Heteroalleles in Common Wheat: Multiple Differences between Allelic Variants of the Gli-B1 Locus
Abstract
1. Introduction
2. Results
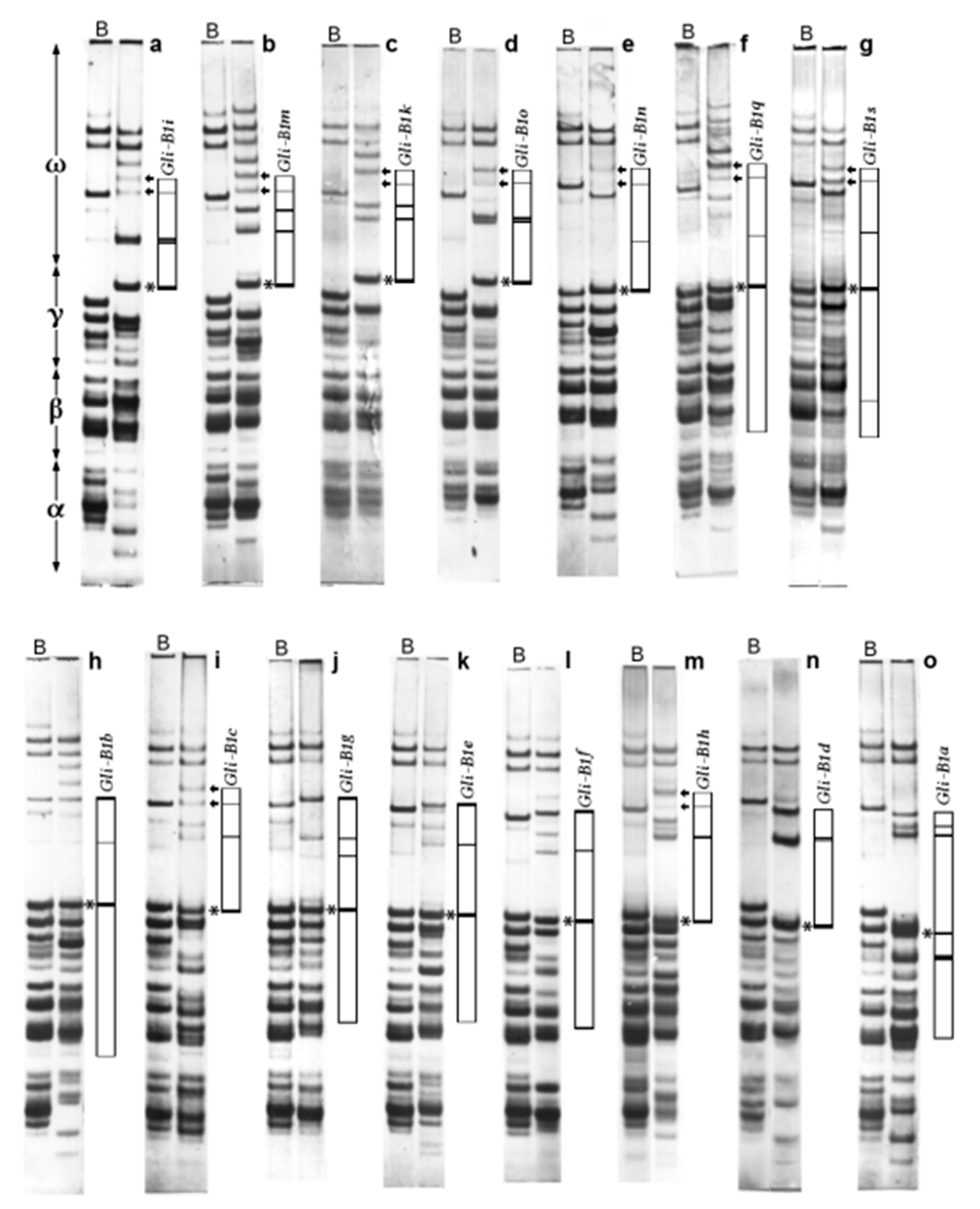
2.1. Alleles at the Gli-B1 Locus Identified by APAGE
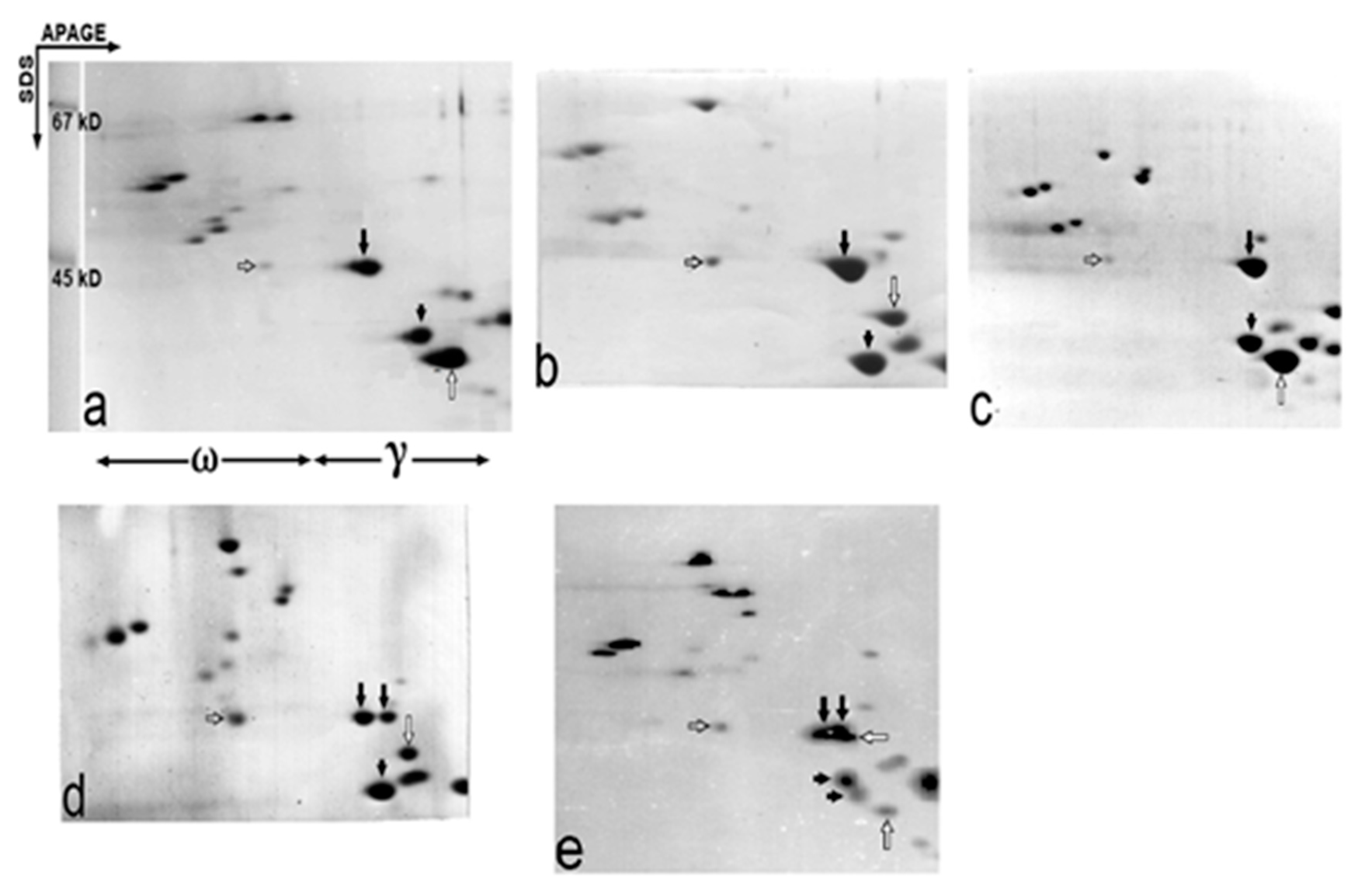
2.2. Apparent Molecular Weight of the Gli-B1-Encoded γ-Gliadins as Studied by Two-Dimensional APAGE × SDS Electrophoresis
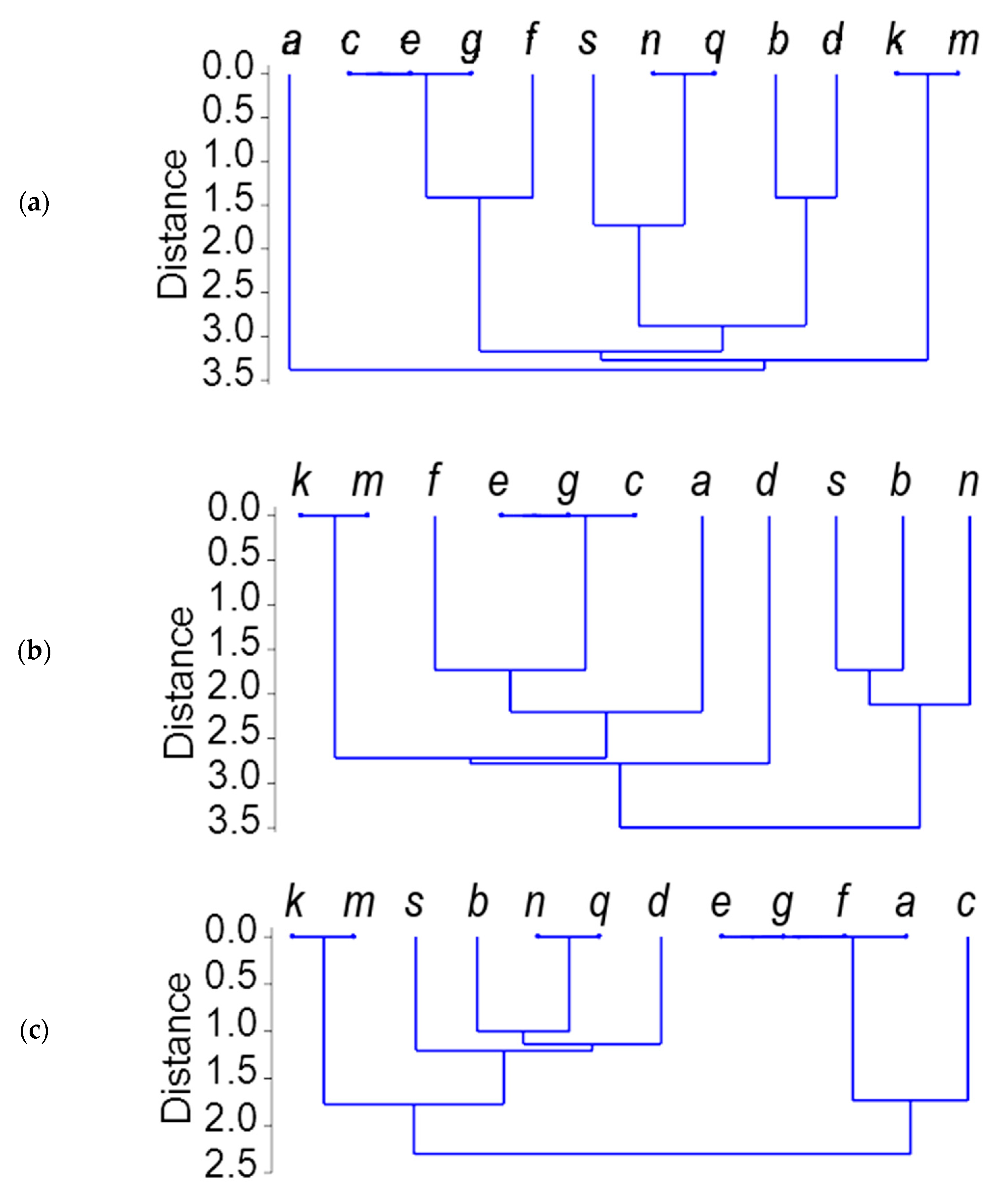
2.3. Gli-B1 Alleles Represented by RFLP Patterns
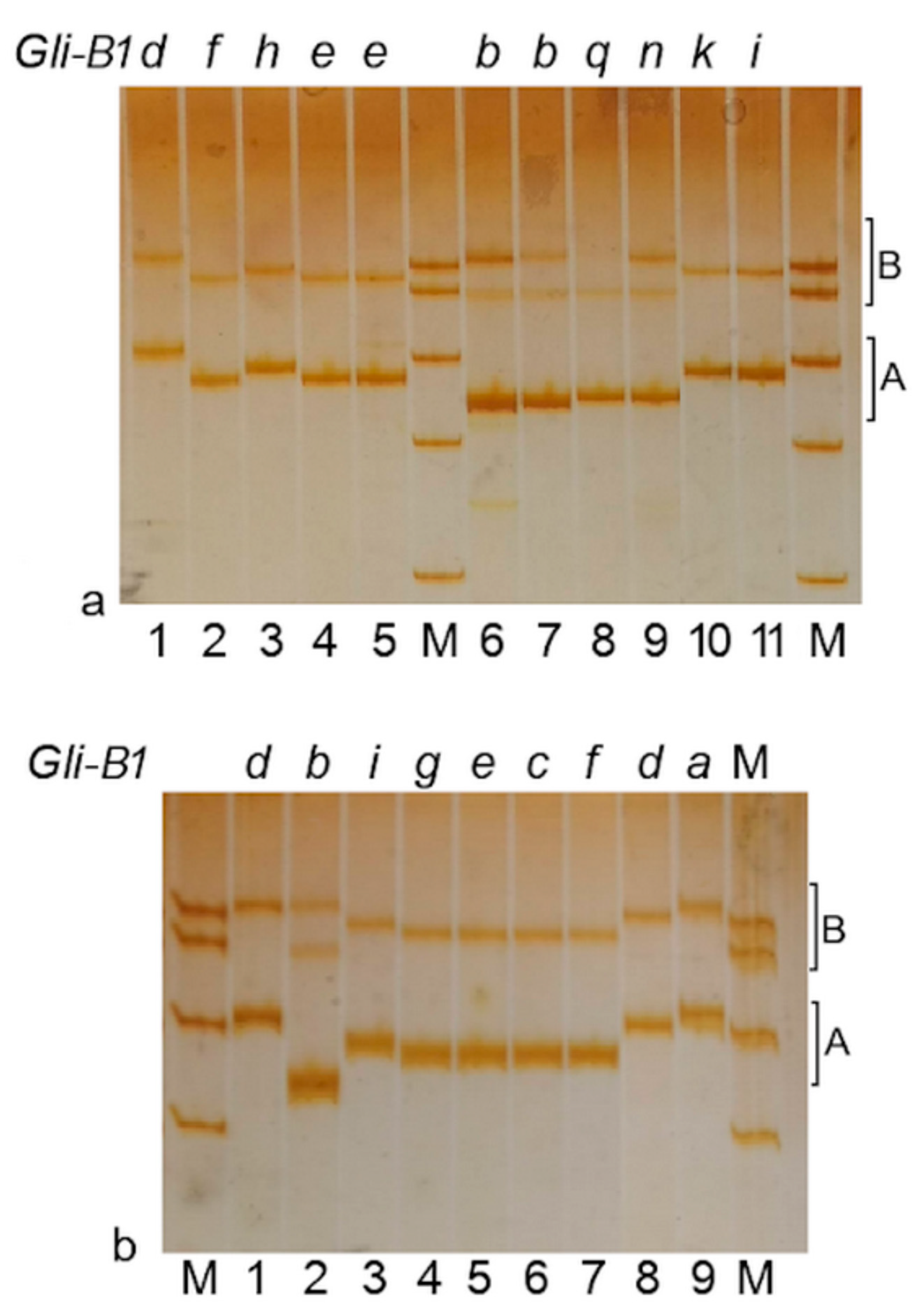
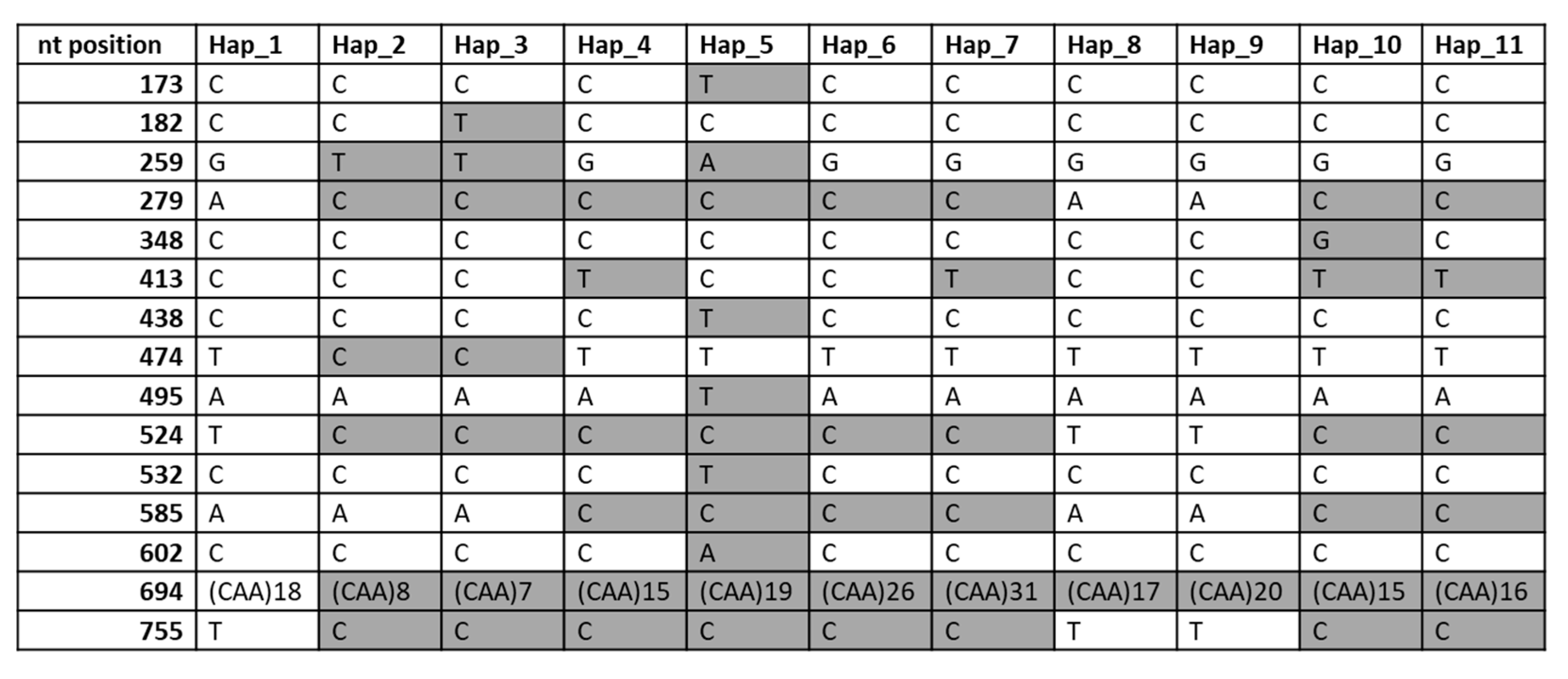
2.4. Comparison of Alleles at the Gli-B1 Locus Using γ-Gliadin Pseudogene SNP Markers
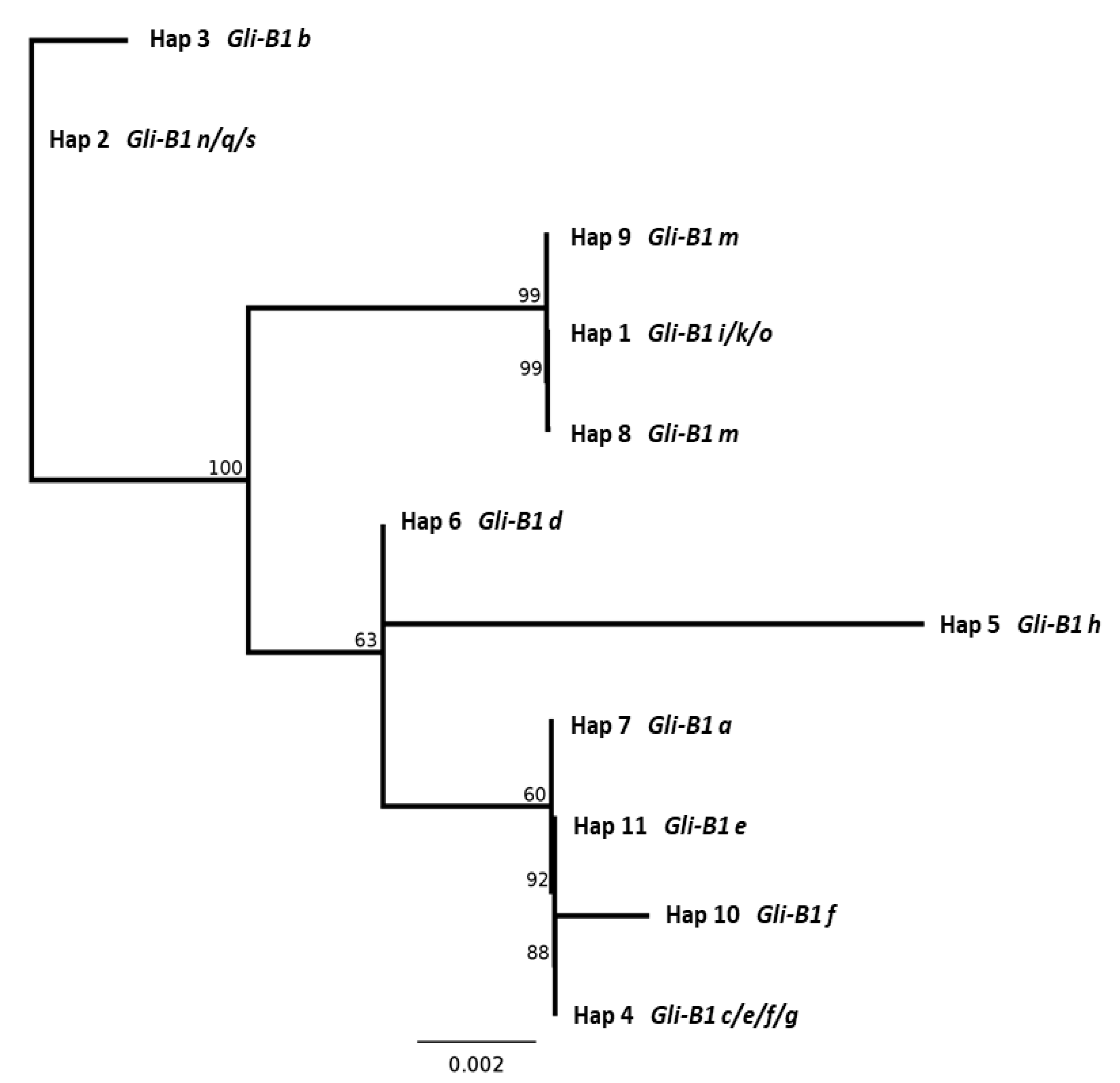
2.5. DNA Sequence of the γ-Gliadin Pseudogene GAG56B in Allelic Variants of the Gli-B1 Locus
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Molecular Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- FAOSTAT (Food and Agriculture Organization of the United Nations). Available online: http://www.fao.org/faostat/es/#data/QC (accessed on 25 January 2021).
- Marcussen, T.; Sandve, S.R.; Heier, L.; Spannagl, M.; Pfeifer, M.; Jakobsen, K.S.; Wulff, B.B.; Steuernagel, B.; Mayer, K.F.; Olsen, O.A.; et al. Ancient hybridizations among the ancestral genomes of bread wheat. Science 2014, 345, 1250092. [Google Scholar] [CrossRef]
- Redaelli, R.; Metakovsky, E.; Davydov, S.D.; Pogna, N.E. Two-dimensional mapping of gliadins using biotypes and null mutants of common wheat cultivar Saratovskaya 29. Hereditas 1994, 121, 131–137. [Google Scholar] [CrossRef]
- Gobaa, S.; Bancel, E.; Branlard, G.; Kleijer, G.; Stamp, P. Proteomic analysis of wheat recombinant inbred lines: Variations in prolamin and dough rheology. J. Cereal Sci. 2008, 47, 610–619. [Google Scholar] [CrossRef]
- Barak, S.; Mudgil, D.; Khatkar, B.S. Biochemical and functional properties of wheat gliadins: A review. Crit. Rev. Food Sci. Nutr. 2015, 55, 357–368. [Google Scholar] [CrossRef]
- Qi, P.F.; Wei, Y.M.; Ouellet, T.; Chen, Q.; Tan, X.; Zheng, Y.L. The γ-gliadin multigene family in common wheat (Triticum aestivum) and its closely related species. BMC Genom. 2009, 10, 168–181. [Google Scholar] [CrossRef] [PubMed]
- Anderson, O.D.; Huo, N.; Gu, J.Q. The gene space in wheat: The complete γ-gliadin gene family from the wheat cultivar Chinese Spring. Funct. Integr. Genom. 2013, 13, 261–273. [Google Scholar] [CrossRef]
- Payne, P.I.; Holt, L.M.; Lawrence, G.J.; Law, C.N. The genetics of gliadin and glutenin, the major storage proteins in the wheat endosperm. Qual. Plant. Plant Foods Hum. Nutr. 1982, 31, 229–241. [Google Scholar] [CrossRef]
- Metakovsky, E.; Chernakov, V.M.; Upelniek, V.P.; Redaelli, R.; Dardavet, M.; Branlard, G.; Pogna, N.E. Minor ω-gliadin-coding loci on chromosome 1A of common wheat: A revision. J. Genet. Breed. 1996, 50, 277–286. [Google Scholar]
- Sozinov, A.A.; Poperelya, F.A. Genetic classification of prolamins and its use for plant breeding. Ann. Technol. Agric. 1980, 29, 229–245. [Google Scholar]
- Metakovsky, E.; Melnik, V.A.; Rodriguez-Quijano, M.; Upelniek, V.P.; Carrillo, J.M. A catalog of gliadin alleles: Polymorphism of 20th-century common wheat germplasm. Crop J. 2018, 6, 629–641. [Google Scholar] [CrossRef]
- Shewry, P.R.; Halford, N.G. Cereal seed storage proteins: Structures, properties and role in grain utilization. J. Exp. Bot. 2000, 53, 947–958. [Google Scholar] [CrossRef] [PubMed]
- Blatter, R.H.E.; Jacomet, S.; Schlumbaum, A. About the origin of European spelt (Triticum spelta L.): Allelic differentiation of the HMW Glutenin B1-1 and A1-2 subunit genes. Theor. Appl. Genet. 2004, 108, 360–367. [Google Scholar] [CrossRef]
- Huang, X.Q.; Cloutier, S. Molecular characterization and genomic organization of low molecular weight glutenin subunit genes at the Glu-3 loci in hexaploid wheat (Triticum aestivum L.). Theor. Appl. Genet. 2008, 116, 953–966. [Google Scholar] [CrossRef] [PubMed]
- Gill, K.S.; Gill, B.S.; Endo, T.R.; Taylor, T. Identification and high-density mapping of gene-rich regions in chromosome group 1 of wheat. Genetics 1996, 144, 1883–1891. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Gu, Y.Q.; Wu, J.; Coleman-Derr, D.; Huo, N.; Crossman, C.; Jia, J.; Zuo, Q.; Ren, Z.; Anderson, O.D.; et al. Rapid evolution and complex structural organization in genomic regions harboring multiple prolamin genes in the polyploid wheat genome. Plant. Molec. Biol. 2007, 65, 189–203. [Google Scholar] [CrossRef]
- Anderson, O.D.; Litts, J.C.; Greene, F.C. The α-gliadin gene family. 1. Character ization of ten new wheat α-gliadin genomic clones, evidence for limited sequence conservation of flanking DNA, and Southern analysis of the gene family. Theor. Appl. Genet. 1997, 95, 50–58. [Google Scholar] [CrossRef]
- Li, W.; Zhang, P.; Fellers, J.P.; Friebe, B.; Gill, B.S. Sequence composition, organization, and evolution of the core Triticeae genome. Plant J. 2004, 40, 500–511. [Google Scholar] [CrossRef]
- Charles, M.; Belcram, H.; Just, J.; Huneau, C.; Violett, A.; Couloux, A.; Segurens, B.; Carter, M.; Huteau, V.; Coriton, O.; et al. Dynamics and differential proliferation of transposable elements during the evolution of the B and A genomes of wheat. Genetics 2006, 180, 1071–1086. [Google Scholar] [CrossRef]
- van Herpen, T.W.J.M.; Goryunova, S.V.; van der Schoot, J.; Mitreva, M.; Salentijn, E.; Vorst, O.; Schenk, M.F.; van Veelen, P.A.; Koning, F.; van Soest, L.J.M.; et al. Alpha-gliadin genes from the A, B, and D genomes of wheat contain different sets of celiac disease epitopes. BMC Genom. 2006, 7, 1–12. [Google Scholar] [CrossRef]
- Harberd, N.P.; Bartles, D.; Thompson, R.D. Analysis of the gliadin multigene loci in bread wheat using nullisomic-tetrasomic lines. Molec. Gen. Genet. 1985, 198, 234–242. [Google Scholar] [CrossRef]
- Gu, Y.Q.; Crossman, C.; Kong, X.; Luo, M.; You, F.M.; Coleman-Derr, D.; Dubcovsky, J.; Anderson, O.D. Genomic organization of the complex α-gliadin gene loci in wheat. Theor. Appl. Genet. 2004, 109, 648–657. [Google Scholar] [CrossRef] [PubMed]
- Anderson, O.D.; Dong, L.; Huo, N.; Gu, Y.Q. A new class of wheat gliadin genes and proteins. PLoS ONE 2012, 7, e52139. [Google Scholar] [CrossRef] [PubMed]
- Von Büren, M.; Lüthy, J.; Hübner, P. A spelt-specific γ-gliadin gene: Discovery and detection. Theor. Appl. Genet. 2000, 100, 271–279. [Google Scholar] [CrossRef]
- Zhang, W.; Gianibelli, M.C.; Ma, W.; Rampling, L.R.; Gale, K.R. Identification of SNPs and development of allele-specific PCR markers for gamma-gliadin alleles in Triticum aestivum. Theor. Appl. Genet. 2003, 107, 130–138. [Google Scholar] [CrossRef] [PubMed]
- Pogna, N.E.; Metakovsky, E.; Redaelli, R.; Raineri, F.; Dachkevitch, T. Recombination mapping of Gli-5, a new gliadin-coding locus on chromosome 1A and 1B in common wheat. Theor. Appl. Genet. 1993, 87, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Pogna, N.E.; Lafiandra, D.; Peillet, P.; Autran, J.C. Evidence for a direct causal effect of low molecular weight subunits of glutenins on gluten viscoelasticity in durum wheats. J. Cereal. Sci. 1988, 7, 211–214. [Google Scholar] [CrossRef]
- Metakovsky, E.; Melnik, V.A.; Vaccino, P.; Rodriguez-Quijano, M. Comparison of alleles at the Gli-1 loci of common wheat by means of two-dimensional electrophoresis of gliadin and RFLP analysis. Cytol. Genet. Kiev 2018, 52, 11–20. [Google Scholar] [CrossRef]
- Vaccino, P.; Metakovsky, E. Gliadin alleles in DNA RFLP patterns of common wheat: Implication for analysis of organization and evolution of complex loci. Theor. Appl. Genet. 1995, 90, 173–181. [Google Scholar] [CrossRef]
- Endo, T.R.; Gill, B.S. Somatic karyotype, heterochromatin distribution, and nature of chromosome differentiation in common wheat, Triticum aestivum L. em Thell. Chromosome 1984, 89, 361–369. [Google Scholar] [CrossRef]
- Friebe, B.; Gill, B.S. C-band polymorphism and structural rearrangements detected in common wheat (Triticum aestivum). Euphytica 1994, 78, 1–5. [Google Scholar]
- Dvorak, J.; Chen, K.C. Distribution of nonstructural variation between wheat cultivars along chromosome arm 6Bp: Evidence from the linkage map and physical map of the arm. Genetics 1984, 106, 325–333. [Google Scholar] [CrossRef] [PubMed]
- Dvorak, J.; Appels, R. Investigation of homologous crossing over and sister chromatid exchange in the wheat Nor-B2 locus coding for rRNA and Gli-B2 locus coding for gliadins. Genetics 1986, 113, 1037–1056. [Google Scholar] [CrossRef] [PubMed]
- Dvorak, J.; McGuire, P.E. Nonstructural chromosome differentiation among wheat cultivars, with special reference to differentiation of chromosomes in related species. Genetics 1981, 97, 391–414. [Google Scholar]
- Crossway, A.; Dvorak, J. Distribution of nonstructural variation along three chromosome arms between wheat cultivars Chinese Spring and Cheyenne. Genetics 1984, 106, 309–324. [Google Scholar] [CrossRef] [PubMed]
- Dvorak, J.; Yang, Z.L.; You, F.M.; Luo, M.C. Deletion polymorphism in wheat chromosome regions with contrasting recombination rates. Genetics 2004, 168, 1665–1675. [Google Scholar] [CrossRef]
- Wicker, T.; Yahiaoui, N.; Guyot, R.; Schlagenhauf, E.; Liu, Z.D.; Dubcovsky, J.; Keller, B. Rapid genome divergence at orthologous low molecular weight glutenin loci of the A and A(m) genomes of wheat. Plant Cell 2003, 15, 1186–1197. [Google Scholar] [CrossRef]
- Ravel, C.; Praud, S.; Murigneux, A.; Canaguier, A.; Sapet, F.; Samson, D.; Balfourier, F.; Dufour, P.; Chalhoub, B.; Brunel, D.; et al. Single-nucleotide polymorphism frequency in a set of selected lines of bread wheat (Triticum aestivum L.). Genome 2006, 49, 1131–1139. [Google Scholar] [CrossRef] [PubMed]
- Ram, S.; Sharma, S.; Verma, A.; Tyagi, B.S.; Peña, R.J. Comparative analyses of LMW glutenin alleles in bread wheat using allele-specific PCR and SDS-PAGE. J. Cereal Sci. 2011, 54, 488–493. [Google Scholar] [CrossRef]
- Matsuoka, Y. Evolution of polyploid Triticum wheats under cultivation: The role of domestication, natural hybridization and allopolyploid speciation in their diversification. Plant Cell Physiol. 2011, 52, 750–764. [Google Scholar] [CrossRef] [PubMed]
- Metakovsky, E.; Kudryavtsev, A.M.; Iakobashvili, Z.A.; Novoselskaya, A.Y. Analysis of phylogenetic relations of durum, carthlicum and common wheats by means of comparison of alleles of gliadin loci. Theor. Appl. Genet. 1989, 77, 881–887. [Google Scholar] [CrossRef] [PubMed]
- Dvorak, J.; Luo, M.C.; Akhunov, E.D.N.I. Vavilov theory of centres of diversity in the light of current understanding of wheat diversity, domestication and evolution. Czech J. Genet. Plant. Breed. 2011, 47, S20–S27. [Google Scholar] [CrossRef]
- Dvorak, J.; Akhunov, E.D.; Akhunova, A.R.; Deal, K.; Luo, N.C. Molecular characterization of a diagnostic DNA marker for domesticated tetraploid wheat provides evidence for gene flow from wild tetraploid wheat to hexaploid wheat. Molec. Biol. Evol. 2006, 23, 1386–1396. [Google Scholar] [CrossRef]
- Walkowiak, S.; Gao, L.; Monat, C.; Haberer, G.; Kassa, M.T.; Brinton, J.; Ramirez-Gonzalez, R.H.; Kolodziej, M.C.; Delorean, E.; Thambugala, D.; et al. Multiple wheat genomes reveal global variation in modern breeding. Nature 2020. [Google Scholar] [CrossRef] [PubMed]
- Metakovsky, E.; Melnik, V.A.; Pascual, L.; Wrigley, C.W. Gliadin genotypes worldwide for spring wheats (Triticum aestivum L.). 2. Strong differentiation of polymorphism between countries and regions of origin. J. Cereal. Sci. 2019, 87, 311–317. [Google Scholar] [CrossRef]
- Fu, Y.B.; Peterson, G.W.; Yu, J.K.; Gao, L.; Jia, J.; Richards, K.W. Impact of plant breeding on genetic diversity of the Canadian hard red spring wheat germplasm as revealed by EST-derived SSR markers. Theor. Appl. Genet. 2006, 112, 1239–1247. [Google Scholar] [CrossRef] [PubMed]
- GRIS Database (CIMMYT International Maize and Wheat Improvement Center). Available online: http://wheatpedigree.net/ (accessed on 11 February 2021).
- Murray, M.; Thompson, W.F. Rapid isolation of high-molecular-weight plant DNA. Nucleic Acid Res. 1980, 8, 4321–4325. [Google Scholar] [CrossRef]
- Gebhardt, C.; Ritter, E.; Debener, T.; Schachtschabel, U.; Walkemeier, B.; Uhrig, H.; Salamini, F. RFLP analysis and linkage mapping in Solanum tuberosum. Theor. Appl. Genet. 1989, 78, 65–75. [Google Scholar] [CrossRef]
- Bartels, D.; Altosaar, I.; Harberd, N.P.; Barker, R.F.; Thompson, R.D. Molecular analysis of gamma-gliadin gene families at the complex Gli-1 locus of bread wheat (T. aestivum L.). Theor. Appl. Genet. 1986, 72, 845–853. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. Isolation of plant DNA from fresh tissue. Focus 1990, 12, 13–15. [Google Scholar]
- Thompson, J.D.; Higgins, J.G.; Gibson, T.J. Clustal W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl. Acid Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
| Cultivar | Country | Year | Allele | Primer | Length (bp) |
|---|---|---|---|---|---|
| Bezostaya-1 | Russia | 1959 | b | GliB1.1 | 369 |
| Gabo | Australia | 1942 | b | GliB1.1 | 369 |
| Marquis | Canada | 1907 | b | GliB1.1 | 369 |
| Insignia | Australia | 1946 | i | GliB1.1 | 400 |
| Mentana | Italy | 1913 | k | GliB1.1 | 400 |
| Titien | France | 1985 | m | GliB1.1 | 400 |
| Aragon-03 | Spain | 1940 | o | GliB1.1 | 400 |
| Intensivnaya | Kyrgyzstan | 1978 | n | GliB1.1 | 372 |
| Goelent | France | 1985 | q | GliB1.1 | 372 |
| Salmone | Italy | 1980 | s | GliB1.1 | 372 |
| Cartaya | Spain | 1984 | l | GliB1.1 or GliB1.2 | - |
| Chinese-Spring | China | - | a | GliB1.2 | 415 |
| Pavon-F-76 | Mexico | 1971 | d | GliB1.2 | 409 |
| Laura | Canada | 1986 | d | GliB1.2 | 409 |
| Rinconada | Spain | 1981 | d | GliB1.2 | 409 |
| Suneca | Australia | 1981 | d | GliB1.2 | 409 |
| Prinqual | France | 1978 | c | GliB1.2 | 397 |
| Escualo | Spain | 1981 | e | GliB1.2 | 397 |
| Glenlea | Canada | 1972 | e | GliB1.2 | 397 |
| Arminda | Netherlands | 1976 | f | GliB1.2 | 397 |
| Cappelle-Desprez | France | 1946 | f | GliB1.2 | 397 |
| Galahad | UK | 1983 | g | GliB1.2 | 397 |
| Ardec | Belgium | 1979 | h | GliB1.2 | 402 |
| Caia | Portugal | - | h | GliB1.2 | 403 |
| Cultivar | Country | Year | Allele | Haplotype (Figure 5) | EM of γ-Gliadin 1 |
|---|---|---|---|---|---|
| Insignia | Australia | 1946 | i | 1 | I |
| Pane-247 | Spain | 1960 | k | 1 | I |
| Etoile-de-Choisy | France | 1950 | m | 1 | I |
| Aragon-03 | Spain | 1940 | o | 1 | I |
| Intensivnaya | Kyrgyzstan | 1978 | n | 2 | II |
| Spada | Italy | 1985 | n | 2 | II |
| Lesostepka-75 | Ukraine | 1945 | q | 2 | II |
| Salmone | Italy | 1980 | s | 2* | II |
| Alcalá | Spain | 1984 | b | 3 | III |
| Anza | Mexico, USA | 1971 | b | 3 | III |
| Glenlea | Canada | 1972 | e | 4 | IV |
| Marius | France | 1976 | f | 4 | IV |
| Pernel | France | 1983 | f | 4 | IV |
| Adalid | Spain | 1987 | g | 4 | IV |
| Calodine | Italy | 1991 | g | 4 | IV |
| Diego | Spain | - | c | 4 | VIII |
| Prinqual | France | 1978 | c | 4 | VIII |
| Siete-Cerros-66 | Mexico | 1966 | c | 4 | VIII |
| Ardec | Belgium | 1979 | h | 5 | V |
| Caia | Portugal | - | h | 5 | V |
| Pepital | Netherlands | 1989 | h | 5 | V |
| Cajeme-71 | Mexico | 1971 | d | 6 | VI |
| Chopin | France | 1984 | d | 6 | VI |
| Katepwa | Canada | 1981 | d | 6 | VI |
| Chinese-Spring | China | - | a | 7 | VII |
| Pyrotrix-28 | Kazakhstan | 1973 | m | 8 | I |
| Titien | France | 1985 | m | 9 | I |
| Astral | France | 1972 | f | 10 | IV |
| Floreal | France | 1984 | f | 10 | V |
| Saratovskaya-39 | Russia | 1968 | e | 11 | IV |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Metakovsky, E.; Pascual, L.; Vaccino, P.; Melnik, V.; Rodriguez-Quijano, M.; Popovych, Y.; Chebotar, S.; Rogers, W.J. Heteroalleles in Common Wheat: Multiple Differences between Allelic Variants of the Gli-B1 Locus. Int. J. Mol. Sci. 2021, 22, 1832. https://doi.org/10.3390/ijms22041832
Metakovsky E, Pascual L, Vaccino P, Melnik V, Rodriguez-Quijano M, Popovych Y, Chebotar S, Rogers WJ. Heteroalleles in Common Wheat: Multiple Differences between Allelic Variants of the Gli-B1 Locus. International Journal of Molecular Sciences. 2021; 22(4):1832. https://doi.org/10.3390/ijms22041832
Chicago/Turabian StyleMetakovsky, Eugene, Laura Pascual, Patrizia Vaccino, Viktor Melnik, Marta Rodriguez-Quijano, Yulia Popovych, Sabina Chebotar, and William John Rogers. 2021. "Heteroalleles in Common Wheat: Multiple Differences between Allelic Variants of the Gli-B1 Locus" International Journal of Molecular Sciences 22, no. 4: 1832. https://doi.org/10.3390/ijms22041832
APA StyleMetakovsky, E., Pascual, L., Vaccino, P., Melnik, V., Rodriguez-Quijano, M., Popovych, Y., Chebotar, S., & Rogers, W. J. (2021). Heteroalleles in Common Wheat: Multiple Differences between Allelic Variants of the Gli-B1 Locus. International Journal of Molecular Sciences, 22(4), 1832. https://doi.org/10.3390/ijms22041832







