Evidence for 2-Methoxyestradiol-Mediated Inhibition of Receptor Tyrosine Kinase RON in the Management of Prostate Cancer
Abstract
1. Introduction
2. Results
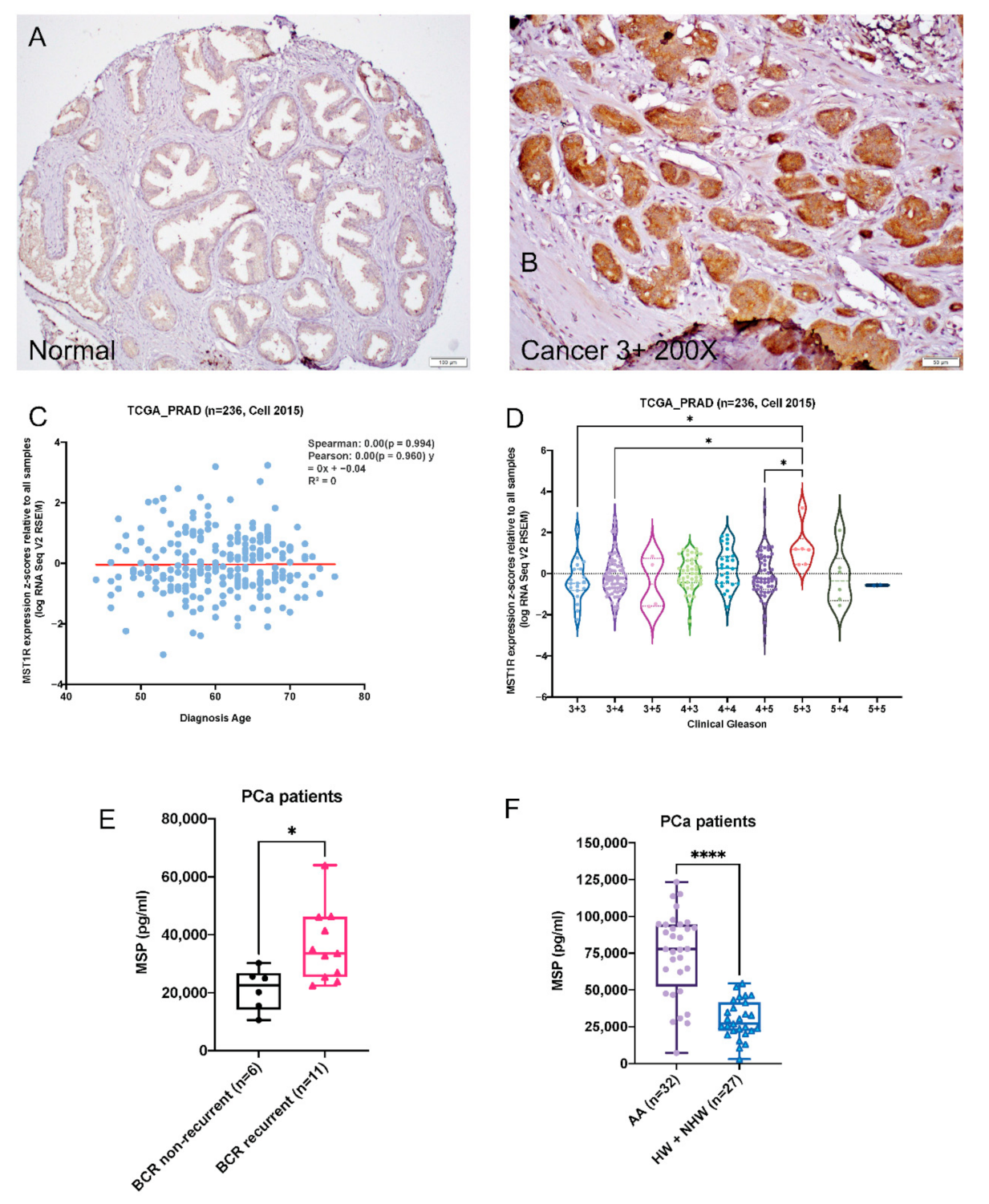
2.1. RON Levels Are Significantly Elevated in Prostate Adenocarcinoma
2.2. Serum MSP Level Is Associated with Biochemical Recurrence
2.3. 2-ME2-Mediated Proliferation Inhibition Is Associated with Decreased Levels, Expression and Activity of RON in Advanced PCa Cell Lines
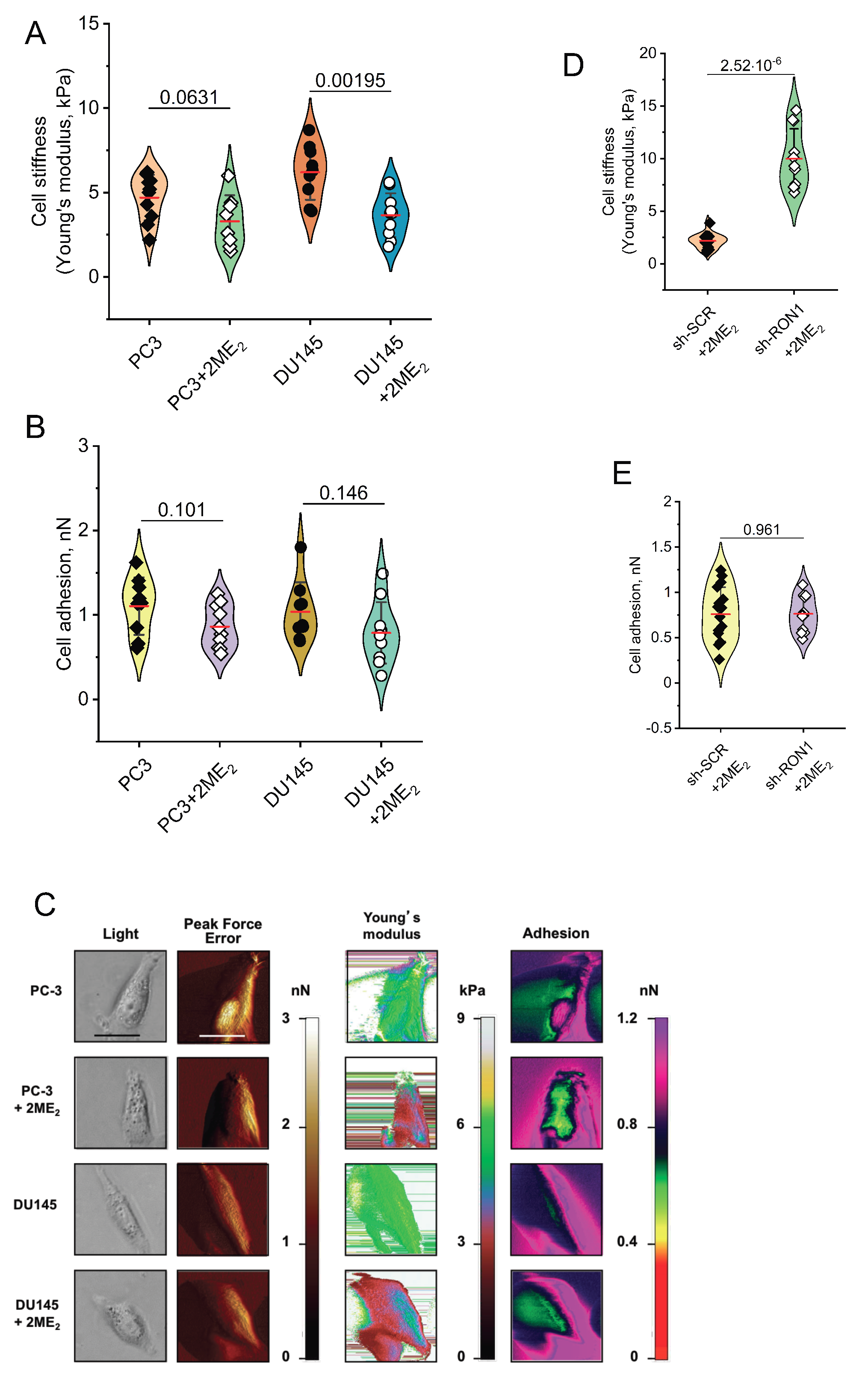
2.4. 2-ME2 Affects Mechanical Properties of PCa Cells
2.5. Heterogeneous Response to Nano-Coated 2-ME2 Intervention for 10 Weeks on Castrate Resistant Prostate Tumor Development
3. Discussion
4. Materials and Methods
4.1. Cell Lines
4.2. Animal Study
4.3. RNA and qPCR
4.4. RNA Interference and Plasmid Transfection
4.5. Western Blotting
4.6. Immunohistochemistry
4.7. Atomic Force Microscopy (AFM)
4.8. Metabolomic Profiling
4.9. Metabolite Identification and Data Analysis
4.10. RON Kinase Activity
4.11. MSP in Serum Samples
4.12. Bioinformatic Analysis
4.13. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef]
- American Cancer Society. Cancer Facts & Figures 2020; American Cancer Society: Atlanta, GA, USA, 2020; pp. 1–76. [Google Scholar]
- Litwin, M.S.; Tan, H.-J. The Diagnosis and Treatment of Prostate Cancer. JAMA 2017, 317, 2532. [Google Scholar] [CrossRef] [PubMed]
- Iida, M.; Brand, T.M.; Campbell, D.A.; Li, C.; Wheeler, D.L. Yes and Lyn play a role in nuclear translocation of the epidermal growth factor receptor. Oncogene 2013, 32, 759–767. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Shen, J.; Hsu, J.L.; Han, Z.; Hsu, M.-C.; Yang, C.-C.; Kuo, H.-P.; Wang, Y.-N.; Yamaguchi, H.; Miller, S.A.; et al. Syntaxin 6-mediated Golgi translocation plays an important role in nuclear functions of EGFR through microtubule-dependent trafficking. Oncogene 2014, 33, 756–770. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Lu, W.; Liu, S.; Yang, Q.; Carver, B.S.; Li, E.; Wang, Y.; Fazli, L.; Gleave, M.; Chen, Z. Crosstalk between Nuclear MET and SOX9/β-Catenin Correlates with Castration Resistant Prostate Cancer. Mol. Endocrinol. 2014, 28, 2014-1078. [Google Scholar] [CrossRef] [PubMed]
- Batth, I.; Yun, H.; Hussain, S.; Meng, P.; Osmulski, P.; Huang, T.H.-M.; Bedolla, R.; Profit, A.; Reddick, R.; Kumar, A. Crosstalk between RON and androgen receptor signaling in the development of castration resistant prostate cancer. Oncotarget 2016, 7, 14048–14063. [Google Scholar] [CrossRef]
- Yao, H.-P.; Zhou, Y.-Q.; Zhang, R.; Wang, M.-H. MSP-RON signalling in cancer: Pathogenesis and therapeutic potential. Nat. Rev. Cancer 2013, 13, 466–481. [Google Scholar] [CrossRef]
- Thobe, M.N.; Gray, J.K.; Gurusamy, D.; Paluch, A.M.; Wagh, P.K.; Pathrose, P.; Lentsch, A.B.; Waltz, S.E. The Ron receptor promotes prostate tumor growth in the TRAMP mouse model. Oncogene 2011, 30, 4990–4998. [Google Scholar] [CrossRef][Green Version]
- Thobe, M.N.; Gurusamy, D.; Pathrose, P.; Waltz, S.E. The Ron receptor tyrosine kinase positively regulates angiogenic chemokine production in prostate cancer cells. Oncogene 2010, 29, 214–226. [Google Scholar] [CrossRef]
- Peace, B.E.; Toney-Earley, K.; Collins, M.H.; Waltz, S.E. Ron receptor signaling augments mammary tumor formation and metastasis in a murine model of breast cancer. Cancer Res. 2005, 65, 1285–1293. [Google Scholar] [CrossRef]
- Zhao, S.; Cao, L.; Freeman, J.W. Knockdown of RON receptor kinase delays but does not prevent tumor progression while enhancing HGF/MET signaling in pancreatic cancer cell lines. Oncogenesis 2013, 2, e76. [Google Scholar] [CrossRef]
- Gray, J.K.; Paluch, A.M.; Stuart, W.D.; Waltz, S.E. Ron receptor overexpression in the murine prostate induces prostate intraepithelial neoplasia. Cancer Lett. 2012, 314, 92–101. [Google Scholar] [CrossRef]
- Vasiliauskas, J.; Nashu, M.A.; Pathrose, P.; Starnes, S.L.; Waltz, S.E. Hepatocyte growth factor-like protein is required for prostate tumor growth in the TRAMP mouse model. Oncotarget 2014, 5, 5547–5558. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Brown, N.E.; Paluch, A.M.; Nashu, M.A.; Komurov, K.; Waltz, S.E. Tumor Cell Autonomous RON Receptor Expression Promotes Prostate Cancer Growth Under Conditions of Androgen Deprivation. Neoplasia 2018, 20, 917–929. [Google Scholar] [CrossRef] [PubMed]
- Ganapathy, M.; Ghosh, R.; Jianping, X.; Zhang, X.; Bedolla, R.; Schoolfield, J.; Yeh, I.; Troyer, D.A.; Olumi, A.F.; Kumar, A.P. Involvement of FLIP in 2-methoxyestradiol-induced tumor regression in transgenic adenocarcinoma of mouse prostate model. Clin. Cancer Res. 2009, 15, 1601–1611. [Google Scholar] [CrossRef]
- Garcia, G.E.; Wisniewski, H.-G.; Lucia, M.S.; Arevalo, N.; Slaga, T.J.; Kraft, S.L.; Strange, R.; Kumar, A.P. 2-Methoxyestradiol inhibits prostate tumor development in transgenic adenocarcinoma of mouse prostate: Role of tumor necrosis factor-alpha-stimulated gene 6. Clin. Cancer Res. 2006, 12, 980–988. [Google Scholar] [CrossRef] [PubMed]
- Mabjeesh, N.J.; Escuin, D.; LaVallee, T.M.; Pribluda, V.S.; Swartz, G.M.; Johnson, M.S.; Willard, M.T.; Zhong, H.; Simons, J.W.; Giannakakou, P. 2ME2 inhibits tumor growth and angiogenesis by disrupting microtubules and dysregulating HIF. Cancer Cell 2003, 3, 363–375. [Google Scholar] [CrossRef]
- Ricker, J.L.; Chen, Z.; Yang, X.P.; Pribluda, V.S.; Swartz, G.M.; Van Waes, C. 2-methoxyestradiol inhibits hypoxia-inducible factor 1alpha, tumor growth, and angiogenesis and augments paclitaxel efficacy in head and neck squamous cell carcinoma. Clin. Cancer Res. 2004, 10, 8665–8673. [Google Scholar] [CrossRef]
- Van Veldhuizen, P.J.; Ray, G.; Banerjee, S.; Dhar, G.; Kambhampati, S.; Dhar, A.; Banerjee, S.K. 2-Methoxyestradiol modulates beta-catenin in prostate cancer cells: A possible mediator of 2-methoxyestradiol-induced inhibition of cell growth. Int. J. Cancer 2008, 122, 567–571. [Google Scholar] [CrossRef]
- Poch, A.; Villanelo, F.; Henriquez, S.; Kohen, P.; Muñoz, A.; Strauss, J.F.; Devoto, L. Molecular modelling predicts that 2-methoxyestradiol disrupts HIF function by binding to the PAS-B domain. Steroids 2019, 144, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhang, X.; Chen, T.; Jiang, X.; Wang, X.; Lei, H.; Wang, Y. Inhibition of immunoproteasome promotes angiogenesis via enhancing hypoxia-inducible factor-1α abundance in rats following focal cerebral ischaemia. Brain Behav. Immun. 2018, 73, 167–179. [Google Scholar] [CrossRef] [PubMed]
- Thangasamy, A.; Rogge, J.; Ammanamanchi, S. Recepteur d’origine nantais tyrosine kinase is a direct target of hypoxia-inducible factor-1alpha-mediated invasion of breast carcinoma cells. J. Biol. Chem. 2009, 284, 14001–14010. [Google Scholar] [CrossRef] [PubMed]
- Hunt, B.G.; Wicker, C.A.; Bourn, J.R.; Lower, E.E.; Takiar, V.; Waltz, S.E. MST1R (RON) expression is a novel prognostic biomarker for metastatic progression in breast cancer patients. Breast Cancer Res. Treat. 2020, 181, 529–540. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, C.; Brown, N.E.; Vasiliauskas, J.; Pathrose, P.; Starnes, S.L.; Waltz, S.E. Prostate Epithelial RON Signaling Promotes M2 Macrophage Activation to Drive Prostate Tumor Growth and Progression. Mol. Cancer Res. 2020, 18, 1244–1254. [Google Scholar] [CrossRef]
- Yin, B.; Liu, Z.; Wang, Y.; Wang, X.; Liu, W.; Yu, P.; Duan, X.; Liu, C.; Chen, Y.; Zhang, Y.; et al. RON and c-Met facilitate metastasis through the ERK signaling pathway in prostate cancer cells. Oncol. Rep. 2017, 37, 3209–3218. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network. The molecular taxonomy of primary prostate cancer. Cell 2015, 163, 1011–1025. [Google Scholar] [CrossRef]
- Bedolla, R.G.; Shah, D.P.; Huang, S.-B.; Reddick, R.L.; Ghosh, R.; Kumar, A.P. Receptor tyrosine kinase recepteur d’origine nantais as predictive marker for aggressive prostate cancer in African Americans. Mol. Carcinog. 2019, 58, 854–861. [Google Scholar] [CrossRef]
- Wang, N.; Zhang, M.; Chang, Y.; Niu, N.; Guan, Y.; Ye, M.; Li, C.; Tang, J. Directly observing alterations of morphology and mechanical properties of living cancer cells with atomic force microscopy. Talanta 2019, 191, 461–468. [Google Scholar] [CrossRef]
- Ba, M.; Duan, Y. Advance of 2-methoxyestradiol as a promising anticancer agent for cancer therapy. Future Med. Chem. 2020, 12, 273–275. [Google Scholar] [CrossRef]
- Kambhampati, S.; Rajewski, R.A.; Tanol, M.; Haque, I.; Das, A.; Banerjee, S.K.S.; Jha, S.; Burns, D.; Borrego-Diaz, E.; Van Veldhuizen, P.J. A second-generation 2-Methoxyestradiol prodrug is effective against Barrett’s adenocarcinoma in a mouse xenograft model. Mol. Cancer Ther. 2013, 12, 255–263. [Google Scholar] [CrossRef]
- Kumar, A.P.; Garcia, G.E.; Slaga, T.J. 2-methoxyestradiol blocks cell-cycle progression at G(2)/M phase and inhibits growth of human prostate cancer cells. Mol. Carcinog. 2001, 31, 111–124. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Li, G.; Zhu, H.; Dong, X.; Zhao, D.; Jiang, X.; Li, J.; Qiao, H.; Ni, S.; Sun, X. 2-Methoxyestradiol synergizes with sorafenib to suppress hepatocellular carcinoma by simultaneously dysregulating hypoxia-inducible factor-1 and -2. Cancer Lett. 2014, 355, 96–105. [Google Scholar] [CrossRef]
- Sheng, L.-X.; Zhang, J.-Y.; Li, L.; Xie, X.; Wen, X.-A.; Cheng, K.-G. Design, Synthesis, and Evaluation of Novel 2-Methoxyestradiol Derivatives as Apoptotic Inducers through an Intrinsic Apoptosis Pathway. Biomolecules 2020, 10, 123. [Google Scholar] [CrossRef] [PubMed]
- Mueck, A.O.; Seeger, H. 2-Methoxyestradiol—Biology and mechanism of action. Steroids 2010, 75, 625–631. [Google Scholar] [CrossRef] [PubMed]
- Singla, M.; Kumar, A.; Bal, A.; Sarkar, S.; Bhattacharyya, S. Epithelial to mesenchymal transition induces stem cell like phenotype in renal cell carcinoma cells. Cancer Cell Int. 2018, 18, 57. [Google Scholar] [CrossRef]
- Satelli, A.; Batth, I.; Brownlee, Z.; Mitra, A.; Zhou, S.; Noh, H.; Rojas, C.R.; Li, H.; Meng, Q.H.; Li, S. EMT circulating tumor cells detected by cell-surface vimentin are associated with prostate cancer progression. Oncotarget 2017, 8, 49329–49337. [Google Scholar] [CrossRef]
- Khan, T.; Scott, K.F.; Becker, T.M.; Lock, J.; Nimir, M.; Ma, Y.; De Souza, P. The Prospect of Identifying Resistance Mechanisms for Castrate-Resistant Prostate Cancer Using Circulating Tumor Cells: Is Epithelial-to-Mesenchymal Transition a Key Player? Prostate Cancer 2020, 2020, 1–16. [Google Scholar] [CrossRef]
- Nauseef, J.T.; Henry, M.D. Epithelial-to-mesenchymal transition in prostate cancer: Paradigm or puzzle? Nat. Rev. Urol. 2011, 8, 1–12. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Ribatti, D.; Tamma, R.; Annese, T. Epithelial-Mesenchymal Transition in Cancer: A Historical Overview. Transl. Oncol. 2020, 13, 100773. [Google Scholar] [CrossRef]
- Gingrich, J.R.; Barrios, R.J.; Kattan, M.W.; Nahm, H.S.; Finegold, M.J.; Greenberg, N.M. Androgen-independent prostate cancer progression in the TRAMP model. Cancer Res. 1997, 57, 4687–4691. [Google Scholar] [PubMed]
- Gingrich, J.R.; Barrios, R.J.; Morton, R.A.; Boyce, B.F.; DeMayo, F.J.; Finegold, M.J.; Angelopoulou, R.; Rosen, J.M.; Greenberg, N.M. Metastatic prostate cancer in a transgenic mouse. Cancer Res. 1996, 56, 4096–4102. [Google Scholar] [PubMed]
- Tevaarwerk, A.J.; Holen, K.D.; Alberti, D.B.; Sidor, C.; Arnott, J.; Quon, C.; Wilding, G.; Liu, G. Phase I trial of 2-methoxyestradiol NanoCrystal dispersion in advanced solid malignancies. Clin. Cancer Res. 2009, 15, 1460–1465. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Mondul, A.M.; Weinstein, S.J.; Karoly, E.D.; Sampson, J.N.; Albanes, D. Prospective serum metabolomic profile of prostate cancer by size and extent of primary tumor. Oncotarget 2017, 8, 45190–45199. [Google Scholar] [CrossRef]
- Saylor, P.J.; Karoly, E.D.; Smith, M.R. Prospective Study of Changes in the Metabolomic Profiles of Men during Their First Three Months of Androgen Deprivation Therapy for Prostate Cancer. Clin. Cancer Res. 2012, 18, 3677–3685. [Google Scholar] [CrossRef] [PubMed]
- Weiner, A.B.; Matulewicz, R.S.; Eggener, S.E.; Schaeffer, E.M. Increasing incidence of metastatic prostate cancer in the United States (2004–2013). Prostate Cancer Prostatic Dis. 2016, 19, 395–397. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. The hallmarks of cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef]
- Ostman, A.; Böhmer, F.D. Regulation of receptor tyrosine kinase signaling by protein tyrosine phosphatases. Trends Cell Biol. 2001, 11, 258–266. [Google Scholar] [CrossRef]
- Read, J.; Ingram, A.; Al Saleh, H.A.; Platko, K.; Gabriel, K.; Kapoor, A.; Pinthus, J.; Majeed, F.; Qureshi, T.; Al-Nedawi, K. Nuclear transportation of exogenous epidermal growth factor receptor and androgen receptor via extracellular vesicles. Eur. J. Cancer 2017, 70, 62–74. [Google Scholar] [CrossRef]
- Brand, T.M.; Iida, M.; Corrigan, K.L.; Braverman, C.M.; Coan, J.P.; Flanigan, B.G.; Stein, A.P.; Salgia, R.; Rolff, J.; Kimple, R.J.; et al. The receptor tyrosine kinase AXL mediates nuclear translocation of the epidermal growth factor receptor. Sci. Signal. 2017, 10, eaag1064. [Google Scholar] [CrossRef]
- Hsu, S.-C.; Miller, S.A.; Wang, Y.; Hung, M.-C. Nuclear EGFR is required for cisplatin resistance and DNA repair. Am. J. Transl. Res. 2009, 1, 249–258. [Google Scholar] [PubMed]
- Liu, H.-S.; Hsu, P.-Y.; Lai, M.-D.; Chang, H.-Y.; Ho, C.-L.; Cheng, H.-L.; Chen, H.-T.; Lin, Y.-J.; Wu, T.-J.; Tzai, T.-S.; et al. An unusual function of RON receptor tyrosine kinase as a transcriptional regulator in cooperation with EGFR in human cancer cells. Carcinogenesis 2010, 31, 1456–1464. [Google Scholar] [CrossRef]
- Chang, H.-Y.; Liu, H.-S.; Lai, M.-D.; Tsai, Y.-S.; Tzai, T.-S.; Cheng, H.-L.; Chow, N.-H. Hypoxia promotes nuclear translocation and transcriptional function in the oncogenic tyrosine kinase RON. Cancer Res. 2014, 74, 4549–4562. [Google Scholar] [CrossRef] [PubMed]
- Batth, I.S.; Yun, H.; Kumar, A.P. Recepteur d’origine nantais (RON), more than a kinase: Role in castrate-resistant prostate cancer. Mol. Carcinog. 2015, 54, 937–946. [Google Scholar] [CrossRef] [PubMed]
- Howlader, N.; Noone, A.M.; Krapcho, M.; Miller, D.; Brest, A.; Yu, M.; Ruhl, J.; Tatalovich, Z.; Mariotto, A.; Lewis, D.R.; et al. (Eds.) SEER Cancer Statistics Review, 1975–2017; National Cancer Institute: Bethesda, MD, USA, 2019.
- Smith, Z.L.; Eggener, S.E.; Murphy, A.B. African-American Prostate Cancer Disparities. Curr. Urol. Rep. 2017, 18, 81. [Google Scholar] [CrossRef]
- McGinley, K.F.; Tay, K.J.; Moul, J.W. Prostate cancer in men of African origin. Nat. Rev. Urol. 2016, 13, 99–107. [Google Scholar] [CrossRef]
- Attard, G.; Parker, C.; Eeles, R.A.; Schröder, F.; Tomlins, S.A.; Tannock, I.; Drake, C.G.; de Bono, J.S. Prostate cancer. Lancet 2016, 387, 70–82. [Google Scholar] [CrossRef]
- Chang, A.J.; Autio, K.A.; Roach, M.; Scher, H.I. High-risk prostate cancer-Classification and therapy. Nat. Rev. Clin. Oncol. 2014. [Google Scholar] [CrossRef] [PubMed]
- Reddy, B.S.; Watanabe, K.; Weisburger, J.H.; Wynder, E.L. Promoting Effect of Bile Acids in Colon Carcinogenesis in Germ-free and Conventional F344 Rats. Cancer Res. 1977, 37, 3238–3242. [Google Scholar]
- McGarr, S.E.; Ridlon, J.M.; Hylemon, P.B. Diet, anaerobic bacterial metabolism, and colon cancer: A review of the literature. J. Clin. Gastroenterol. 2005, 39, 98–109. [Google Scholar] [CrossRef]
- Pai, R.; Tarnawski, A.S.; Tran, T. Deoxycholic Acid Activates β-Catenin Signaling Pathway and Increases Colon Cell Cancer Growth and Invasiveness. Mol. Biol. Cell 2004, 15, 2156–2163. [Google Scholar] [CrossRef] [PubMed]
- Schöffski, P.; Gordon, M.; Smith, D.C.; Kurzrock, R.; Daud, A.; Vogelzang, N.J.; Lee, Y.; Scheffold, C.; Shapiro, G.I. Phase II randomised discontinuation trial of cabozantinib in patients with advanced solid tumours. Eur. J. Cancer 2017, 86, 296–304. [Google Scholar] [CrossRef] [PubMed]
- Qiao, Y.; Feng, F.Y.; Wang, Y.; Cao, X.; Han, S.; Wilder-Romans, K.; Navone, N.M.; Logothetis, C.; Taichman, R.S.; Keller, E.T.; et al. Mechanistic Support for Combined MET and AR Blockade in Castration-Resistant Prostate Cancer. Neoplasia 2016, 18, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Hussain, S.S.; Huang, S.-B.; Bedolla, R.G.; Rivas, P.; Basler, J.W.; Swanson, G.P.; Hui-Ming Huang, T.; Narayanasamy, G.; Papanikolaou, N.; Miyamoto, H.; et al. Suppression of ribosomal protein RPS6KB1 by Nexrutine increases sensitivity of prostate tumors to radiation. Cancer Lett. 2018, 433, 232–241. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Rivas, P.; Bedolla, R.; Thapa, D.; Reddick, R.L.; Ghosh, R.; Kumar, A.P. Dietary resveratrol prevents development of high-grade prostatic intraepithelial neoplastic lesions: Involvement of SIRT1/S6K axis. Cancer Prev. Res. 2013, 6, 27–39. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, R.; Ganapathy, M.; Alworth, W.L.; Chan, D.C.; Kumar, A.P. Combination of 2-methoxyestradiol (2-ME2) and eugenol for apoptosis induction synergistically in androgen independent prostate cancer cells. J. Steroid Biochem. Mol. Biol. 2009, 113, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Sneddon, I.N. The relation between load and penetration in the axisymmetric boussinesq problem for a punch of arbitrary profile. Int. J. Eng. Sci. 1965, 3, 47–57. [Google Scholar] [CrossRef]
- Lekka, M.; Wiltowska-Zuber, J. Biomedical applications of AFM. J. Phys. Conf. Ser. 2009, 146, 012023. [Google Scholar] [CrossRef]
- Lekka, M.; Laidler, P. Applicability of AFM in cancer detection. Nat. Nanotechnol. 2009, 4, 72. [Google Scholar] [CrossRef]
- Sreekumar, A.; Poisson, L.M.; Rajendiran, T.M.; Khan, A.P.; Cao, Q.; Yu, J.; Laxman, B.; Mehra, R.; Lonigro, R.J.; Li, Y.; et al. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature 2009, 457, 910–914. [Google Scholar] [CrossRef]
- Boudonck, K.J.; Mitchell, M.W.; Német, L.; Keresztes, L.; Nyska, A.; Shinar, D.; Rosenstock, M. Discovery of Metabolomics Biomarkers for Early Detection of Nephrotoxicity. Toxicol. Pathol. 2009, 37, 280–292. [Google Scholar] [CrossRef]
- Lawton, K.A.; Berger, A.; Mitchell, M.; Milgram, K.E.; Evans, A.M.; Guo, L.; Hanson, R.W.; Kalhan, S.C.; Ryals, J.A.; Milburn, M.V. Analysis of the adult human plasma metabolome. Pharmacogenomics 2008, 9, 383–397. [Google Scholar] [CrossRef] [PubMed]
- Evans, A.M.; DeHaven, C.D.; Barrett, T.; Mitchell, M.; Milgram, E. Integrated, Nontargeted Ultrahigh Performance Liquid Chromatography/Electrospray Ionization Tandem Mass Spectrometry Platform for the Identification and Relative Quantification of the Small-Molecule Complement of Biological Systems. Anal. Chem. 2009, 81, 6656–6667. [Google Scholar] [CrossRef] [PubMed]
- Storey, J.D.; Tibshirani, R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 2003, 100, 9440–9445. [Google Scholar] [CrossRef] [PubMed]
- Hoadley, K.A.; Yau, C.; Hinoue, T.; Wolf, D.M.; Lazar, A.J.; Drill, E.; Shen, R.; Taylor, A.M.; Cherniack, A.D.; Thorsson, V.; et al. Cell-of-origni pattterns dominate the molecular classification of 10,000 tumors from 33 types of cancer. Cell 2018, 173, 1291–1304.e6. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of comlex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pI1. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring mutlidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef]
- Ghandi, M.; Huang, F.W.; Jané-Valbuena, J.; Kryukov, G.V.; Lo, C.C.; McDonald, E.R.; Barretina, J.; Gelfand, E.T.; Bielski, C.M.; Li, H.; et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature 2019, 569, 503–508. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Batth, I.S.; Huang, S.-B.; Villarreal, M.; Gong, J.; Chakravarthy, D.; Keppler, B.; Jayamohan, S.; Osmulski, P.; Xie, J.; Rivas, P.; et al. Evidence for 2-Methoxyestradiol-Mediated Inhibition of Receptor Tyrosine Kinase RON in the Management of Prostate Cancer. Int. J. Mol. Sci. 2021, 22, 1852. https://doi.org/10.3390/ijms22041852
Batth IS, Huang S-B, Villarreal M, Gong J, Chakravarthy D, Keppler B, Jayamohan S, Osmulski P, Xie J, Rivas P, et al. Evidence for 2-Methoxyestradiol-Mediated Inhibition of Receptor Tyrosine Kinase RON in the Management of Prostate Cancer. International Journal of Molecular Sciences. 2021; 22(4):1852. https://doi.org/10.3390/ijms22041852
Chicago/Turabian StyleBatth, Izhar Singh, Shih-Bo Huang, Michelle Villarreal, Jingjing Gong, Divya Chakravarthy, Brian Keppler, Sridharan Jayamohan, Pawel Osmulski, Jianping Xie, Paul Rivas, and et al. 2021. "Evidence for 2-Methoxyestradiol-Mediated Inhibition of Receptor Tyrosine Kinase RON in the Management of Prostate Cancer" International Journal of Molecular Sciences 22, no. 4: 1852. https://doi.org/10.3390/ijms22041852
APA StyleBatth, I. S., Huang, S.-B., Villarreal, M., Gong, J., Chakravarthy, D., Keppler, B., Jayamohan, S., Osmulski, P., Xie, J., Rivas, P., Bedolla, R., Liss, M. A., Yeh, I.-T., Reddick, R., Miyamoto, H., Ghosh, R., & Kumar, A. P. (2021). Evidence for 2-Methoxyestradiol-Mediated Inhibition of Receptor Tyrosine Kinase RON in the Management of Prostate Cancer. International Journal of Molecular Sciences, 22(4), 1852. https://doi.org/10.3390/ijms22041852








