Analysis of blaCHDL Genes and Insertion Sequences Related to Carbapenem Resistance in Acinetobacter baumannii Clinical Strains Isolated in Warsaw, Poland
Abstract
1. Introduction
2. Results
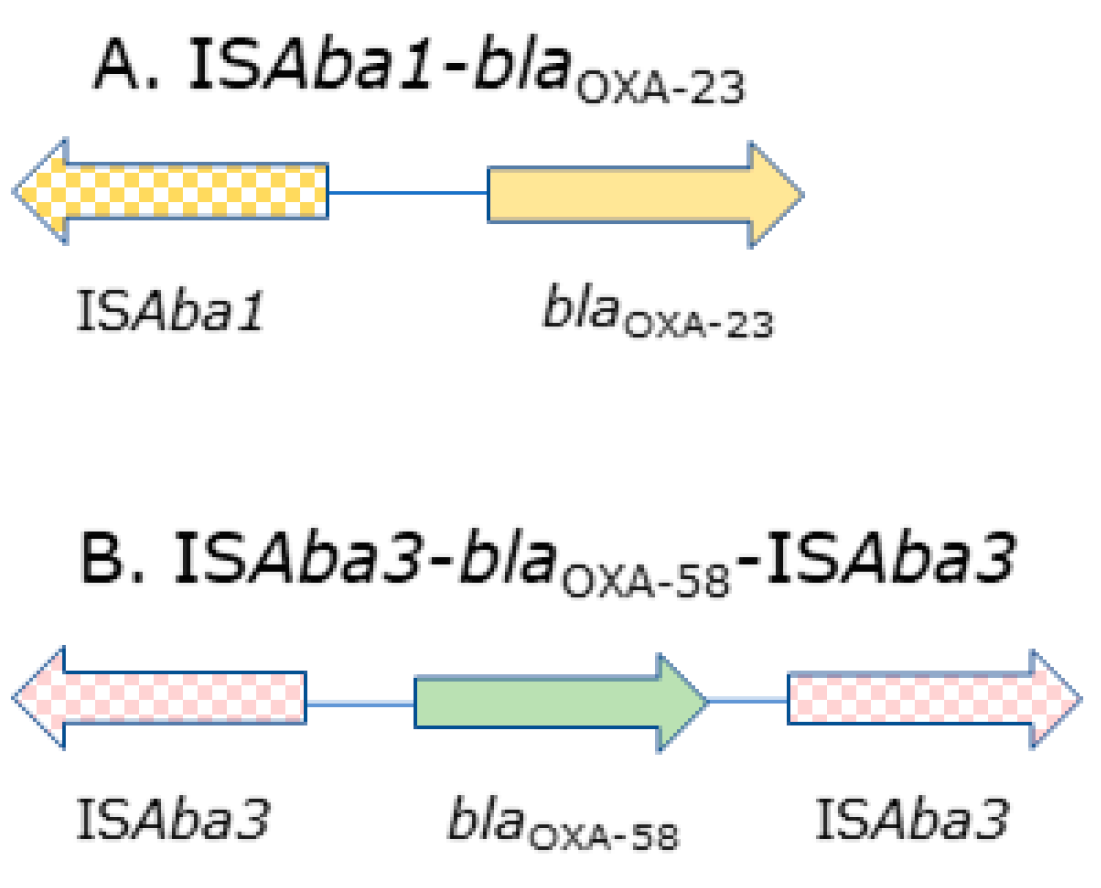
2.1. Distribution of blaCHDL Genes and Insertion Sequences
2.2. Antimicrobial Agent Resistance Profiles in Relation to blaCHDL Genes Presence
2.3. Phenotypic Detection of Carbapenemases
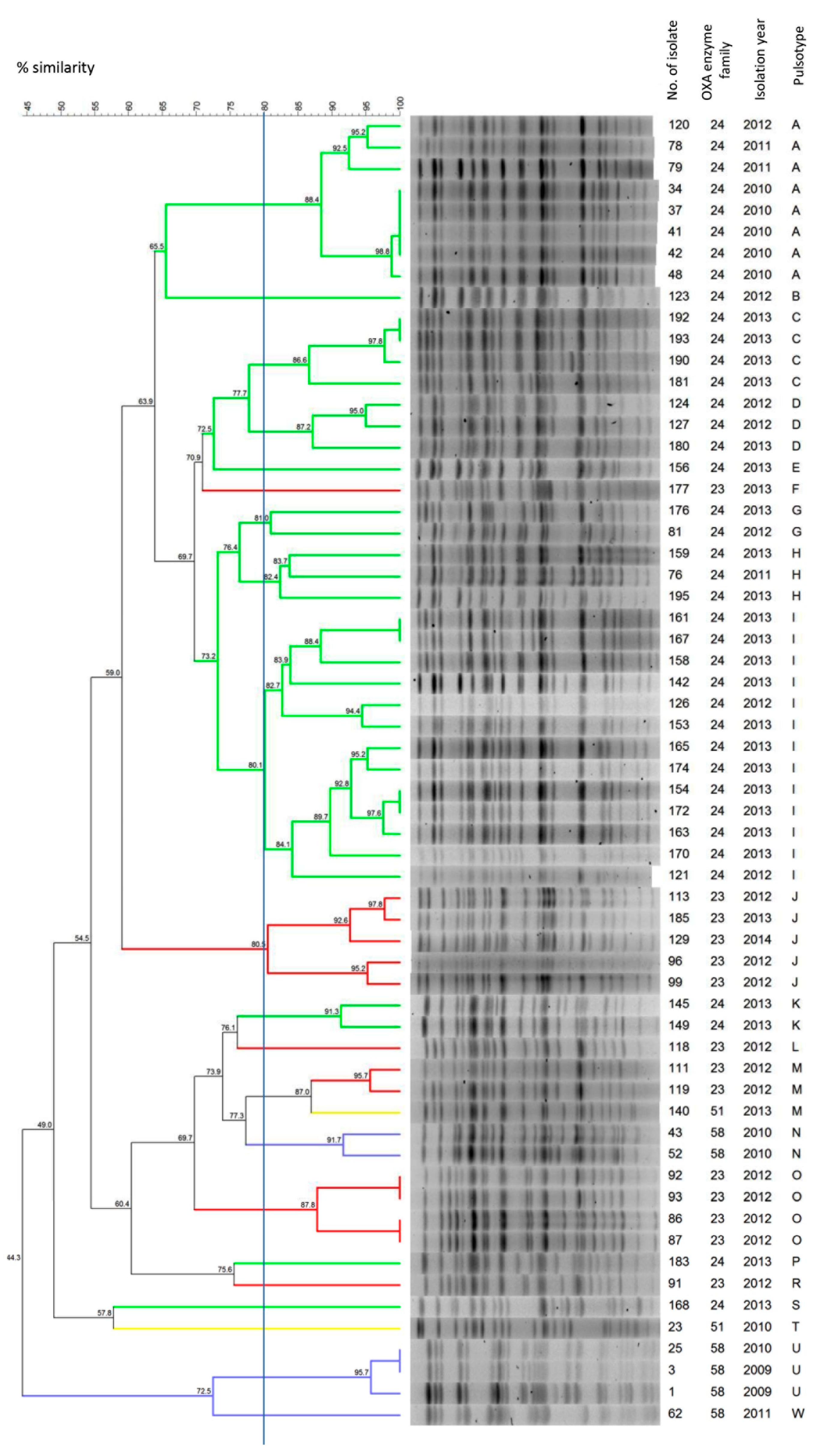
2.4. Molecular Typing and Genome Analysis of the Isolates
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains
4.2. Antimicrobial Susceptibility Testing
4.3. Phenotypic Detection of Carbapenemase Production
4.3.1. Detection of Metallo-β-Lactamases (MBL)
4.3.2. CarbAcineto NP Test
4.4. Molecular Detection of blaOXA-like Genes and ISs Upstream of blaCHDL Genes
4.5. Pulsed Field Gel Electrophoresis (PFGE)
4.6. Whole Genome Sequencing (WGS) and Multi-Locus Sequence Typing (MLST)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Maragakis, L.L.; Perl, T.M. Acinetobacter baumannii: Epidemiology, antimicrobial resistance, and treatment options. Clin. Infect. Dis. 2008, 46, 1254–1263. [Google Scholar] [CrossRef] [PubMed]
- Potron, A.; Poirel, L.; Nordmann, P. Emerging broad-spectrum resistance in Pseudomonas aeruginosa and Acinetobacter baumannii: Mechanisms and epidemiology. Int. J. Antimicrob. Agents 2015, 45, 568–585. [Google Scholar] [CrossRef]
- Poirel, L.; Naas, T.; Nordmann, P. Diversity, epidemiology, and genetics of class D beta-lactamases. Antimicrob. Agents Chemother. 2010, 54, 24–38. [Google Scholar] [CrossRef]
- El-Shazly, S.; Dashti, A.; Vali, L.; Bolaris, M.; Ibrahim, A.S. Molecular epidemiology and characterization of multiple-drug resistant (MDR) clinical isolates of Acinetobacter baumannii. Int. J. Infect. Dis. 2015. [Google Scholar] [CrossRef]
- Evans, B.A.; Amyes, S.G. OXA beta-lactamases. Clin. Microbiol. Rev. 2014, 27, 241–263. [Google Scholar] [CrossRef] [PubMed]
- Poirel, L.; Nordmann, P. Carbapenem resistance in Acinetobacter baumannii: Mechanisms and epidemiology. Clin. Microbiol. Infect. 2006, 12, 826–836. [Google Scholar] [CrossRef]
- Poirel, L.; Nordmann, P. Genetic structures at the origin of acquisition and expression of the carbapenem-hydrolyzing oxacillinase gene bla (OXA-58) in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2006, 50, 1442–1448. [Google Scholar] [CrossRef] [PubMed]
- European Centre for Disease Prevention and Control. Rapid Risk Assessment: Carbapenem-Resistant Acinetobacter Baumannii in Healthcare Settings—8 December 2016. Available online: https://www.ecdc.europa.eu/en/publications-data/rapid-risk-assessment-carbapenem-resistant-acinetobacter-baumannii-healthcare (accessed on 25 February 2021).
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Figueiredo, S.; Poirel, L.; Papa, A.; Koulourida, V.; Nordmann, P. Overexpression of the naturally occurring blaOXA-51 gene in Acinetobacter baumannii mediated by novel insertion sequence ISAba9. Antimicrob. Agents Chemother. 2009, 53, 4045–4047. [Google Scholar] [CrossRef]
- Pagano, M.; Martins, A.F.; Barth, A.L. Mobile genetic elements related to carbapenem resistance in Acinetobacter baumannii. Braz. J. Microbiol. 2016, 47, 785–792. [Google Scholar] [CrossRef]
- Karah, N.; Giske, C.G.; Sundsfjord, A.; Samuelsen, O. A diversity of OXA-carbapenemases and class 1 integrons among carbapenem-resistant Acinetobacter baumannii clinical isolates from Sweden belonging to different international clonal lineages. Microb. Drug Resist. 2011, 17, 545–549. [Google Scholar] [CrossRef] [PubMed]
- Adams, M.D.; Bishop, B.; Wright, M.S. Quantitative assessment of insertion sequence impact on bacterial genome architecture. Microb. Genom. 2016, 2, e000062. [Google Scholar] [CrossRef]
- Castanheira, M.; Costello, S.E.; Woosley, L.N.; Deshpande, L.M.; Davies, T.A.; Jones, R.N. Evaluation of clonality and carbapenem resistance mechanisms among Acinetobacter baumannii-Acinetobacter calcoaceticus complex and Enterobacteriaceae isolates collected in European and Mediterranean countries and detection of two novel beta-lactamases, GES-22 and VIM-35. Antimicrob. Agents Chemother. 2014, 58, 7358–7366. [Google Scholar] [CrossRef] [PubMed]
- Mugnier, P.D.; Poirel, L.; Naas, T.; Nordmann, P. Worldwide dissemination of the bla (OXA-23) carbapenemase gene of Acinetobacter baumannii. Emerg. Infect. Dis. 2010, 16, 35–40. [Google Scholar] [CrossRef]
- Liu, C.P.; Lu, H.P.; Luor, T. Clonal relationship and the association of the ST218 strain harboring blaOXA-72 gene to mortality in carbapenem-resistant Acinetobacter baumannii bacteremia. J. Microbiol. Immunol. Infect. 2019, 52, 297–303. [Google Scholar] [CrossRef] [PubMed]
- Hujer, A.M.; Hujer, K.M.; Leonard, D.A.; Powers, R.A.; Wallar, B.J.; Mack, A.R.; Taracila, M.A.; Rather, P.N.; Higgins, P.G.; Prati, F.; et al. A comprehensive and contemporary “snapshot” of beta-lactamases in carbapenem resistant Acinetobacter baumannii. Diagn. Microbiol. Infect. Dis. 2021, 99, 115242. [Google Scholar] [CrossRef] [PubMed]
- Mosqueda, N.; Espinal, P.; Cosgaya, C.; Viota, S.; Plasensia, V.; Alvarez-Lerma, F.; Montero, M.; Gómez, J.; Horcajada, J.P.; Vila, J.; et al. Globally expanding carbapenemase finally appears in Spain: Nosocomial outbreak of Acinetobacter baumannii producing plasmid-encoded OXA-23 in Barcelona, Spain. Antimicrob. Agents Chemother. 2013, 57, 5155–5157. [Google Scholar] [CrossRef] [PubMed]
- Chmielarczyk, A.; Pilarczyk-Zurek, M.; Kaminska, W.; Pobiega, M.; Romaniszyn, D.; Ziolkowski, G.; Wojkowska-Mach, J.; Bulanda, M. Molecular epidemiology and drug resistance of Acinetobacter baumannii isolated from hospitals in southern Poland: ICU as a risk factor for XDR strains. Microb. Drug Resist. 2016, 22, 328–335. [Google Scholar] [CrossRef]
- Nowak, P.; Paluchowska, P.; Budak, A. Distribution of bla (OXA) genes among carbapenem-resistant Acinetobacter baumannii nosocomial strains in Poland. New Microbiol. 2012, 35, 317–325. [Google Scholar] [PubMed]
- Todorova, B.; Velinov, T.; Ivanov, I.; Dobreva, E.; Kantardjiev, T. First detection of OXA-24 carbapenemase-producing Acinetobacter baumannii isolates in Bulgaria. World J. Microbiol. Biotechnol. 2014, 30, 1427–1430. [Google Scholar] [CrossRef]
- Heritier, C.; Poirel, L.; Lambert, T.; Nordmann, P. Contribution of acquired carbapenem-hydrolyzing oxacillinases to carbapenem resistance in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2005, 49, 3198–3202. [Google Scholar] [CrossRef]
- Afzal-Shah, M.; Woodford, N.; Livermore, D.M. Characterization of OXA-25, OXA-26, and OXA-27, molecular class D beta-lactamases associated with carbapenem resistance in clinical isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 2001, 45, 583–588. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.F.; Lan, C.Y. Antimicrobial resistance in Acinetobacter baumannii: From bench to bedside. World J. Clin. Cases 2014, 2, 787–814. [Google Scholar] [CrossRef] [PubMed]
- Saavedra, S.Y.; Cayo, R.; Gales, A.C.; Leal, A.L.; Saavedra, C.H. Early dissemination of OXA-72-producing Acinetobacter baumannii strain in Colombia: A case report. Braz. J. Infect. Dis. 2014, 18, 678–680. [Google Scholar] [CrossRef] [PubMed]
- Villalon, P.; Valdezate, S.; Medina-Pascual, M.J.; Carrasco, G.; Vindel, A.; Saez-Nieto, J.A. Epidemiology of the Acinetobacter-derived cephalosporinase, carbapenem-hydrolysing oxacillinase and metallo-beta-lactamase genes, and of common insertion sequences, in epidemic clones of Acinetobacter baumannii from Spain. J. Antimicrob. Chemother. 2013, 68, 550–553. [Google Scholar] [CrossRef] [PubMed]
- Lopes, B.S.; Evans, B.A.; Amyes, S.G. Disruption of the blaOXA-51-like gene by ISAba16 and activation of the blaOXA-58 gene leading to carbapenem resistance in Acinetobacter baumannii Ab244. J. Antimicrob. Chemother. 2012, 67, 59–63. [Google Scholar] [CrossRef]
- Izdebski, R.; Fiett, J.; Hryniewicz, W.; Gniadkowski, M. Molecular analysis of Acinetobacter baumannii isolates from invasive infections in 2009 in Poland. J. Clin. Microbiol. 2012, 50, 3813–3815. [Google Scholar] [CrossRef] [PubMed]
- Chmielarczyk, A.; Pobiega, M.; Romaniszyn, D.; Wojkowska-Mach, J. Multi-locus sequence typing (MLST) of non-fermentative Gram-negative bacilli isolated from bloodstream infections in southern Poland. Folia Microbiol. 2018, 63, 191–196. [Google Scholar] [CrossRef]
- Palmieri, M.; D’Andrea, M.M.; Pelegrin, A.C.; Perrot, N.; Mirande, C.; Blanc, B.; Legakis, N.; Goossens, H.; Rossolini, G.M.; van Belkum, A. Abundance of colistin-resistant, OXA-23- and ArmA-producing Acinetobacter baumannii belonging to international clone 2 in Greece. Front. Microbiol. 2020, 11, 668. [Google Scholar] [CrossRef] [PubMed]
- Wareth, G.; Brandt, C.; Sprague, L.D.; Neubauer, H.; Pletz, M.W. Spatio-temporal distribution of Acinetobacter baumannii in Germany—a comprehensive systematic review of studies on resistance development in humans (2000–2018). Microorganisms 2020, 8, 375. [Google Scholar] [CrossRef]
- Pfeifer, Y.; Hunfeld, K.P.; Borgmann, S.; Maneg, D.; Blobner, W.; Werner, G.; Higgins, P.G. Carbapenem-resistant Acinetobacter baumannii ST78 with OXA-72 carbapenemase and ESBL gene blaCTX-M-115. J. Antimicrob. Chemother. 2016, 71, 1426–1428. [Google Scholar] [CrossRef]
- Pfeifer, Y.; Trifonova, A.; Pietsch, M.; Brunner, M.; Todorova, I.; Gergova, I.; Wilharm, G.; Werner, G.; Savov, E. Clonal transmission of Gram-negative bacteria with carbapenemases NDM-1, VIM-1, and OXA-23/72 in a Bulgarian hospital. Microb. Drug Resist. 2017, 23, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Ladavac, R.; Bedenic, B.; Vranic-Ladavac, M.; Barisic, N.; Karcic, N.; Pompe, K.; Ferencic, A.; Stojanovic, A.; Seifert, H.; Katic, S.; et al. Emergence of different Acinetobacter baumannii clones in a Croatian hospital and correlation with antibiotic susceptibility. J. Glob. Antimicrob. Resist. 2017, 10, 213–218. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chen, Q.; Zhou, J.W.; Fan, J.Z.; Wu, S.H.; Xu, L.H.; Jiang, Y.; Ruan, Z.; Yu, Y.S.; Yu, D.J.; Wang, X.J. Simultaneous emergence and rapid spread of three OXA-23 producing Acinetobacter baumannii ST208 strains in intensive care units confirmed by whole genome sequencing. Infect. Genet. Evol. 2018, 58, 243–250. [Google Scholar] [CrossRef]
- Leung, E.C.; Leung, P.H.; Lai, R.W. Emergence of carbapenem-resistant Acinetobacter baumannii ST195 harboring blaOXA-23 isolated from bacteremia in Hong Kong. Microb. Drug Resist. 2019, 25, 1199–1203. [Google Scholar] [CrossRef]
- Jia, H.; Chen, Y.; Wang, J.; Xie, X.; Ruan, Z. Emerging challenges of whole-genome-sequencing-powered epidemiological surveillance of globally distributed clonal groups of bacterial infections, giving Acinetobacter baumannii ST195 as an example. Int. J. Med. Microbiol. 2019, 309, 151339. [Google Scholar] [CrossRef]
- Fang, Y.; Quan, J.; Hua, X.; Feng, Y.; Li, X.; Wang, J.; Ruan, Z.; Shang, S.; Yu, Y. Complete genome sequence of Acinetobacter baumannii XH386 (ST208), a multi-drug resistant bacteria isolated from pediatric hospital in China. Genom. Data 2016, 7, 269–274. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, X.; Du, Z.; Huang, W.; Zhang, X.; Zhou, Y. Outbreak of multidrug-resistant Acinetobacter baumannii ST208 producing OXA-23-Like carbapenemase in a children’s hospital in Shanghai, China. Microb. Drug Resist. 2020. [Google Scholar] [CrossRef]
- Kim, M.H.; Jeong, H.; Sim, Y.M.; Lee, S.; Yong, D.; Ryu, C.M.; Choi, J.Y. Using comparative genomics to understand molecular features of carbapenem-resistant Acinetobacter baumannii from South Korea causing invasive infections and their clinical implications. PLoS ONE 2020, 15, e0229416. [Google Scholar] [CrossRef]
- Dortet, L.; Poirel, L.; Errera, C.; Nordmann, P. CarbAcineto NP test for rapid detection of carbapenemase-producing Acinetobacter. spp. J. Clin. Microbiol. 2014, 52, 2359–2364. [Google Scholar] [CrossRef]
- Literacka, E.; Herda, M.; Baraniak, A.; Zabicka, D.; Hryniewicz, W.; Skoczynska, A.; Gniadkowski, M. Evaluation of the Carba NP test for carbapenemase detection in Enterobacteriaceae, Pseudomonas. spp. and Acinetobacter. spp., and its practical use in the routine work of a national reference laboratory for susceptibility testing. Eur. J. Clin. Microbiol. Infect. Dis. 2017, 36, 2281–2287. [Google Scholar] [CrossRef]
- Higgins, P.G.; Wisplinghoff, H.; Krut, O.; Seifert, H. A PCR-based method to differentiate between Acinetobacter baumannii and Acinetobacter genomic species 13TU. Clin. Microbiol. Infect. 2007, 13, 1199–1201. [Google Scholar] [CrossRef]
- Higgins, P.G.; Lehmann, M.; Wisplinghoff, H.; Seifert, H. gyrB multiplex PCR to differentiate between Acinetobacter calcoaceticus and Acinetobacter genomic species 3. J. Clin. Microbiol. 2010, 48, 4592–4594. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing, 28th ed.; supplement M100; CLSI: Wayne, PA, USA, 2018. [Google Scholar]
- Clinical and Laboratory Standards Institute. Methods for Dilution Antimicrobial Susceptibility Test for Bacteria that Grow Aerobically, 9th ed.; approved standard Document M07-A9; CLSI: Wayne, PA, USA, 2012. [Google Scholar]
- Lee, K.; Lim, Y.S.; Yong, D.; Yum, J.H.; Chong, Y. Evaluation of the Hodge test and the imipenem-EDTA double-disk synergy test for differentiating metallo-beta-lactamase-producing isolates of Pseudomonas spp. and Acinetobacter spp. J. Clin. Microbiol. 2003, 41, 4623–4629. [Google Scholar] [CrossRef]
- Woodford, N.; Ellington, M.J.; Coelho, J.M.; Turton, J.F.; Ward, M.E.; Brown, S.; Amyes, S.G.; Livermore, D.M. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int. J. Antimicrob. Agents 2006, 27, 351–353. [Google Scholar] [CrossRef]
- Seifert, H.; Dolzani, L.; Bressan, R.; van der Reijden, T.; van Strijen, B.; Stefanik, D.; Heersma, H.; Dijkshoorn, L. Standardization and interlaboratory reproducibility assessment of pulsed-field gel electrophoresis-generated fingerprints of Acinetobacter baumannii. J. Clin. Microbiol. 2005, 43, 4328–4335. [Google Scholar] [CrossRef]
- Hunter, S.B.; Vauterin, P.; Lambert-Fair, M.A.; Van Duyne, M.S.; Kubota, K.; Graves, L.; Wrigley, D.; Barrett, T.; Ribot, E. Establishment of a universal size standard strain for use with the PulseNet standardized pulsed-field gel electrophoresis protocols: Converting the national databases to the new size standard. J. Clin. Microbiol. 2005, 43, 1045–1050. [Google Scholar] [CrossRef]
- Tenover, F.C.; Arbeit, R.D.; Goering, R.V.; Mickelsen, P.A.; Murray, B.E.; Persing, D.H.; Swaminathan, B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: Criteria for bacterial strain typing. J. Clin. Microbiol. 1995, 33, 2233–2239. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Afgan, E.; Baker, D.; van den Beek, M.; Blankenberg, D.; Bouvier, D.; Cech, M.; Chilton, J.; Clements, D.; Coraor, N.; Eberhard, C.; et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 2016, 44, W3–W10. [Google Scholar] [CrossRef] [PubMed]
- Public Databases for Molecular Typing and Microbial Genome Diversity. Available online: https://pubmlst.org/organisms/acinetobacter-baumannii (accessed on 25 February 2021).
| Antimicrobial Agent | No. of Isolates with the Indicated Susceptibility Carrying blaCHDL Genes | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ISAba1- blaOXA-51-like (n = 2) | blaOXA-51-like | |||||||||||
| ISAba3- blaOXA-58-like (n = 6) | ISAba1-blaOXA-23-like (n = 14) | blaOXA-24-like (n = 39) | ||||||||||
| R | I | S | R | I | S | R | I | S | R | I | S | |
| Ampicillin/sulbactam | 1 | 0 | 1 | 0 | 1 | 5 | 3 | 8 | 3 | 3 | 4 | 32 |
| Cefepime | 0 | 1 | 1 | 0 | 4 | 2 | 12 | 2 | 0 | 10 | 28 | 1 |
| Ceftazidime | 2 | 0 | 0 | 2 | 4 | 0 | 14 | 0 | 0 | 39 | 0 | 0 |
| Imipenem | 1 | 1 | 0 | 6 | 0 | 0 | 14 | 0 | 0 | 39 | 0 | 0 |
| Meropenem | 2 | 0 | 0 | 0 | 6 | 0 | 14 | 0 | 0 | 39 | 0 | 0 |
| Piperacillin | 2 | 0 | 0 | 6 | 0 | 0 | 14 | 0 | 0 | 39 | 0 | 0 |
| Piperacillin/tazobactam | 2 | 0 | 0 | 6 | 0 | 0 | 14 | 0 | 0 | 39 | 0 | 0 |
| Ciprofloxacin | 2 | 0 | 0 | 6 | 0 | 0 | 14 | 0 | 0 | 39 | 0 | 0 |
| Levofloxacin | 1 | 1 | 0 | 2 | 4 | 0 | 14 | 0 | 0 | 16 | 23 | 0 |
| Gentamicin | 2 | 0 | 0 | 2 | 0 | 4 | 8 | 0 | 6 | 34 | 3 | 2 |
| Tobramicin | 1 | 0 | 1 | 2 | 0 | 4 | 11 | 0 | 3 | 32 | 5 | 2 |
| Colistin | 0 | 0 | 2 | 0 | 0 | 6 | 0 | 0 | 14 | 0 | 0 | 39 |
| Trimethoprim/Sulfamethoxazole | 2 | 0 | 0 | 6 | 0 | 0 | 14 | 0 | 0 | 34 | 0 | 5 |
| Groups of Isolates Carrying the following Genes | Carbapenem | No. of Isolates with the Indicated MIC Values | ||||||
|---|---|---|---|---|---|---|---|---|
| 4 mg/L | 8 mg/L | 16 mg/L | 32 mg/L | 64 mg/L | 128 mg/L | |||
| ISAba1-blaOXA-51-like (n = 2) | Imipenem | 1 | 1 | 0 | 0 | 0 | 0 | |
| Meropenem | 0 | 1 | 1 | 0 | 0 | 0 | ||
| blaOXA-51-like | ISAba3-blaOXA-58-like (n = 6) | Imipenem | 0 | 2 | 4 | 0 | 0 | 0 |
| Meropenem | 0 | 5 | 0 | 0 | 1 | 0 | ||
| ISAba1-blaOXA-23-like (n = 14) | Imipenem | 0 | 1 | 9 | 4 | 0 | 0 | |
| Meropenem | 0 | 2 | 3 | 4 | 5 | 0 | ||
| blaOXA-24-like (n = 39) | Imipenem | 0 | 2 | 14 | 12 | 11 | 0 | |
| Meropenem | 0 | 0 | 0 | 2 | 17 | 20 | ||
| Oxford ST * | Acquired CHDLs Enzyme (Family) | Other β-lactamases | Isolate Number | Isolation Year | Clinical Material |
|---|---|---|---|---|---|
| 195 | OXA-23 (OXA-23-like) | ADC-30, TEM-1 | 96 | 2012 | Respiratory tract sample |
| 195 | OXA-23 (OXA-23-like) | ADC-30, TEM-1 | 185 | 2013 | Urine |
| 195 | OXA-23 (OXA-23-like) | ADC-30, TEM-1 | 129 | 2014 | Urine |
| 208 | OXA-72 (OXA-24-like) | ADC-30 | 176 | 2013 | Urine |
| 208/1806 | OXA-58 (OXA-58-like) | ADC-30 | 43 | 2010 | Fistula |
| 208/1806 | OXA-58 (OXA-58-like) | ADC-30 | 52 | 2010 | Respiratory tract sample |
| 348 | OXA-72 (OXA-24-like) | ADC-30 | 76 | 2011 | Wound |
| 348 | OXA-72 (OXA-24-like) | ADC-30 | 81 | 2012 | Wound |
| 348 | OXA-72 (OXA-24-like) | ADC-30 | 159 | 2013 | Urine |
| 348 | OXA-72 (OXA-24-like) | ADC-30 | 165 | 2013 | Respiratory tract sample |
| 348 | OXA-72 (OXA-24-like) | ADC-30 | 195 | 2013 | Wound |
| 348 | OXA-23 (OXA-23-like) | ADC-30 | 113 | 2012 | Urine |
| 348 | OXA-23 (OXA-23-like) | ADC-30 | 118 | 2012 | Rectal swab |
| 425 | OXA-23 (OXA-23-like) | ADC-30, TEM-1 | 86 | 2012 | Urine |
| 425 | OXA-23 (OXA-23-like) | ADC-30, TEM-1 | 87 | 2012 | Urine |
| Target Genes | Primer | Sequence (5’ → 3’) | Product Length (bp) | Reference |
|---|---|---|---|---|
| blaOXA-51-like | 51/F | taatgctttgatcggccttg | 352 | [49] |
| 51/R | tggattgcacttcatcttgg | |||
| blaISAba1+OXA-51-like | ISAba1/F | aatcacaagcatgatgagcg | 880 | this study |
| 51/R | tggattgcacttcatcttgg | [49] | ||
| blaOXA-58-like | 58/F | aagtattggggcttgtgctg | 598 | [49] |
| 58/R | cctctgcgctctacatac | this study | ||
| blaISAba3+OXA-58-like | ISAba3/F | aggcaggttggacatttgat | 755 | this study |
| 58/R | cctctgcgctctacatac | this study | ||
| blaOXA-23-like | 23/F | gatcggattggagaaccaga | 501 | [49] |
| 23/R | catttctgaccgcatttccat | this study | ||
| blaISAba1+OXA-23-like | ISAba1/F | aatcacaagcatgatgagcg | 962 | this study |
| 23/R | catttctgaccgcatttccat | this study | ||
| blaOXA-24-like | 24/F | ggttagttggcccccttaaa | 248 | [49] |
| 24/R | agttgagcgaaaaggggatt | |||
| blaISAba1+OXA-24-like | ISAba1/F | aatcacaagcatgatgagcg | 448 | this study |
| 24/R | agttgagcgaaaaggggatt | [49] | ||
| blaOXA-143-like | 143/F | cagtgcatgctcatctattc | 460 | this study |
| 143/R | ggccaaccaaccagaagtt | this study |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Słoczyńska, A.; Wand, M.E.; Tyski, S.; Laudy, A.E. Analysis of blaCHDL Genes and Insertion Sequences Related to Carbapenem Resistance in Acinetobacter baumannii Clinical Strains Isolated in Warsaw, Poland. Int. J. Mol. Sci. 2021, 22, 2486. https://doi.org/10.3390/ijms22052486
Słoczyńska A, Wand ME, Tyski S, Laudy AE. Analysis of blaCHDL Genes and Insertion Sequences Related to Carbapenem Resistance in Acinetobacter baumannii Clinical Strains Isolated in Warsaw, Poland. International Journal of Molecular Sciences. 2021; 22(5):2486. https://doi.org/10.3390/ijms22052486
Chicago/Turabian StyleSłoczyńska, Alicja, Matthew E. Wand, Stefan Tyski, and Agnieszka E. Laudy. 2021. "Analysis of blaCHDL Genes and Insertion Sequences Related to Carbapenem Resistance in Acinetobacter baumannii Clinical Strains Isolated in Warsaw, Poland" International Journal of Molecular Sciences 22, no. 5: 2486. https://doi.org/10.3390/ijms22052486
APA StyleSłoczyńska, A., Wand, M. E., Tyski, S., & Laudy, A. E. (2021). Analysis of blaCHDL Genes and Insertion Sequences Related to Carbapenem Resistance in Acinetobacter baumannii Clinical Strains Isolated in Warsaw, Poland. International Journal of Molecular Sciences, 22(5), 2486. https://doi.org/10.3390/ijms22052486





