1 and UCP3 by purine nucleotides and phosphate. Biochim. Biophys. Acta Biomembr. 2018, 1860, 664–672. [Google Scholar] [CrossRef] [PubMed]
- Zackova, M.; Jezek, P. Reconstitution of novel mitochondrial uncoupling proteins UCP2 and UCP3. Biosci. Rep. 2002, 22, 33–46. [Google Scholar] [CrossRef]
- Hoang, T.; Smith, M.D.; Jelokhani-Niaraki, M. Toward understanding the mechanism of ion transport activity of neuronal uncoupling proteins UCP2, UCP4, and UCP5. Biochemistry 2012, 51, 4004–4014. [Google Scholar] [CrossRef]
- Pohl, E.E.; Rupprecht, A.; Macher, G.; Hilse, K.E. Important Trends in UCP3 Investigation. Front. Physiol. 2019, 10, 470. [Google Scholar] [CrossRef] [PubMed]
- Heldt, H.W.; Jacobs, H.; Klingenberg, M. Endogenous Adp of Mitochondria, an Early Phosphate Acceptor of Oxidative Phosphorylation as Disclosed by Kinetic Studies with C14 Labelled Adp and Atp and with Atractyloside. Biochem. Biophys. Res. Commun. 1965, 18, 174–179. [Google Scholar] [CrossRef]
- Pfaff, E.; Klingenberg, M.; Heldt, H.W. Unspecific permeation and specific exchange of adenine nucleotides in liver mitochondria. Biochim. Biophys. Acta 1965, 104, 312–315. [Google Scholar] [CrossRef]
- Vignais, P.V. The mitochondrial adenine nucleotide translocator. J. Bioenerg. 1976, 8, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Kunji, E.R.; Aleksandrova, A.; King, M.S.; Majd, H.; Ashton, V.L.; Cerson, E.; Springett, R.; Kibalchenko, M.; Tavoulari, S.; Crichton, P.G.; et al. The transport mechanism of the mitochondrial ADP/ATP carrier. Biochim. Biophys. Acta 2016, 1863, 2379–2393. [Google Scholar] [CrossRef]
- Klingenberg, M. The ADP and ATP transport in mitochondria and its carrier. Biochim. Biophys. Acta 2008, 1778, 1978–2021. [Google Scholar] [CrossRef] [PubMed]
- Andreyev, A.Y.; Bondareva, T.O.; Dedukhova, V.I.; Mokhova, E.N.; Skulachev, V.P.; Volkov, N.I. Carboxyatractylate inhibits the uncoupling effect of free fatty acids. FEBS Lett. 1988, 226, 265–269. [Google Scholar] [CrossRef]
- Andreyev, A.Y.; Bondareva, T.O.; Dedukhova, V.I.; Mokhova, E.N.; Skulachev, V.P.; Tsofina, L.M.; Volkov, N.I.; Vygodina, T.V. The ATP/ADP-antiporter is involved in the uncoupling effect of fatty acids on mitochondria. Eur. J. Biochem. 1989, 182, 585–592. [Google Scholar] [CrossRef]
- Brustovetsky, N.; Klingenberg, M. The reconstituted ADP/ATP carrier can mediate H+ transport by free fatty acids, which is further stimulated by mersalyl. J. Biol. Chem. 1994, 269, 27329–27336. [Google Scholar] [CrossRef]
- Shabalina, I.G.; Kramarova, T.V.; Nedergaard, J.; Cannon, B. Carboxyatractyloside effects on brown-fat mitochondria imply that the adenine nucleotide translocator isoforms Ant1 and Ant2 may be responsible for basal and fatty acid-induced uncoupling, respectively. Biochem. J. 2006, 399, 405–414. [Google Scholar] [CrossRef]
- Brand, M.D.; Pakay, J.L.; Ocloo, A.; Kokoszka, J.; Wallace, D.C.; Brookes, P.S.; Cornwall, E.J. The basal proton conductance of mitochondria depends on adenine nucleotide translocase content. Biochem. J. 2005, 392 Pt 2, 353–362. [Google Scholar] [CrossRef]
- Bertholet, A.M.; Chouchani, E.T.; Kazak, L.; Angelin, A.; Fedorenko, A.; Long, J.Z.; Vidoni, S.; Garrity, R.; Cho, J.; Terada, N.; et al. H(+) transport is an integral function of the mitochondrial ADP/ATP carrier. Nature 2019, 571, 515–520. [Google Scholar] [CrossRef] [PubMed]
- Kamp, F.; Hamilton, J.A. pH gradients across phospholipid membranes caused by fast flip-flop of un-ionized fatty acids. Proc. Natl. Acad. Sci. USA 1992, 89, 11367–11370. [Google Scholar] [CrossRef] [PubMed]
- Pohl, E.E.; Peterson, U.; Sun, J.; Pohl, P. Changes of intrinsic membrane potentials induced by flip-flop of long-chain fatty acids. Biochemistry 2000, 39, 1834–1839. [Google Scholar] [CrossRef] [PubMed]
- Skulachev, V.P. Fatty acid circuit as a physiological mechanism of uncoupling of oxidative phosphorylation. FEBS Lett. 1991, 294, 158–162. [Google Scholar] [CrossRef]
- Fedorenko, A.; Lishko, P.V.; Kirichok, Y. Mechanism of fatty-acid-dependent UCP1 uncoupling in brown fat mitochondria. Cell 2012, 151, 400–413. [Google Scholar] [CrossRef] [PubMed]
- Kreiter, J.; Beitz, E.; Pohl, E.E. A Fluorescence-Based Method to Measure ADP/ATP Exchange of Recombinant Adenine Nucleotide Translocase in Liposomes. Biomolecules 2020, 10, 685. [Google Scholar] [CrossRef] [PubMed]
- Kramer, R.; Klingenberg, M. Electrophoretic control of reconstituted adenine nucleotide translocation. Biochemistry 1982, 21, 1082–1089. [Google Scholar] [CrossRef] [PubMed]
- Heidkamper, D.; Muller, V.; Nelson, D.R.; Klingenberg, M. Probing the role of positive residues in the ADP/ATP carrier from yeast. The effect of six arginine mutations on transport and the four ATP versus ADP exchange modes. Biochemistry 1996, 35, 16144–16152. [Google Scholar] [CrossRef] [PubMed]
- King, M.S.; Kerr, M.; Crichton, P.G.; Springett, R.; Kunji, E.R.S. Formation of a cytoplasmic salt bridge network in the matrix state is a fundamental step in the transport mechanism of the mitochondrial ADP/ATP carrier. Biochim. Biophys. Acta 2016, 1857, 14–22. [Google Scholar] [CrossRef] [PubMed]
- Rupprecht, A.; Sokolenko, E.A.; Beck, V.; Ninnemann, O.; Jaburek, M.; Trimbuch, T.; Klishin, S.S.; Jezek, P.; Skulachev, V.P.; Pohl, E.E. Role of the transmembrane potential in the membrane proton leak. Biophys. J 2010, 98, 1503–1511. [Google Scholar] [CrossRef] [PubMed]
- Kreiter, J.; Pohl, E.E. A Micro-agar Salt Bridge Electrode for Analyzing the Proton Turnover Rate of Recombinant Membrane Proteins. J. Vis. Exp. 2019, 143, e58552. [Google Scholar] [CrossRef]
- Erokhova, L.; Horner, A.; Kugler, P.; Pohl, P. Monitoring single-channel water permeability in polarized cells. J. Biol. Chem. 2011, 286, 39926–39932. [Google Scholar] [CrossRef]
- Nicholls, D.G. The effective proton conductance of the inner membrane of mitochondria from brown adipose tissue. Dependency on proton electrochemical potential gradient. Eur. J. Biochem. 1977, 77, 349–356. [Google Scholar] [CrossRef] [PubMed]
- Klingenberg, M.; Winkler, E. The reconstituted isolated uncoupling protein is a membrane potential driven H+ translocator. EMBO J. 1985, 4, 3087–3092. [Google Scholar] [CrossRef]
- Urbankova, E.; Voltchenko, A.; Pohl, P.; Jezek, P.; Pohl, E.E. Transport kinetics of uncoupling proteins. Analysis of UCP1 reconstituted in planar lipid bilayers. J. Biol. Chem. 2003, 278, 32497–32500. [Google Scholar] [CrossRef] [PubMed]
- Mifsud, J.; Ravaud, S.; Krammer, E.M.; Chipot, C.; Kunji, E.R.; Pebay-Peyroula, E.; Dehez, F. The substrate specificity of the human ADP/ATP carrier AAC1. Mol. Membr. Biol. 2013, 30, 160–168. [Google Scholar] [CrossRef] [PubMed]
- Krammer, E.M.; Ravaud, S.; Dehez, F.; Frelet-Barrand, A.; Pebay-Peyroula, E.; Chipot, C. High-chloride concentrations abolish the binding of adenine nucleotides in the mitochondrial ADP/ATP carrier family. Biophys. J. 2009, 97, L25–L27. [Google Scholar] [CrossRef]
- Pashkovskaya, A.A.; Vazdar, M.; Zimmermann, L.; Jovanovic, O.; Pohl, P.; Pohl, E.E. Mechanism of Long-Chain Free Fatty Acid Protonation at the Membrane-Water Interface. Biophys. J. 2018, 114, 2142–2151. [Google Scholar] [CrossRef] [PubMed]
- Ruprecht, J.J.; Kunji, E.R. Structural changes in the transport cycle of the mitochondrial ADP/ATP carrier. Curr. Opin. Struct. Biol. 2019, 57, 135–144. [Google Scholar] [CrossRef] [PubMed]
- Pebay-Peyroula, E.; Dahout-Gonzalez, C.; Kahn, R.; Trezeguet, V.; Lauquin, G.J.; Brandolin, G. Structure of mitochondrial ADP/ATP carrier in complex with carboxyatractyloside. Nature 2003, 426, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Jaburek, M.; Varecha, M.; Jezek, P.; Garlid, K.D. Alkylsulfonates as probes of uncoupling protein transport mechanism. Ion pair transport demonstrates that direct H(+) translocation by UCP1 is not necessary for uncoupling. J. Biol. Chem. 2001, 276, 31897–31905. [Google Scholar]
- Jezek, P.; Holendova, B.; Garlid, K.D.; Jaburek, M. Mitochondrial Uncoupling Proteins: Subtle Regulators of Cellular Redox Signaling. Antioxid. Redox Signal. 2018, 29, 667–714. [Google Scholar] [PubMed]
- Wang, Y.; Tajkhorshid, E. Electrostatic funneling of substrate in mitochondrial inner membrane carriers. Proc. Natl. Acad. Sci. USA 2008, 105, 9598–9603. [Google Scholar] [CrossRef] [PubMed]
- Dehez, F.; Pebay-Peyroula, E.; Chipot, C. Binding of ADP in the mitochondrial ADP/ATP carrier is driven by an electrostatic funnel. J. Am. Chem. Soc. 2008, 130, 12725–12733. [Google Scholar] [CrossRef]
- Pietropaolo, A.; Pierri, C.L.; Palmieri, F.; Klingenberg, M. The switching mechanism of the mitochondrial ADP/ATP carrier explored by free-energy landscapes. Biochim. Biophys. Acta 2016, 1857, 772–781. [Google Scholar] [CrossRef]
- Škulj, S.; Brkljača, Z.; Vazdar, M. Molecular Dynamics Simulations of the Elusive Matrix-Open State of Mitochondrial ADP/ATP Carrier. Isr. J. Chem. 2020, 60, 735–743. [Google Scholar] [CrossRef]
- Samartsev, V.N.; Smirnov, A.V.; Zeldi, I.P.; Markova, O.V.; Mokhova, E.N.; Skulachev, V.P. Involvement of aspartate/glutamate antiporter in fatty acid-induced uncoupling of liver mitochondria. Biochim. Biophys. Acta 1997, 1319, 251–257. [Google Scholar] [CrossRef]
- Wieckowski, M.R.; Wojtczak, L. Involvement of the dicarboxylate carrier in the protonophoric action of long-chain fatty acids in mitochondria. Biochem. Biophys. Res. Commun. 1997, 232, 414–417. [Google Scholar] [PubMed]
- Engstova, H.; Zackova, M.; Ruzicka, M.; Meinhardt, A.; Hanus, J.; Kramer, R.; Jezek, P. Natural and azido fatty acids inhibit phosphate transport and activate fatty acid anion uniport mediated by the mitochondrial phosphate carrier. J. Biol. Chem. 2001, 276, 4683–4691. [Google Scholar] [CrossRef]
- Yu, X.X.; Lewin, D.A.; Zhong, A.; Brush, J.; Schow, P.W.; Sherwood, S.W.; Pan, G.; Adams, S.H. Overexpression of the human 2-oxoglutarate carrier lowers mitochondrial membrane potential in HEK-293 cells: Contrast with the unique cold-induced mitochondrial carrier CGI-69. Biochem. J. 2001, 353 Pt 2, 369–375. [Google Scholar] [CrossRef]
- Demine, S.; Renard, P.; Arnould, T. Mitochondrial Uncoupling: A Key Controller of Biological Processes in Physiology and Diseases. Cells 2019, 8, 795. [Google Scholar] [CrossRef]
- Beck, V.; Jaburek, M.; Breen, E.P.; Porter, R.K.; Jezek, P.; Pohl, E.E. A new automated technique for the reconstitution of hydrophobic proteins into planar bilayer membranes. Studies of human recombinant uncoupling protein 1. Biochim. Biophys. Acta 2006, 1757, 474–479. [Google Scholar] [CrossRef] [PubMed]
- Knyazev, D.G.; Lents, A.; Krause, E.; Ollinger, N.; Siligan, C.; Papinski, D.; Winter, L.; Horner, A.; Pohl, P. The bacterial translocon SecYEG opens upon ribosome binding. J. Biol. Chem. 2013, 288, 17941–17946. [Google Scholar] [CrossRef] [PubMed]
- Majima, H.J.; Oberley, T.D.; Furukawa, K.; Mattson, M.P.; Yen, H.C.; Szweda, L.I.; St Clair, D.K. Prevention of mitochondrial injury by manganese superoxide dismutase reveals a primary mechanism for alkaline-induced cell death. J. Biol. Chem. 1998, 273, 8217–8224. [Google Scholar] [CrossRef] [PubMed]
- Magde, D.; Elson, E.L.; Webb, W.W. Fluorescence correlation spectroscopy. II. An experimental realization. Biopolymers 1974, 13, 29–61. [Google Scholar] [CrossRef]
- Boscia, A.L.; Treece, B.W.; Mohammadyani, D.; Klein-Seetharaman, J.; Braun, A.R.; Wassenaar, T.A.; Klosgen, B.; Tristram-Nagle, S. X-ray structure, thermodynamics, elastic properties and MD simulations of cardiolipin/dimyristoylphosphatidylcholine mixed membranes. Chem. Phys. Lipids 2014, 178, 1–10. [Google Scholar] [CrossRef] [PubMed]
- De Vries, A.H.; Mark, A.E.; Marrink, S.J. The Binary Mixing Behavior of Phospholipids in a Bilayer: A Molecular Dynamics Study. J. Phys. Chem. B 2004, 108, 2454–2463. [Google Scholar] [CrossRef]
- Webb, B.; Sali, A. Comparative Protein Structure Modeling Using MODELLER. Curr. Protoc. Bioinform. 2014, 47, 5.6.1–5.6.37. [Google Scholar] [CrossRef]
- Jo, S.; Lim, J.B.; Klauda, J.B.; Im, W. CHARMM-GUI Membrane Builder for mixed bilayers and its application to yeast membranes. Biophys. J. 2009, 97, 50–58. [Google Scholar] [CrossRef]
- Wu, E.L.; Cheng, X.; Jo, S.; Rui, H.; Song, K.C.; Davila-Contreras, E.M.; Qi, Y.; Lee, J.; Monje-Galvan, V.; Venable, R.M.; et al. CHARMM-GUI Membrane Builder toward realistic biological membrane simulations. J. Comput. Chem. 2014, 35, 1997–2004. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Cheng, X.; Swails, J.M.; Yeom, M.S.; Eastman, P.K.; Lemkul, J.A.; Wei, S.; Buckner, J.; Jeong, J.C.; Qi, Y.; et al. CHARMM-GUI Input Generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM Simulations Using the CHARMM36 Additive Force Field. J. Chem. Theory Comput. 2016, 12, 405–413. [Google Scholar] [CrossRef] [PubMed]
- Jo, S.; Kim, T.; Im, W. Automated Builder and Database of Protein/Membrane Complexes for Molecular Dynamics Simulations. PLoS ONE 2007, 2, e880. [Google Scholar] [CrossRef]
- Nosé, S. A molecular dynamics method for simulations in the canonical ensemble. Mol. Phys. 1984, 52, 255–268. [Google Scholar] [CrossRef]
- Parrinello, M.; Rahman, A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar] [CrossRef]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef]
- Huang, J.; Rauscher, S.; Nawrocki, G.; Ran, T.; Feig, M.; de Groot, B.L.; Grubmuller, H.; MacKerell, A.D., Jr. CHARMM36m: An improved force field for folded and intrinsically disordered proteins. Nat. Methods 2017, 14, 71–73. [Google Scholar] [CrossRef]
- Aksimentiev, A.; Schulten, K. Imaging alpha-hemolysin with molecular dynamics: Ionic conductance, osmotic permeability, and the electrostatic potential map. Biophys. J. 2005, 88, 3745–3761. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kreiter, J.; Rupprecht, A.; Škulj, S.; Brkljača, Z.; Žuna, K.; Knyazev, D.G.; Bardakji, S.; Vazdar, M.; Pohl, E.E. ANT1 Activation and Inhibition Patterns Support the Fatty Acid Cycling Mechanism for Proton Transport. Int. J. Mol. Sci. 2021, 22, 2490. https://doi.org/10.3390/ijms22052490
Kreiter J, Rupprecht A, Škulj S, Brkljača Z, Žuna K, Knyazev DG, Bardakji S, Vazdar M, Pohl EE. ANT1 Activation and Inhibition Patterns Support the Fatty Acid Cycling Mechanism for Proton Transport. International Journal of Molecular Sciences. 2021; 22(5):2490. https://doi.org/10.3390/ijms22052490
Chicago/Turabian StyleKreiter, Jürgen, Anne Rupprecht, Sanja Škulj, Zlatko Brkljača, Kristina Žuna, Denis G. Knyazev, Sarah Bardakji, Mario Vazdar, and Elena E. Pohl. 2021. "ANT1 Activation and Inhibition Patterns Support the Fatty Acid Cycling Mechanism for Proton Transport" International Journal of Molecular Sciences 22, no. 5: 2490. https://doi.org/10.3390/ijms22052490
APA StyleKreiter, J., Rupprecht, A., Škulj, S., Brkljača, Z., Žuna, K., Knyazev, D. G., Bardakji, S., Vazdar, M., & Pohl, E. E. (2021). ANT1 Activation and Inhibition Patterns Support the Fatty Acid Cycling Mechanism for Proton Transport. International Journal of Molecular Sciences, 22(5), 2490. https://doi.org/10.3390/ijms22052490







