DNA Methyltransferase 1 (DNMT1) Shapes Neuronal Activity of Human iPSC-Derived Glutamatergic Cortical Neurons
Abstract
1. Introduction
2. Results
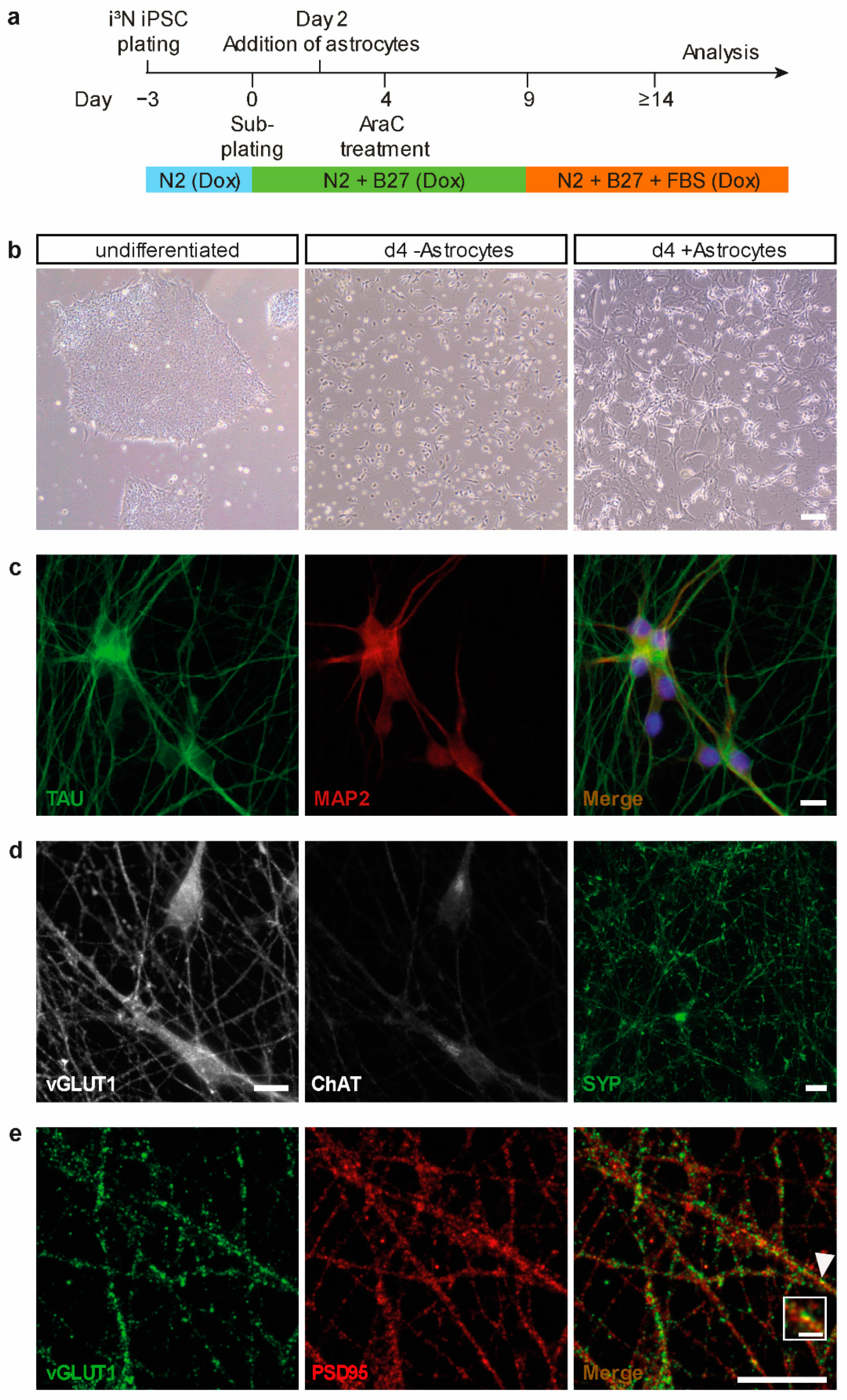
2.1. Generation of Cortical Neurons from Human Induced Pluripotent Stem Cells (iPSCs)
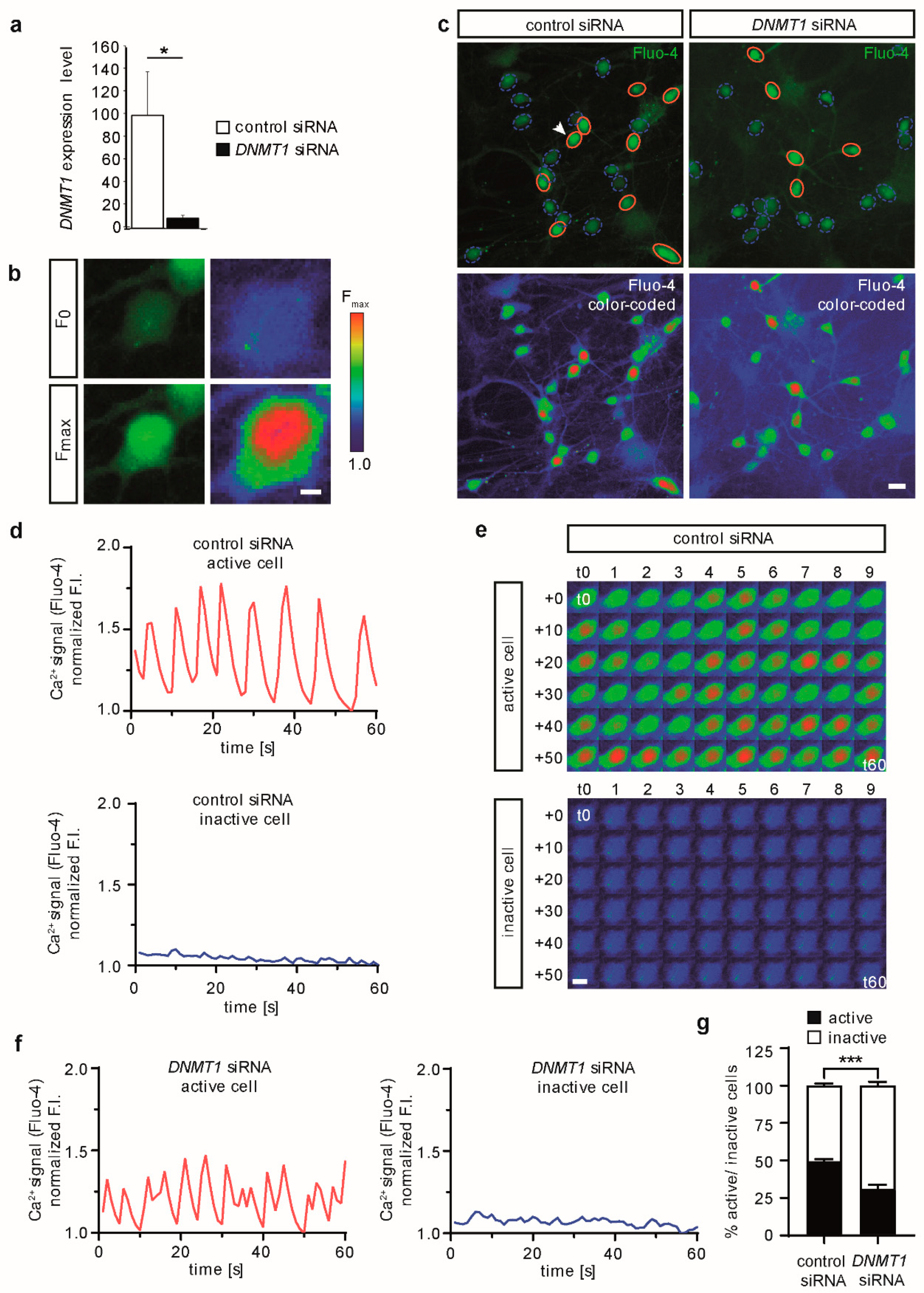
2.2. DNMT1 Depletion Reduces Spontaneous Neuronal Activity in Human iPSC-Derived Cortical Excitatory Neurons
3. Discussion
4. Materials and Methods
4.1. Isolation and Cultivation of Primary Murine Astrocytes
4.2. iPSC Maintenance and Differentiation into Cortical Neurons
4.3. Immunocytochemistry of iPSC-Derived Neuronal Cultures
4.4. siRNA Oligo Transfection
4.5. Live-Cell Calcium Imaging
4.6. Cultivation Conditions and Treatment of HEK Cells
4.7. RNA Isolation and Quantitative Reverse Transcription PCR (qRT-PCR)
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Campbell, R.R.; Wood, M.A. How the epigenome integrates information and reshapes the synapse. Nat. Rev. Neurosci. 2019, 20, 133–147. [Google Scholar] [CrossRef]
- Alberini, C.M. Transcription factors in long-term memory and synaptic plasticity. Physiol. Rev. 2009, 89, 121–145. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, P.J.; Abel, T. The role of protein synthesis in memory consolidation: Progress amid decades of debate. Neurobiol. Learn. Mem. 2008, 89, 293–311. [Google Scholar] [CrossRef]
- Guo, J.U.; Ma, D.K.; Mo, H.; Ball, M.P.; Jang, M.H.; Bonaguidi, M.A.; Balazer, J.A.; Eaves, H.L.; Xie, B.; Ford, E.; et al. Neuronal activity modifies the DNA methylation landscape in the adult brain. Nat. Neurosci. 2011, 14, 1345–1351. [Google Scholar] [CrossRef]
- Barry, G.; Briggs, J.A.; Vanichkina, D.P.; Poth, E.M.; Beveridge, N.J.; Ratnu, V.S.; Nayler, S.P.; Nones, K.; Hu, J.; Bredy, T.W.; et al. The long noncoding RNA Gomafu is acutely regulated in response to neuronal activation and involved in schizophrenia-associated alternative splicing. Mol. Psychiatry 2014, 19, 486–494. [Google Scholar] [CrossRef]
- Lipovich, L.; Dachet, F.; Cai, J.; Bagla, S.; Balan, K.; Jia, H.; Loeb, J.A. Activity-dependent human brain coding/noncoding gene regulatory networks. Genetics 2012, 192, 1133–1148. [Google Scholar] [CrossRef]
- Sweatt, J.D. Dynamic DNA methylation controls glutamate receptor trafficking and synaptic scaling. Neurochem. J. 2016, 137, 312–330. [Google Scholar] [CrossRef]
- Guo, H.; Zhu, P.; Yan, L.; Li, R.; Hu, B.; Lian, Y.; Yan, J.; Ren, X.; Lin, S.; Li, J.; et al. The DNA methylation landscape of human early embryos. Nature 2014, 511, 606–610. [Google Scholar] [CrossRef]
- Ito, S.; Shen, L.; Dai, Q.; Wu, S.C.; Collins, L.B.; Swenberg, J.A.; He, C. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 2011, 333, 1300–1303. [Google Scholar] [CrossRef] [PubMed]
- Kohli, R.M.; Zhang, Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature 2013, 502, 472–479. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Zhang, Y. TET-mediated active DNA demethylation: Mechanism, function and beyond. Nat. Rev. Genet. 2017, 18, 517–534. [Google Scholar] [CrossRef]
- Sharma, A.; Klein, S.L.; Barboza, L.; Lodhi, N.; Toth, M. Principles governing DNA methylation during neuronal lineage and subtype specification. Neurosci. J. 2016, 36, 1711–1722. [Google Scholar] [CrossRef] [PubMed]
- Lister, R.; Mukamel, E.A.; Nery, J.R.; Urich, M.; Puddifoot, C.A.; Johnson, N.D.; Lucero, J.; Huang, Y.; Dwork, A.J.; Schultz, M.D.; et al. Global epigenomic reconfiguration during mammalian brain development. Science 2013, 341. [Google Scholar] [CrossRef]
- Mo, A.; Mukamel, E.A.; Davis, F.P.; Luo, C.; Henry, G.L.; Picard, S.; Urich, M.A.; Nery, J.R.; Sejnowski, T.J.; Lister, R.; et al. Epigenomic Signatures of Neuronal Diversity in the Mammalian Brain. Neuron 2015, 86, 1369–1384. [Google Scholar] [CrossRef] [PubMed]
- Pensold, D.; Symmank, J.; Hahn, A.; Lingner, T.; Salinas-Riester, G.; Downie, B.R.; Ludewig, F.; Rotzsch, A.; Haag, N.; Andreas, N.; et al. The DNA Methyltransferase 1 (DNMT1) Controls the Shape and Dynamics of Migrating POA-Derived Interneurons Fated for the Murine Cerebral Cortex. Cereb. Cortex 2017, 27, 5696–5714. [Google Scholar] [CrossRef]
- Levenson, J.M.; Roth, T.L.; Lubin, F.D.; Miller, C.A.; Huang, I.C.; Desai, P.; Malone, L.M.; Sweatt, J.D. Evidence That DNA (Cytosine-5) Methyltransferase Regulates Synaptic Plasticity in the Hippocampus. Biol. J. Chem. 2006, 281, 15763–15773. [Google Scholar] [CrossRef]
- Nelson, E.D.; Kavalali, E.T.; Monteggia, L.M. Activity-dependent suppression of miniature neurotransmission through the regulation of DNA methylation. Neurosci. J. 2008, 28, 395–406. [Google Scholar] [CrossRef] [PubMed]
- Meadows, J.P.; Guzman-Karlsson, M.C.; Phillips, S.; Holleman, C.; Posey, J.L.; Day, J.J.; Hablitz, J.J. DNA methylation regulates neuronal glutamatergic synaptic scaling. Sci. Signal. 2015, 8, ra61. [Google Scholar] [CrossRef] [PubMed]
- Halder, R.; Hennion, M.; Vidal, R.O.; Shomroni, O.; Rahman, R.U.; Rajput, A.; Centeno, T.P.; Van Bebber, F.; Capece, V.; Vizcaino, J.C.G.; et al. DNA methylation changes in plasticity genes accompany the formation and maintenance of memory. Nat. Neurosci. 2016, 19, 102–110. [Google Scholar] [CrossRef] [PubMed]
- Pensold, D.; Reichard, J.; Van Loo, K.M.; Ciganok, N.; Hahn, A.; Bayer, C.; Liebmann, L.; Groß, J.; Tittelmeier, J.; Lingner, T.; et al. DNA Methylation-Mediated Modulation of Endocytosis as Potential Mechanism for Synaptic Function Regulation in Murine Inhibitory Cortical Interneurons. Cereb. Cortex 2020, 30, 3921–3937. [Google Scholar] [CrossRef]
- Oliveira, M.M. DNA methylation: A permissive mark in memory formation and maintenance. Learn. Mem. 2016, 23, 587–593. [Google Scholar] [CrossRef]
- Bayraktar, G.; Yuanxiang, P.; Confettura, A.D.; Gomes, G.M.; Raza, S.A.; Stork, O.; Tajima, S.; Suetake, I.; Karpova, A.; Yildirim, F.; et al. Synaptic control of DNA methylation involves activity-dependent degradation of DNMT3A1 in the nucleus. Neuropsychopharmacology 2020, 45, 2120–2130. [Google Scholar] [CrossRef]
- Zhu, Q.; Wang, L.; Zhang, Y.; Zhao, F.H.; Luo, J.; Xiao, Z.; Chen, G.J.; Wang, X.F. Increased expression of DNA methyltransferase 1 and 3a in human temporal lobe epilepsy. Mol. J. Neurosci. 2012, 46, 420–426. [Google Scholar] [CrossRef] [PubMed]
- Miyaoka, Y.; Chan, A.H.; Judge, L.M.; Yoo, J.; Huang, M.; Nguyen, T.D.; Lizarraga, P.P.; So, P.L.; Conklin, B.R. Isolation of single-base genome-edited human iPS cells without antibiotic selection. Nat. Methods 2014, 11, 291–293. [Google Scholar] [CrossRef]
- Wang, C.; Ward, M.E.; Chen, R.; Liu, K.; Tracy, T.E.; Chen, X.; Xie, M.; Sohn, P.D.; Ludwig, C.; Meyer-Franke, A.; et al. Scalable Production of iPSC-Derived Human Neurons to Identify Tau-Lowering Compounds by High-Content Screening. Stem Cell Rep. 2017, 9, 1221–1233. [Google Scholar] [CrossRef] [PubMed]
- Nunez, J. Immature and mature variants of MAP2 and tau proteins and neuronal plasticity. Trends Neurosci. 1988, 11, 477–479. [Google Scholar] [CrossRef]
- Bernhardt, R.; Matus, A. Light and electron microscopic studies of the distribution of microtubule-associated protein 2 in rat brain: A difference between dendritic and axonal cytoskeletons. Comp. J. Neurol. 1984, 226, 203–221. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Pak, C.; Han, Y.; Ahlenius, H.; Zhang, Z.; Chanda, S.; Marro, S.; Patzke, C.; Acuna, C.; Covy, J.; et al. Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron 2013, 78, 785–798. [Google Scholar] [CrossRef]
- He, S.; Wang, F.; Yang, L.; Guo, C.; Wan, R.; Ke, A.; Xu, L.; Hu, G.; Xu, X.; Shen, J.; et al. Expression of DNMT1 and DNMT3a Are Regulated by GLI1 in Human Pancreatic Cancer. PLoS ONE 2011, 6, e27684. [Google Scholar] [CrossRef]
- Roy, J.G.; McElhaney, J.E.; Verschoor, C.P. Reliable reference genes for the quantification of mRNA in human T-cells and PBMCs stimulated with live influenza virus. BMC Immunol. 2020, 21, 1–7. [Google Scholar] [CrossRef]
- Symmank, J.; Bayer, C.; Schmidt, C.; Hahn, A.; Pensold, D.; Zimmer-Bensch, G. DNMT1 modulates interneuron morphology by regulating Pak6 expression through crosstalk with histone modifications. Epigenetics 2018, 13, 536–556. [Google Scholar] [CrossRef] [PubMed]
- Bayer, C.; Pitschelatow, G.; Hannemann, N.; Linde, J.; Reichard, J.; Pensold, D.; Zimmer-Bensch, G. DNA Methyltransferase 1 (DNMT1) Acts on Neurodegeneration by Modulating Proteostasis-Relevant Intracellular Processes. Int. Mol. J. Sci. 2020, 21, 5420. [Google Scholar] [CrossRef]
- Symmank, J.; Bayer, C.; Reichard, J.; Pensold, D.; Zimmer-Bensch, G. Neuronal Lhx1 expression is regulated by DNMT1-dependent modulation of histone marks. Epigenetics 2020, 15, 1259–1274. [Google Scholar] [CrossRef]
- Gao, L.; Emperle, M.; Guo, Y.; Grimm, S.A.; Ren, W.; Adam, S.; Uryu, H.; Zhang, Z.M.; Chen, D.; Yin, J.; et al. Comprehensive structure-function characterization of DNMT3B and DNMT3A reveals distinctive de novo DNA methylation mechanisms. bioRxiv 2020. [Google Scholar] [CrossRef] [PubMed]
- Dukatz, M.; Adam, S.; Biswal, M.; Song, J.; Bashtrykov, P.; Jeltsch, A. Complex DNA sequence readout mechanisms of the DNMT3B DNA methyltransferase. Nucleic Acids Res. 2020, 48, 11495–11509. [Google Scholar] [CrossRef] [PubMed]
- Adam, S.; Anteneh, H.; Hornisch, M.; Wagner, V.; Lu, J.; Radde, N.E.; Bashtrykov, P.; Song, J.; Jeltsch, A. DNA sequence-dependent activity and base flipping mechanisms of DNMT1 regulate genome-wide DNA methylation. Nat. Commun. 2020, 11, 1–15. [Google Scholar] [CrossRef]
- Chalei, V.; Sansom, S.N.; Kong, L.; Lee, S.; Montiel, J.F.; Vance, K.W.; Ponting, C.P. The long noncoding RNA Dali is an epigenetic regulator of neural differentiation. eLife 2014, 3, 1–24. [Google Scholar] [CrossRef]
- Zimmer-Bensch, G. Emerging Roles of Long Non-Coding RNAs as Drivers of Brain Evolution. Cells 2019, 8, 1399. [Google Scholar] [CrossRef]
- Marchese, F.P.; Raimondi, I.; Huarte, M. The multidimensional mechanisms of long noncoding RNA function. Genome Biol. 2017, 18, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Linde, J.; Zimmer-Bensch, G. DNA Methylation-Dependent Dysregulation of GABAergic Interneuron Functionality in Neuropsychiatric Diseases. Front. Neurosci. 2020, 14, 982. [Google Scholar] [CrossRef] [PubMed]
- Zempel, H.; Mandelkow, E.M. Tracking Tau in neurons: How to grow, fix, and stain primary neurons for the investigation of Tau in all developmental stages. In Methods in Molecular Biology; Humana Press Inc.: Totowa, NJ, USA, 2017; pp. 327–334. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bachmann, S.; Linde, J.; Bell, M.; Spehr, M.; Zempel, H.; Zimmer-Bensch, G. DNA Methyltransferase 1 (DNMT1) Shapes Neuronal Activity of Human iPSC-Derived Glutamatergic Cortical Neurons. Int. J. Mol. Sci. 2021, 22, 2034. https://doi.org/10.3390/ijms22042034
Bachmann S, Linde J, Bell M, Spehr M, Zempel H, Zimmer-Bensch G. DNA Methyltransferase 1 (DNMT1) Shapes Neuronal Activity of Human iPSC-Derived Glutamatergic Cortical Neurons. International Journal of Molecular Sciences. 2021; 22(4):2034. https://doi.org/10.3390/ijms22042034
Chicago/Turabian StyleBachmann, Sarah, Jenice Linde, Michael Bell, Marc Spehr, Hans Zempel, and Geraldine Zimmer-Bensch. 2021. "DNA Methyltransferase 1 (DNMT1) Shapes Neuronal Activity of Human iPSC-Derived Glutamatergic Cortical Neurons" International Journal of Molecular Sciences 22, no. 4: 2034. https://doi.org/10.3390/ijms22042034
APA StyleBachmann, S., Linde, J., Bell, M., Spehr, M., Zempel, H., & Zimmer-Bensch, G. (2021). DNA Methyltransferase 1 (DNMT1) Shapes Neuronal Activity of Human iPSC-Derived Glutamatergic Cortical Neurons. International Journal of Molecular Sciences, 22(4), 2034. https://doi.org/10.3390/ijms22042034






