Aptamer-Functionalized Hybrid Nanostructures for Sensing, Drug Delivery, Catalysis and Mechanical Applications
Abstract
1. Introduction
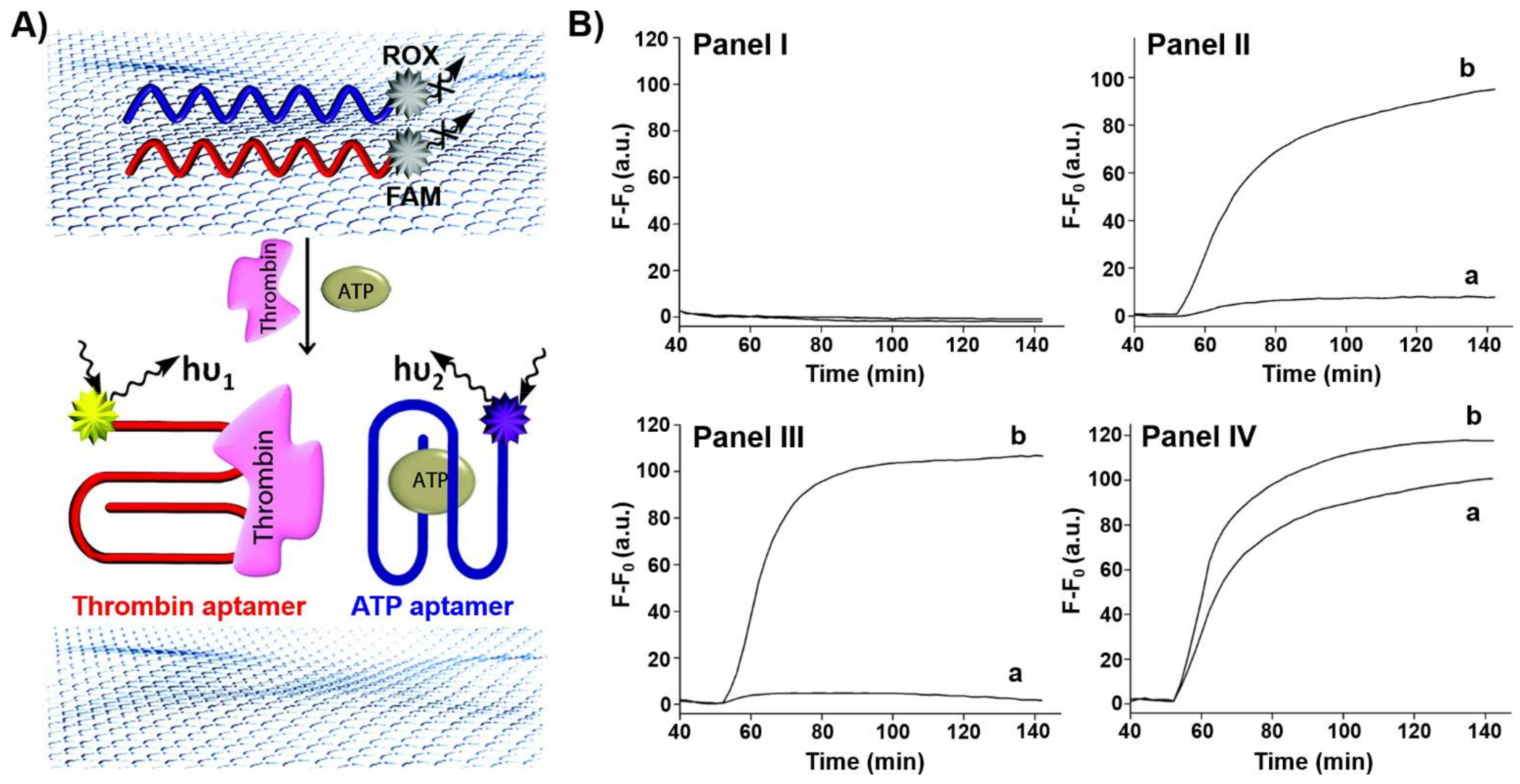
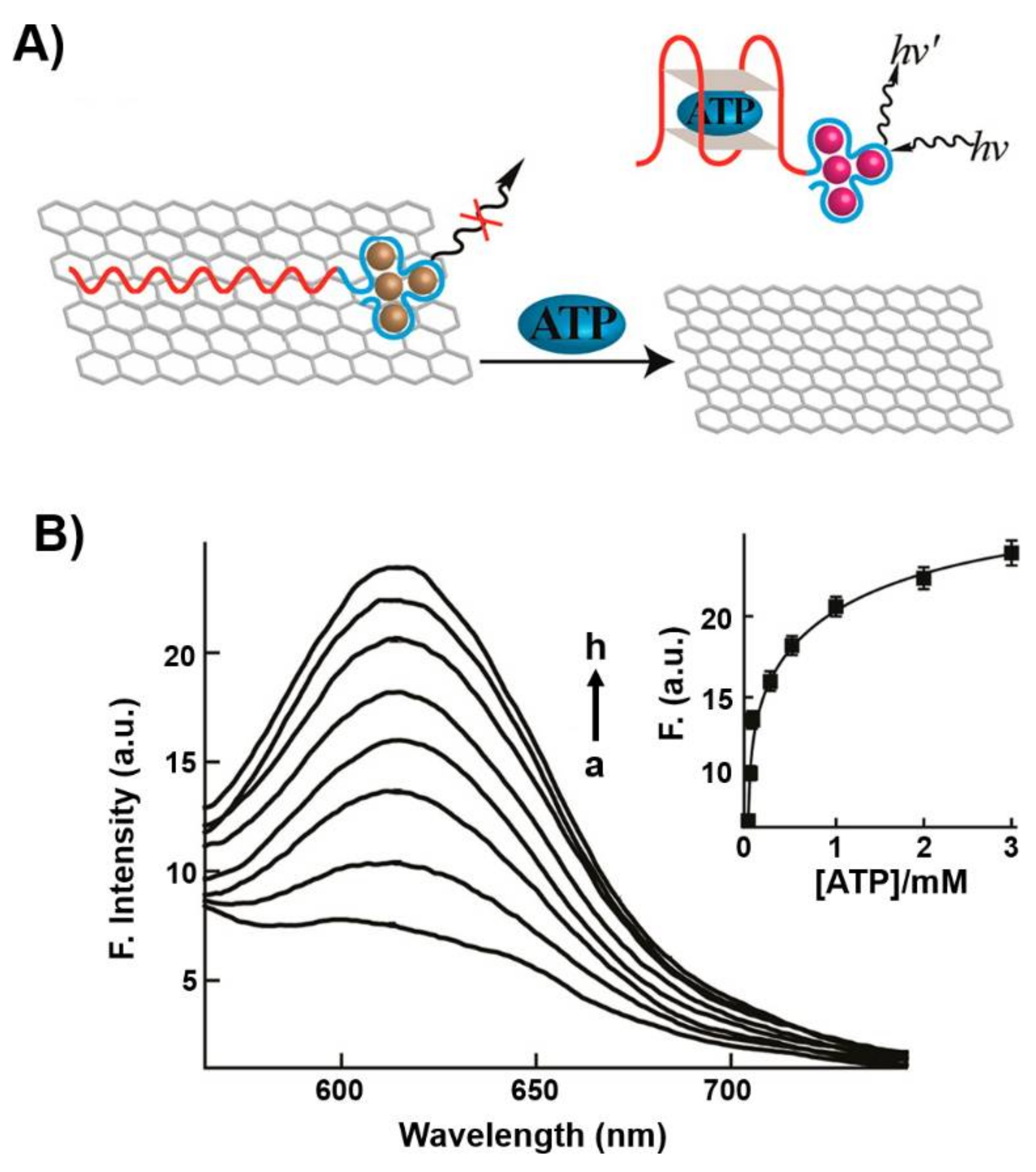
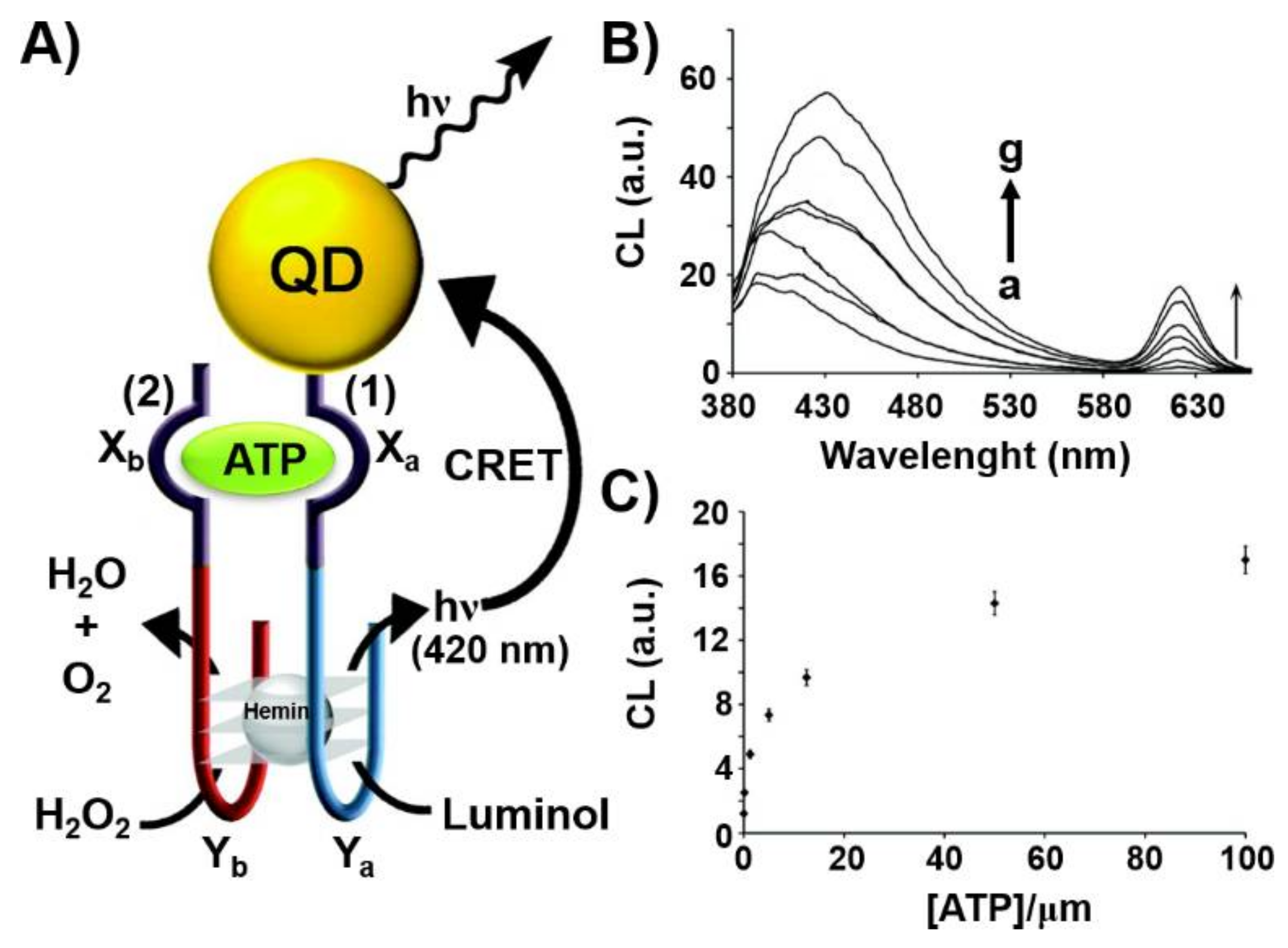
2. Hybrid Aptamer Nanostructures for Sensing Applications
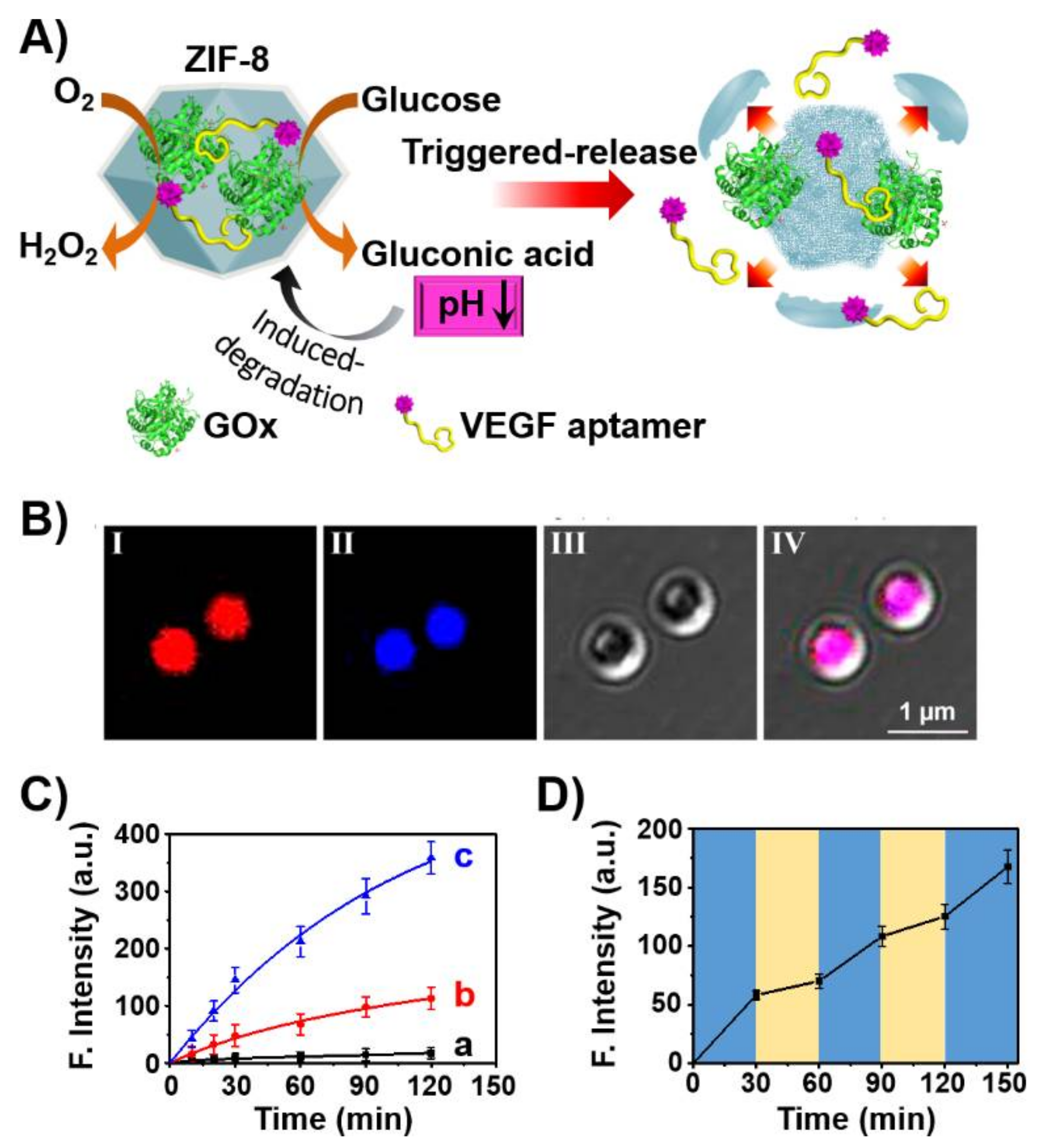
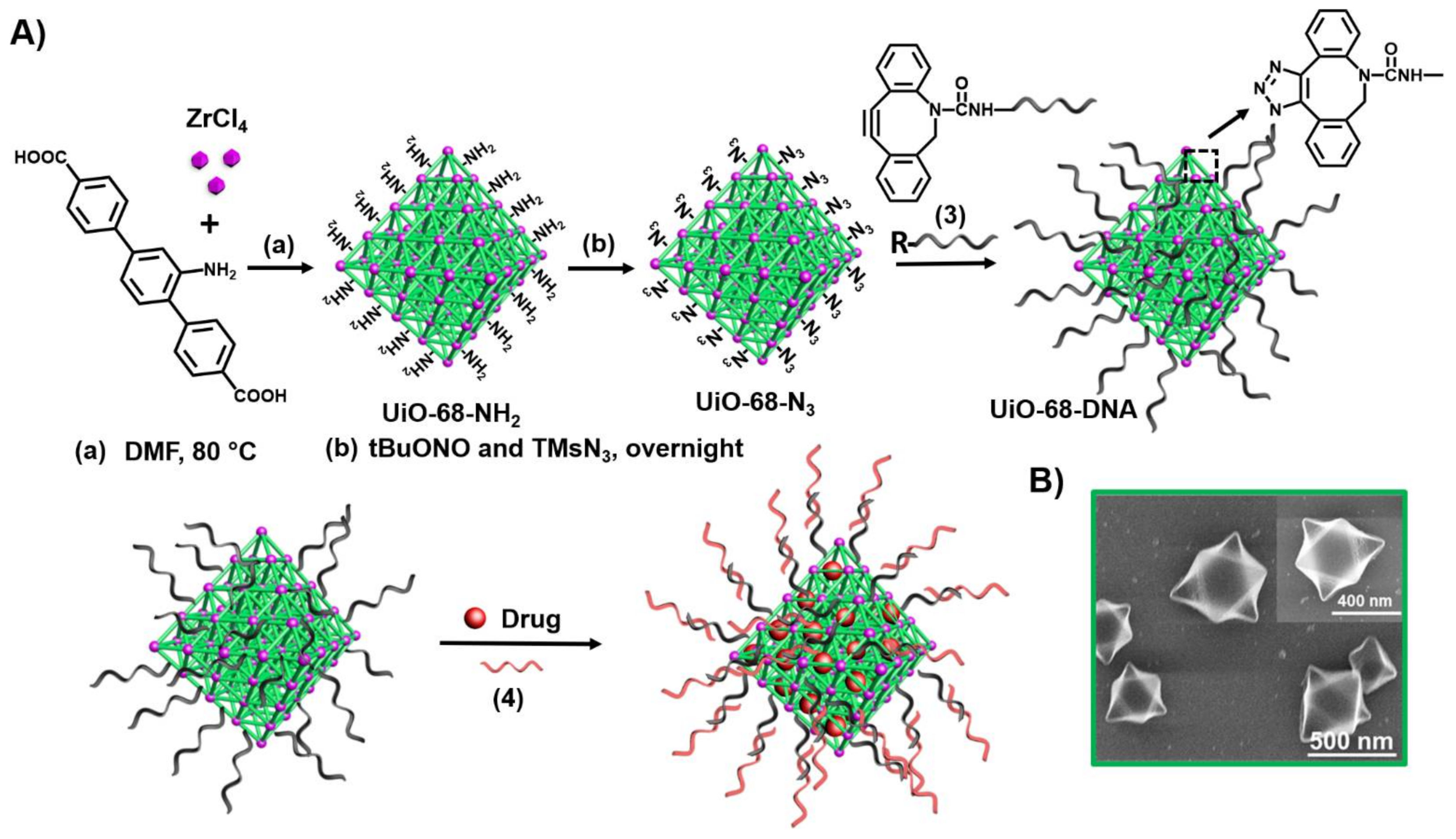
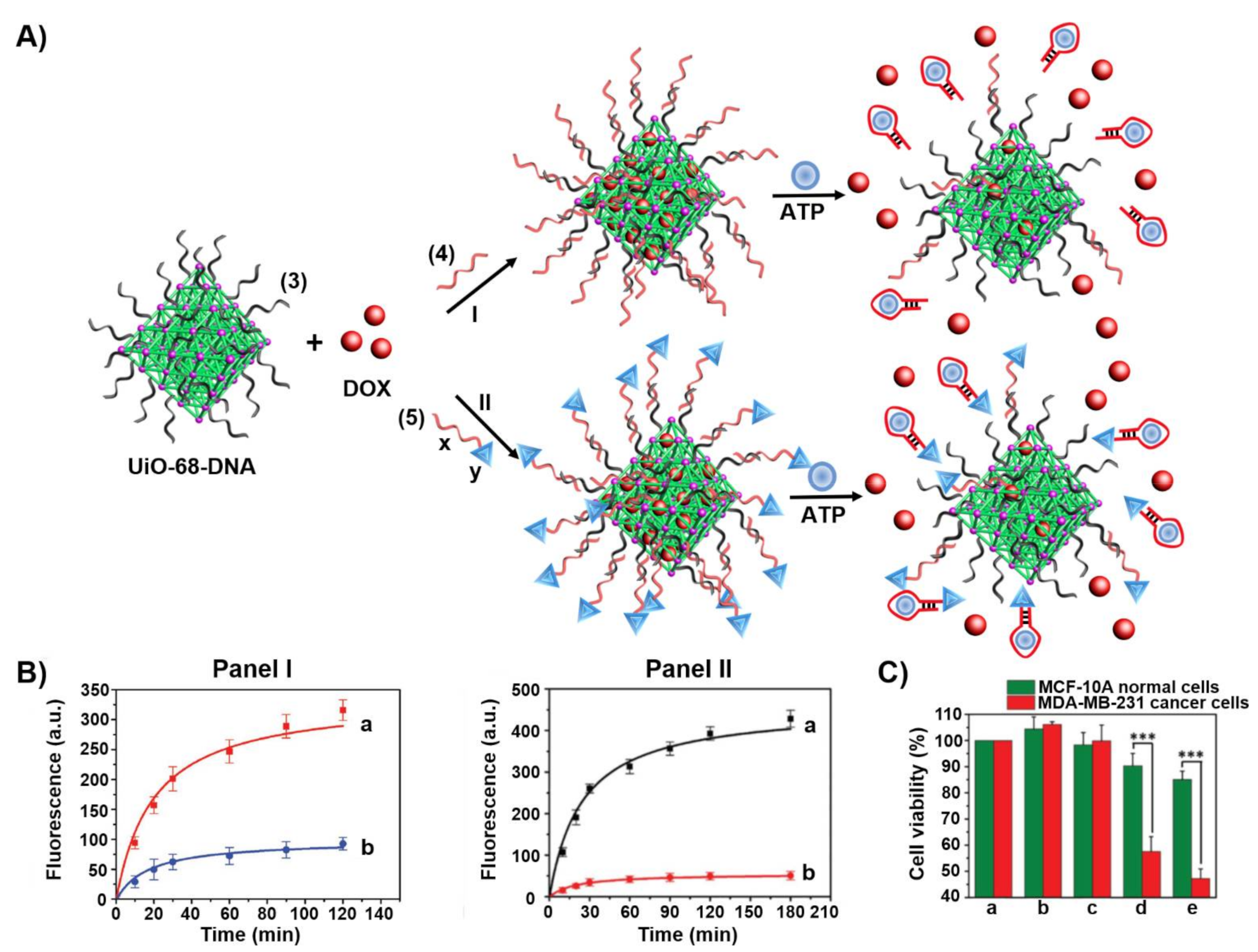
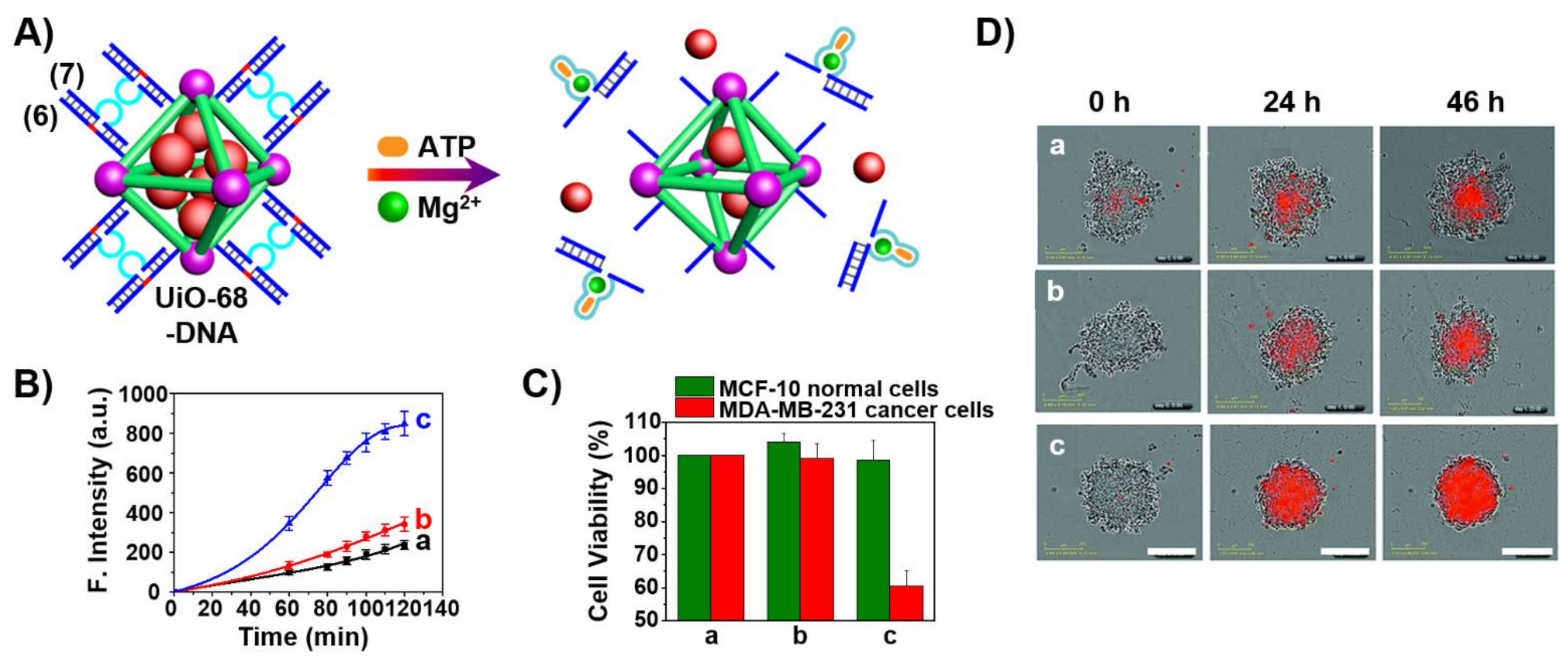
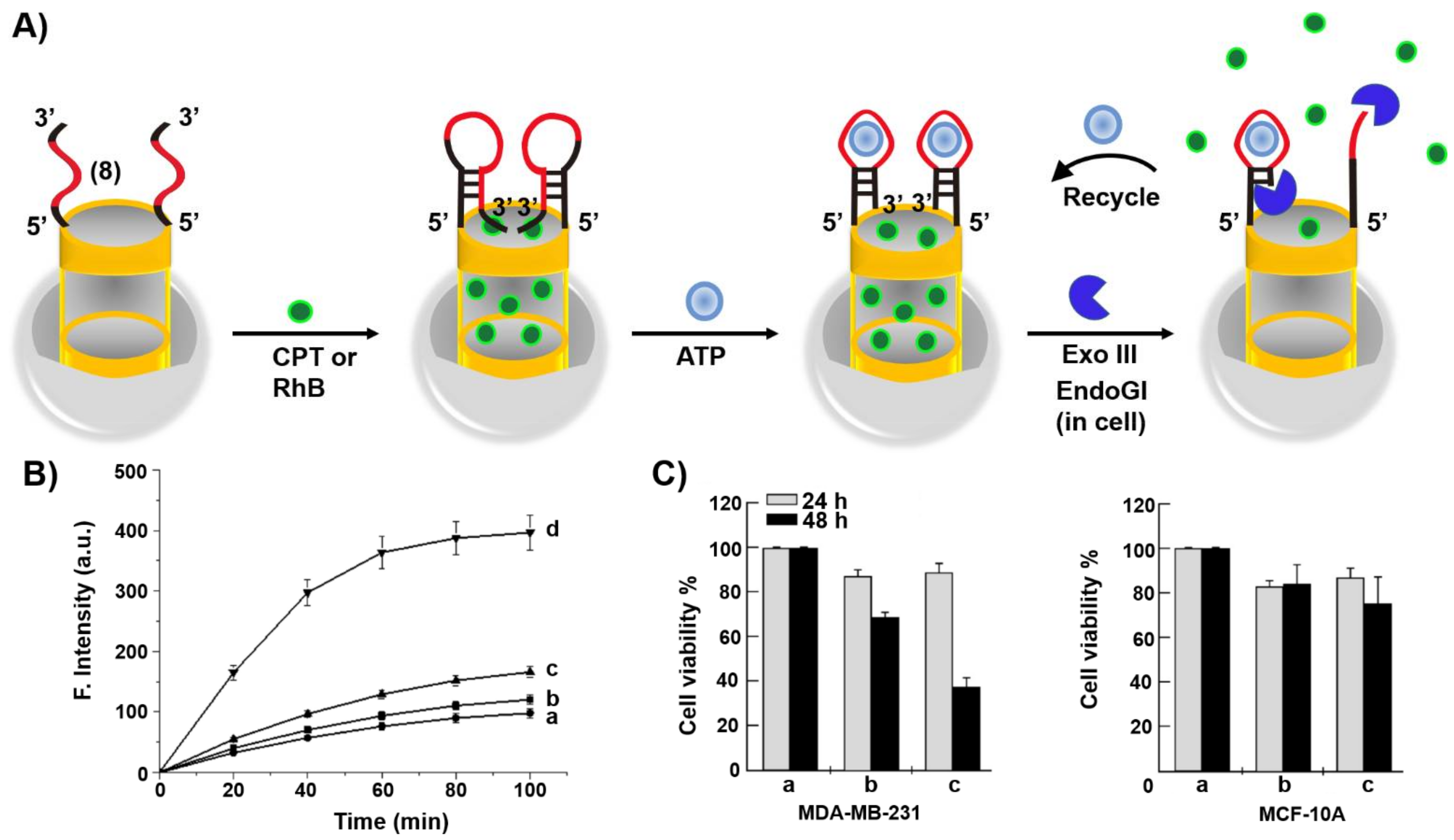
3. Aptamers as Responsive Gates for Nano or Microcarriers
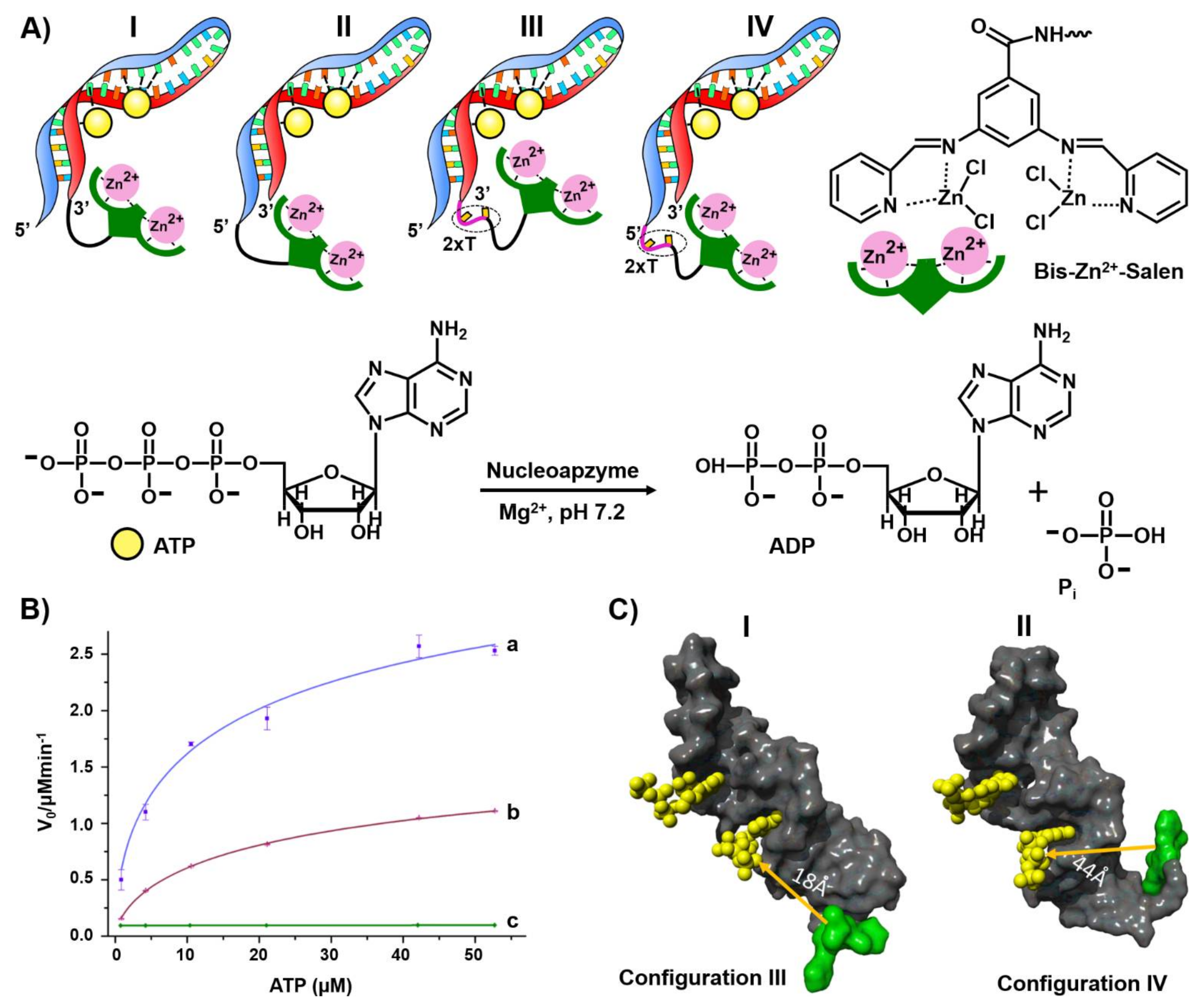
4. Aptamer Nanostructures for Catalysis
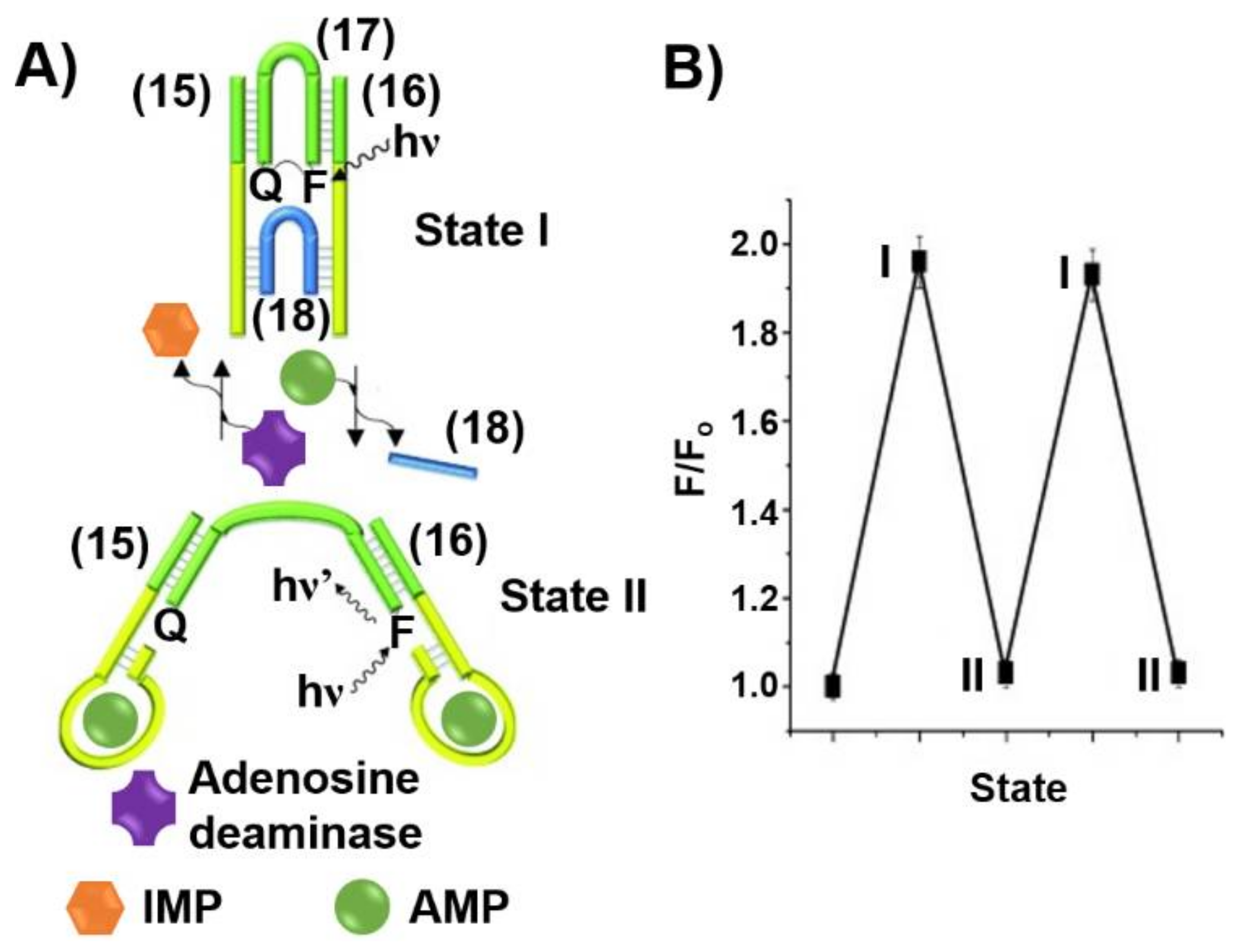
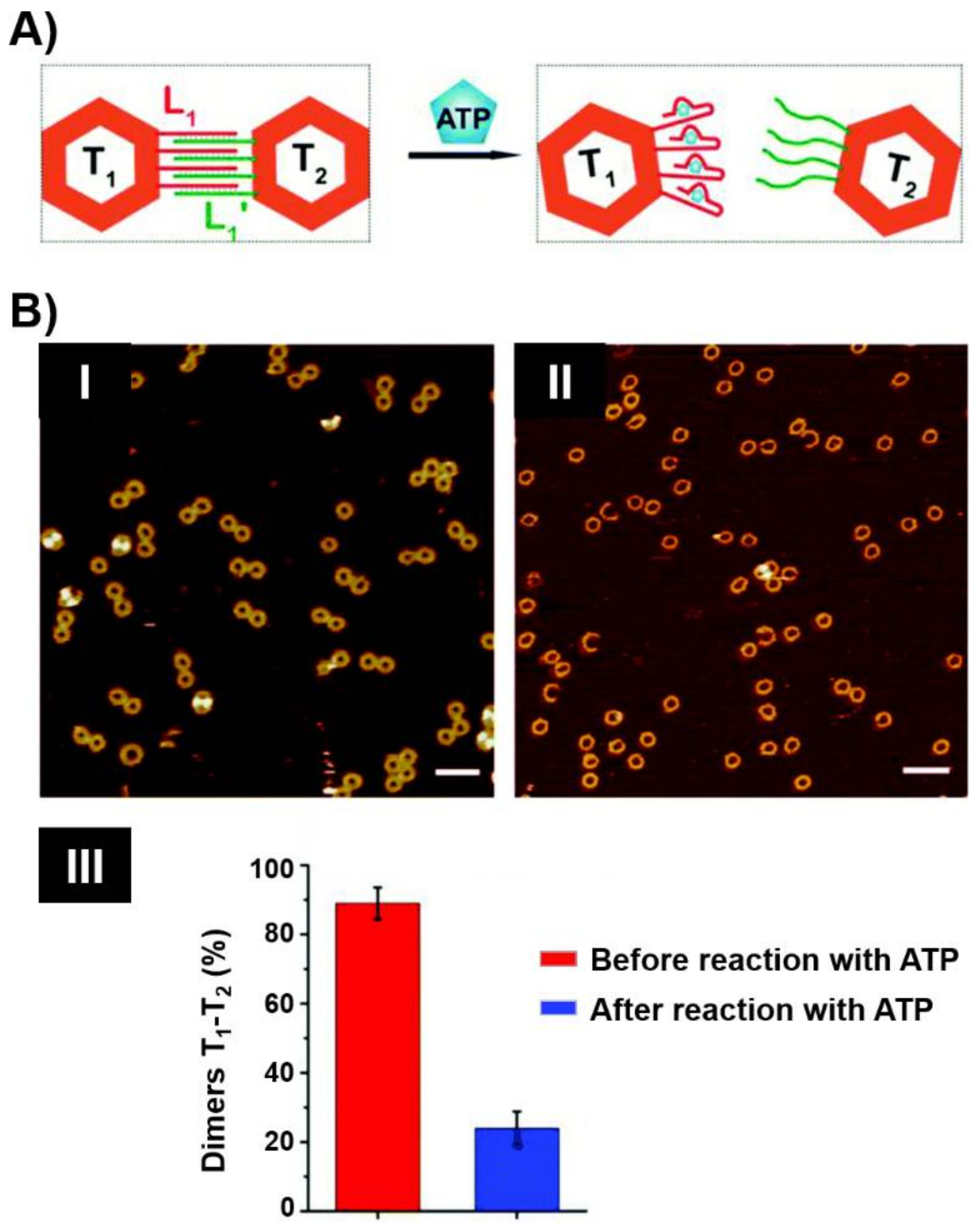
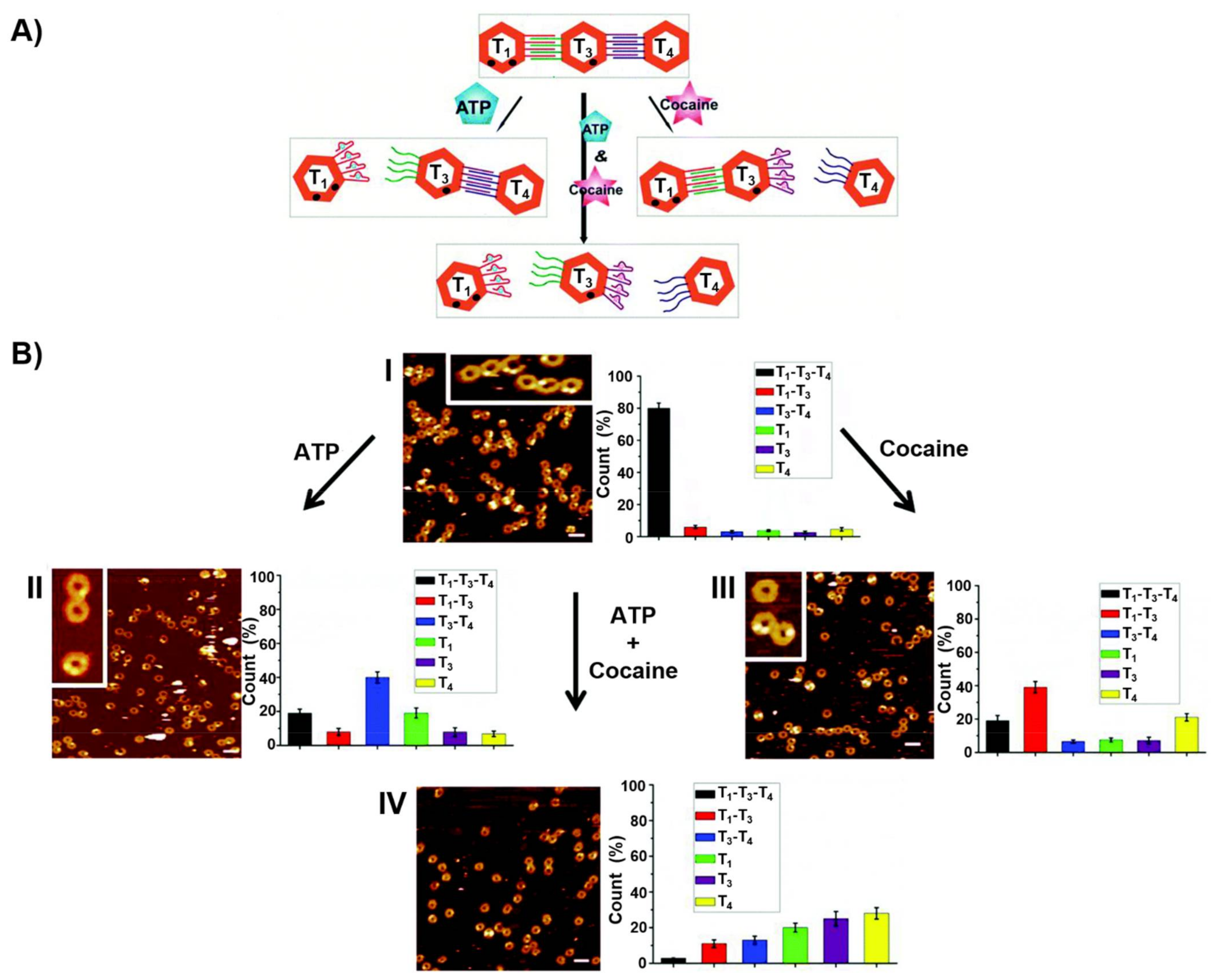
5. DNA Motor Systems Driven by Aptamer-Ligand Complexes
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhong, Y.; Zhao, J.; Li, J.; Liao, X.; Chen, F. Advances of aptamers screened by Cell-SELEX in selection procedure, cancer diagnostics and therapeutics. Anal. Biochem. 2020, 598, 113620. [Google Scholar] [CrossRef] [PubMed]
- Darmostuk, M.; Rimpelova, S.; Gbelcova, H.; Ruml, T. Current approaches in SELEX: An update to aptamer selection technology. Biotechnol. Adv. 2014, 33, 1141–1161. [Google Scholar] [CrossRef]
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef]
- Tan, W.; Donovan, M.J.; Jiang, J. Aptamers from cell-based selection for bioanalytical applications. Chem. Rev. 2013, 113, 2842–2862. [Google Scholar] [CrossRef] [PubMed]
- Mascini, M.; Palchetti, I.; Tombelli, S. Nucleic acid and peptide aptamers: Fundamentals and bioanalytical aspects. Angew. Chem. Int. Ed. 2012, 51, 1316–1332. [Google Scholar] [CrossRef]
- Vázquez-González, M.; Zhou, Z.; Biniuri, Y.; Willner, B.; Willner, I. Mimicking Functions of Native Enzymes or Photosynthetic Reaction Centers by Nucleoapzymes and Photonucleoapzymes. Biochemistry 2020. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Li, Q.; Zuo, X.; Fan, C. Catalytic Nucleic Acids for Bioanalysis. ACS Appl. Bio. Mater. 2020, 3, 2674–2685. [Google Scholar] [CrossRef]
- Famulok, M.; Hartig, J.S.; Mayer, G. Functional aptamers and aptazymes in biotechnology, diagnostics, and therapy. Chem. Rev. 2007, 107, 3715–3743. [Google Scholar] [CrossRef]
- Meng, H.M.; Liu, H.; Kuai, H.; Peng, R.; Mo, L.; Zhang, X.B. Aptamer-integrated DNA nanostructures for biosensing, bioimaging and cancer therapy. Chem. Soc. Rev. 2016, 45, 2583–2602. [Google Scholar] [CrossRef]
- Lao, Y.H.; Phua, K.K.L.; Leong, K.W. Aptamer nanomedicine for cancer therapeutics: Barriers and potential for translation. ACS Nano 2015, 9, 2235–2254. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Jimmy Huang, P.J.; Ding, J.; Liu, J. Aptamer-based biosensors for biomedical diagnostics. Analyst 2014, 139, 2627–2640. [Google Scholar] [CrossRef] [PubMed]
- Famulok, M.; Mayer, G. Aptamer modules as sensors and detectors. Acc. Chem. Res. 2011, 44, 1349–1358. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Li, B.; Wang, E. “Fitting” makes “sensing” simple: Label-free detection strategies based on nucleic acid aptamers. Acc. Chem. Res. 2013, 46, 203–213. [Google Scholar] [CrossRef]
- Yoo, H.; Jo, H.; Oh, S.S. Detection and beyond: Challenges and advances in aptamer-based biosensors. Mater. Adv. 2020, 1, 2663–2687. [Google Scholar] [CrossRef]
- Yu, H.; Chen, Z.; Liu, Y.; Alkhamis, O.; Song, Z.; Xiao, Y. Fabrication of Aptamer-Modified Paper Electrochemical Devices for On-site Biosensing. Angew. Chem. Int. Ed. 2020. [Google Scholar] [CrossRef]
- Ge, L.; Wang, W.; Sun, X.; Hou, T.; Li, F. Affinity-Mediated Homogeneous Electrochemical Aptasensor on a Graphene Platform for Ultrasensitive Biomolecule Detection via Exonuclease-Assisted Target-Analog Recycling Amplification. Anal. Chem. 2016, 88, 2212–2219. [Google Scholar] [CrossRef]
- Li, H.; Arroyo-Currás, N.; Kang, D.; Ricci, F.; Plaxco, K.W. Dual-Reporter Drift Correction To Enhance the Performance of Electrochemical Aptamer-Based Sensors in Whole Blood. J. Am. Chem. Soc. 2016, 138, 15809–15812. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Xiang, Y.; Lu, Y.; Crooks, R.M. Aptamer-based origami paper analytical device for electrochemical detection of adenosine. Angew. Chem. Int. Ed. 2012, 51, 6925–6928. [Google Scholar] [CrossRef]
- Zang, Y.; Lei, J.; Hao, Q.; Ju, H. Signal-On photoelectrochemical sensing strategy based on target-dependent aptamer conformational conversion for selective detection of Lead(II) ion. ACS Appl. Mater. Interfaces 2014, 6, 15991–15997. [Google Scholar] [CrossRef]
- Li, R.; Liu, Y.; Cheng, L.; Yang, C.; Zhang, J. Photoelectrochemical aptasensing of kanamycin using visible light-activated carbon nitride and graphene oxide nanocomposites. Anal. Chem. 2014, 86, 9372–9375. [Google Scholar] [CrossRef]
- Freeman, R.; Girsh, J.; Willner, I. Nucleic acid/quantum dots (QDs) hybrid systems for optical and photoelectrochemical sensing. ACS Appl. Mater. Interfaces 2013, 5, 2815–2834. [Google Scholar] [CrossRef]
- Xiaoru, Z.; Shuguo, L.; Xia, J.; Shusheng, Z. A new photoelectrochemical aptasensor for the detection of thrombin based on functionalized graphene and CdSe nanoparticles multilayers. Chem. Commun. 2011, 47, 4929–4931. [Google Scholar] [CrossRef]
- Zhu, Z.; Feng, M.; Zuo, L.; Zhu, Z.; Wang, F.; Chen, L.; Li, J.; Shan, G.; Luo, S.Z. An aptamer based surface plasmon resonance biosensor for the detection of ochratoxin A in wine and peanut oil. Biosens. Bioelectron. 2015, 65, 320–326. [Google Scholar] [CrossRef]
- Bai, Y.; Feng, F.; Zhao, L.; Wang, C.; Wang, H.; Tian, M.; Qin, J.; Duan, Y.; He, X. Aptamer/thrombin/aptamer-AuNPs sandwich enhanced surface plasmon resonance sensor for the detection of subnanomolar thrombin. Biosens. Bioelectron. 2013, 47, 265–270. [Google Scholar] [CrossRef] [PubMed]
- Kwon, M.J.; Lee, J.; Wark, A.W.; Lee, H.J. Nanoparticle-enhanced surface plasmon resonance detection of proteins at attomolar concentrations: Comparing different nanoparticle shapes and sizes. Anal. Chem. 2012, 84, 1702–1707. [Google Scholar] [CrossRef] [PubMed]
- Ozalp, V.C.; Bayramoglu, G.; Erdem, Z.; Arica, M.Y. Pathogen detection in complex samples by quartz crystal microbalance sensor coupled to aptamer functionalized core-shell type magnetic separation. Anal. Chim. Acta 2015, 853, 533–540. [Google Scholar] [CrossRef] [PubMed]
- Porfirieva, A.; Evtugyn, G.; Hianik, T. Polyphenothiazine modified electrochemical aptasensor for detection of human α-thrombin. Electroanalysis 2007, 19, 1915–1920. [Google Scholar] [CrossRef]
- Minunni, M.; Tombelli, S.; Gullotto, A.; Luzi, E.; Mascini, M. Development of biosensors with aptamers as bio-recognition element: The case of HIV-1 Tat protein. Biosens. Bioelectron. 2004, 20, 1149–1156. [Google Scholar] [CrossRef]
- Pavlov, V.; Xiao, Y.; Shlyahovsky, B.; Willner, I. Aptamer-functionalized Au nanoparticles for the amplified optical detection of thrombin. J. Am. Chem. Soc. 2004, 126, 11768–11769. [Google Scholar] [CrossRef]
- Wei, X.; Zhou, W.; Sanjay, S.T.; Zhang, J.; Jin, Q.; Xu, F.; Dominguez, D.C.; Li, X. Multiplexed Instrument-Free Bar-Chart SpinChip Integrated with Nanoparticle-Mediated Magnetic Aptasensors for Visual Quantitative Detection of Multiple Pathogens. Anal. Chem. 2018, 90, 9888–9896. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Hu, Y.; Dong, N.; Zhu, G.; Zhu, T.; Jiang, N. A novel SERS-based magnetic aptasensor for prostate specific antigen assay with high sensitivity. Biosens. Bioelectron. 2017, 94, 286–291. [Google Scholar] [CrossRef] [PubMed]
- Jie, G.; Yuan, J. Novel magnetic Fe3O4@CdSe composite quantum dot-based electrochemiluminescence detection of thrombin by a multiple DNA cycle amplification strategy. Anal. Chem. 2012, 84, 2811–2817. [Google Scholar] [CrossRef]
- Poturnayova, A.; Castillo, G.; Subjakova, V.; Tatarko, M.; Snejdarkova, M.; Hianik, T. Optimization of cytochrome c detection by acoustic and electrochemical methods based on aptamer sensors. Sens. Actuators B Chem. 2017, 238, 817–827. [Google Scholar] [CrossRef]
- Neves, M.A.D.; Blaszykowski, C.; Bokhari, S.; Thompson, M. Ultra-high frequency piezoelectric aptasensor for the label-free detection of cocaine. Biosens. Bioelectron. 2015, 72, 383–392. [Google Scholar] [CrossRef] [PubMed]
- Chang, K.; Pi, Y.; Lu, W.; Wang, F.; Pan, F.; Li, F.; Jia, S.; Shi, J.; Deng, S.; Chen, M. Label-free and high-sensitive detection of human breast cancer cells by aptamer-based leaky surface acoustic wave biosensor array. Biosens. Bioelectron. 2014, 60, 318–324. [Google Scholar] [CrossRef]
- Li, K.; Qin, W.; Li, F.; Zhao, X.; Jiang, B.; Wang, K.; Deng, S.; Fan, C.; Li, D. Nanoplasmonic imaging of latent fingerprints and identification of cocaine. Angew. Chem. Int. Ed. 2013, 52, 11542–11545. [Google Scholar] [CrossRef]
- Wen, Y.; Pei, H.; Wan, Y.; Su, Y.; Huang, Q.; Song, S.; Fan, C. DNA nanostructure-decorated surfaces for enhanced aptamer-target binding and electrochemical cocaine sensors. Anal. Chem. 2011, 83, 7418–7423. [Google Scholar] [CrossRef]
- Kawano, R.; Osaki, T.; Sasaki, H.; Takinoue, M.; Yoshizawa, S.; Takeuchi, S. Rapid detection of a cocaine-binding aptamer using biological nanopores on a chip. J. Am. Chem. Soc. 2011, 133, 8474–8477. [Google Scholar] [CrossRef]
- Du, Y.; Chen, C.; Yin, J.; Li, B.; Zhou, M.; Dong, S.; Wang, E. Solid-state probe based electrochemical aptasensor for cocaine: A potentially convenient, sensitive, repeatable, and integrated sensing platform for drugs. Anal. Chem. 2010, 82, 1556–1563. [Google Scholar] [CrossRef]
- Sabherwal, P.; Shorie, M.; Pathania, P.; Chaudhary, S.; Bhasin, K.K.; Bhalla, V.; Suri, C.R. Hybrid aptamer-antibody linked fluorescence resonance energy transfer based detection of trinitrotoluene. Anal. Chem. 2014, 86, 7200–7204. [Google Scholar] [CrossRef]
- Priyanka, P.; Shorie, M.; Bhalla, V.; Pathania, P.; Suri, C.R. Nanobioprobe mediated DNA aptamers for explosive detection. Chem. Commun. 2014, 50, 1080–1082. [Google Scholar] [CrossRef]
- Ho, M.Y.; D’Souza, N.; Migliorato, P. Electrochemical aptamer-based sandwich assays for the detection of explosives. Anal. Chem. 2012, 84, 4245–4247. [Google Scholar] [CrossRef]
- Liu, M.; Khan, A.; Wang, Z.; Liu, Y.; Yang, G.; Deng, Y.; He, N. Aptasensors for pesticide detection. Biosens. Bioelectron. 2019, 130, 174–184. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Wang, L.; Tu, Z.; Sun, X.; He, Q.; Lei, Z.; Xu, C.; Liu, Y.; Zhang, X.; Yang, J.; et al. Organophosphorus pesticides detection using broad-specific single-stranded DNA based fluorescence polarization aptamer assay. Biosens. Bioelectron. 2014, 55, 216–219. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Wu, J.; Zheng, L.; Qian, H.; Xue, F.; Wu, Y.; Pan, D.; Adeloju, S.B.; Chen, W. Rolling chain amplification based signal-enhanced electrochemical aptasensor for ultrasensitive detection of ochratoxin A. Anal. Chem. 2013, 85, 10842–10849. [Google Scholar] [CrossRef] [PubMed]
- Schoukroun-Barnes, L.R.; Wagan, S.; White, R.J. Enhancing the analytical performance of electrochemical RNA aptamer-based sensors for sensitive detection of aminoglycoside antibiotics. Anal. Chem. 2014, 86, 1131–1137. [Google Scholar] [CrossRef]
- Song, K.M.; Jeong, E.; Jeon, W.; Cho, M.; Ban, C. Aptasensor for ampicillin using gold nanoparticle based dual fluorescence-colorimetric methods. Anal. Bioanal. Chem. 2012, 402, 2153–2161. [Google Scholar] [CrossRef] [PubMed]
- Rowe, A.A.; Miller, E.A.; Plaxco, K.W. Reagentless measurement of aminoglycoside antibiotics in blood serum via an electrochemical, ribonucleic acid aptamer-based biosensor. Anal. Chem. 2010, 82, 7090–7095. [Google Scholar] [CrossRef]
- Ni, S.; Shen, Z.; Zhang, P.; Liu, G. Enhanced performance of an electrochemical aptasensor for real-time detection of vascular endothelial growth factor (VEGF) by nanofabrication and ratiometric measurement. Anal. Chim. Acta 2020, 1121, 74–82. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, X.; Feng, P.; Han, Q.; Liu, W.; Lu, Y.; Song, C.; Li, F. Photodriven Regeneration of G-Quadruplex Aptasensor for Sensitively Detecting Thrombin. Anal. Chem. 2020, 92, 7419–7424. [Google Scholar] [CrossRef]
- Zhu, J.; Gan, H.; Wu, J.; Ju, H. Molecular Machine Powered Surface Programmatic Chain Reaction for Highly Sensitive Electrochemical Detection of Protein. Anal. Chem. 2018, 90, 5503–5508. [Google Scholar] [CrossRef]
- Yang, J.; Dou, B.; Yuan, R.; Xiang, Y. Aptamer/Protein Proximity Binding-Triggered Molecular Machine for Amplified Electrochemical Sensing of Thrombin. Anal. Chem. 2017, 89, 5138–5143. [Google Scholar] [CrossRef]
- Zhao, J.; Hu, S.; Zhong, W.; Wu, J.; Shen, Z.; Chen, Z.; Li, G. Highly sensitive electrochemical aptasensor based on a ligase-assisted exonuclease III-catalyzed degradation reaction. ACS Appl. Mater. Interfaces 2014, 6, 7070–7075. [Google Scholar] [CrossRef]
- Kwon, O.S.; Park, S.J.; Hong, J.Y.; Han, A.R.; Lee, J.S.; Lee, J.S.; Oh, J.H.; Jang, J. Flexible FET-Type VEGF aptasensor based on nitrogen-doped graphene converted from conducting polymer. ACS Nano 2012, 6, 1486–1493. [Google Scholar] [CrossRef]
- Hu, J.; Wang, T.; Kim, J.; Shannon, C.; Easley, C.J. Quantitation of femtomolar protein levels via direct readout with the electrochemical proximity assay. J. Am. Chem. Soc. 2012, 134, 7066–7072. [Google Scholar] [CrossRef]
- Rotem, D.; Jayasinghe, L.; Salichou, M.; Bayley, H. Protein detection by nanopores equipped with aptamers. J. Am. Chem. Soc. 2012, 134, 2781–2787. [Google Scholar] [CrossRef]
- Kizer, M.; Li, P.; Cress, B.F.; Lin, L.; Jing, T.T.; Zhang, X.; Xia, K.; Linhardt, R.J.; Wang, X. RNA Aptamers with Specificity for Heparosan and Chondroitin Glycosaminoglycans. ACS Omega 2018, 3, 13667–13675. [Google Scholar] [CrossRef]
- Chen, L.; Fu, Y.; Wang, N.; Yang, A.; Li, Y.; Wu, J.; Ju, H.; Yan, F. Organic Electrochemical Transistors for the Detection of Cell Surface Glycans. ACS Appl. Mater. Interfaces 2018, 10, 18470–18477. [Google Scholar] [CrossRef]
- He, Y.; Li, J.; Liu, Y. Reusable and Dual-Potential Responses Electrogenerated Chemiluminescence Biosensor for Synchronously Cytosensing and Dynamic Cell Surface N-Glycan Evaluation. Anal. Chem. 2015, 87, 9777–9785. [Google Scholar] [CrossRef]
- Micura, R.; Höbartner, C. Fundamental studies of functional nucleic acids: Aptamers, riboswitches, ribozymes and DNAzymes. Chem. Soc. Rev. 2020, 49, 7331–7353. [Google Scholar] [CrossRef]
- Lake, R.J.; Yang, Z.; Zhang, J.J.; Lu, Y. DNAzymes as Activity-Based Sensors for Metal Ions: Recent Applications, Demonstrated Advantages, Current Challenges, and Future Directions. Acc. Chem. Res. 2019, 52, 3275–3286. [Google Scholar] [CrossRef] [PubMed]
- Willner, I.; Shlyahovsky, B.; Zayats, M.; Willner, B. DNAzymes for sensing, nanobiotechnology and logic gate applications. Chem. Soc. Rev. 2008, 37, 1153–1165. [Google Scholar] [CrossRef]
- Travascio, P.; Li, Y.; Sen, D. DNA-enhanced peroxidase activity of a DNA-aptamer-hemin complex. Chem. Biol. 1998, 5, 505–517. [Google Scholar] [CrossRef]
- Luo, G.F.; Biniuri, Y.; Vázquez-González, M.; Wulf, V.; Fadeev, M.; Lavi, R.; Willner, I. Metal Ion-Terpyridine-Functionalized L-Tyrosinamide Aptamers: Nucleoapzymes for Oxygen Insertion into C-H Bonds and the Transformation of L-Tyrosinamide into Amidodopachrome. Adv. Funct. Mater. 2019, 29, 1901484. [Google Scholar] [CrossRef]
- Luo, G.F.; Biniuri, Y.; Chen, W.H.; Wang, J.; Neumann, E.; Marjault, H.B.; Nechushtai, R.; Winkler, M.; Happe, T.; Willner, I. Modelling Photosynthesis with ZnII-Protoporphyrin All-DNA G-Quadruplex/Aptamer Scaffolds. Angew. Chem. Int. Ed. 2020, 59, 9163–9170. [Google Scholar] [CrossRef]
- Panigaj, M.; Johnson, M.B.; Ke, W.; McMillan, J.; Goncharova, E.A.; Chandler, M.; Afonin, K.A. Aptamers as Modular Components of Therapeutic Nucleic Acid Nanotechnology. ACS Nano 2019, 13, 12301–12321. [Google Scholar] [CrossRef]
- Zununi Vahed, S.; Fathi, N.; Samiei, M.; Maleki Dizaj, S.; Sharifi, S. Targeted cancer drug delivery with aptamer-functionalized polymeric nanoparticles. J. Drug Target. 2019, 27, 292–299. [Google Scholar] [CrossRef]
- Gopinath, S.C.B.; Lakshmipriya, T.; Chen, Y.; Arshad, M.K.M.; Kerishnan, J.P.; Ruslinda, A.R.; Al-Douri, Y.; Voon, C.H.; Hashim, U. Cell-targeting aptamers act as intracellular delivery vehicles. Appl. Microbiol. Biotechnol. 2016, 100, 6955–6969. [Google Scholar] [CrossRef]
- Ray, P.; White, R.R. Aptamers for targeted drug delivery. Pharmaceuticals 2010, 3, 1761–1778. [Google Scholar] [CrossRef]
- Cerchia, L.; de Franciscis, V. Targeting cancer cells with nucleic acid aptamers. Trends Biotechnol. 2010, 28, 517–525. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Wen, J.; Li, H.; Xu, L.; Liu, Y.; Zhao, N.; Zeng, Z.; Qi, J.; Jiang, W.; Han, W.; et al. Aptamer-Engineered Natural Killer Cells for Cell-Specific Adaptive Immunotherapy. Small 2019, 15, 1900903. [Google Scholar] [CrossRef]
- Parekh, P.; Kamble, S.; Zhao, N.; Zeng, Z.; Portier, B.P.; Zu, Y. Immunotherapy of CD30-expressing lymphoma using a highly stable ssDNA aptamer. Biomaterials 2013, 34, 8909–8917. [Google Scholar] [CrossRef]
- Parekh, P.; Kamble, S.; Zhao, N.; Zeng, Z.; Wen, J.; Yuan, B.; Zu, Y. Biostable ssDNA aptamers specific for Hodgkin lymphoma. Sensors 2013, 13, 14543–14557. [Google Scholar] [CrossRef] [PubMed]
- Steen Burrell, K.A.; Layzer, J.; Sullenger, B.A. A kallikrein-targeting RNA aptamer inhibits the intrinsic pathway of coagulation and reduces bradykinin release. J. Thromb. Haemost. 2017, 15, 1807–1817. [Google Scholar] [CrossRef]
- Woodruff, R.S.; Xu, Y.; Layzer, J.; Wu, W.; Ogletree, M.L.; Sullenger, B.A. Inhibiting the intrinsic pathway of coagulation with a factor XII-targeting RNA aptamer. J. Thromb. Haemost. 2013, 11, 1364–1373. [Google Scholar] [CrossRef]
- Jiang, Z.; Thayumanavan, S. Noncationic Material Design for Nucleic Acid Delivery. Adv. Ther. 2020, 3, 1900206. [Google Scholar] [CrossRef]
- Ding, F.; Huang, X.; Gao, X.; Xie, M.; Pan, G.; Li, Q.; Song, J.; Zhu, X.; Zhang, C. A non-cationic nucleic acid nanogel for the delivery of the CRISPR/Cas9 gene editing tool. Nanoscale 2019, 11, 17211–17215. [Google Scholar] [CrossRef]
- Lu, X.; Jia, F.; Tan, X.; Wang, D.; Cao, X.; Zheng, J.; Zhang, K. Effective Antisense Gene Regulation via Noncationic, Polyethylene Glycol Brushes. J. Am. Chem. Soc. 2016, 138, 9097–9100. [Google Scholar] [CrossRef]
- Biju, V. Chemical modifications and bioconjugate reactions of nanomaterials for sensing, imaging, drug delivery and therapy. Chem. Soc. Rev. 2014, 43, 744–764. [Google Scholar] [CrossRef]
- Bogart, L.K.; Pourroy, G.; Murphy, C.J.; Puntes, V.; Pellegrino, T.; Rosenblum, D.; Peer, D.; Lévy, R. Nanoparticles for imaging, sensing, and therapeutic intervention. ACS Nano 2014, 8, 3107–3122. [Google Scholar] [CrossRef]
- Lee, J.H.; Yigit, M.V.; Mazumdar, D.; Lu, Y. Molecular diagnostic and drug delivery agents based on aptamer-nanomaterial conjugates. Adv. Drug Deliv. Rev. 2010, 62, 592–605. [Google Scholar] [CrossRef]
- Wang, G.; Wang, Y.; Chen, L.; Choo, J. Nanomaterial-assisted aptamers for optical sensing. Biosens. Bioelectron. 2010, 25, 1859–1868. [Google Scholar] [CrossRef]
- Wei, Q.; Nagi, R.; Sadeghi, K.; Feng, S.; Yan, E.; Ki, S.J.; Caire, R.; Tseng, D.; Ozcan, A. Detection and spatial mapping of mercury contamination in water samples using a smart-phone. ACS Nano 2014, 8, 1121–1129. [Google Scholar] [CrossRef]
- Pelossof, G.; Tel-Vered, R.; Liu, X.Q.; Willner, I. Amplified surface plasmon resonance based DNA biosensors, aptasensors, and Hg2+ sensors using hemin/G-quadruplexes and Au nanoparticles. Chem. Eur. J. 2011, 17, 8904–8912. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, F.; Yang, X. Colorimetric biosensing of mercury(II) ion using unmodified gold nanoparticle probes and thrombin-binding aptamer. Biosens. Bioelectron. 2010, 25, 1994–1998. [Google Scholar] [CrossRef] [PubMed]
- Samanta, A.; Medintz, I.L. Nanoparticles and DNA-a powerful and growing functional combination in bionanotechnology. Nanoscale 2016, 8, 9037–9095. [Google Scholar] [CrossRef]
- Liang, H.; Zhang, X.B.; Lv, Y.; Gong, L.; Wang, R.; Zhu, X.; Yang, R.; Tan, W. Functional DNA-containing nanomaterials: Cellular applications in biosensing, imaging, and targeted therapy. Acc. Chem. Res. 2014, 47, 1891–1901. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, B.; Liu, H.; Zhang, X.; Tan, W. Aptamer-conjugated gold nanoparticles for bioanalysis. Nanomedicine 2013, 8, 983–993. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Aizen, R.; Freeman, R.; Yehezkeli, O.; Willner, I. Multiplexed aptasensors and amplified dna sensors using functionalized graphene oxide: Application for logic gate operations. ACS Nano 2012, 6, 3553–3563. [Google Scholar] [CrossRef]
- Liu, X.; Wang, F.; Aizen, R.; Yehezkeli, O.; Willner, I. Graphene oxide/nucleic-acid-stabilized silver nanoclusters: Functional hybrid materials for optical aptamer sensing and multiplexed analysis of pathogenic DNAs. J. Am. Chem. Soc. 2013, 135, 11832–11839. [Google Scholar] [CrossRef]
- Freeman, R.; Liu, X.; Willner, I. Chemiluminescent and chemiluminescence resonance energy transfer (CRET) detection of DNA, metal ions, and aptamer-substrate complexes using hemin/G-quadruplexes and CdSe/ZnS quantum dots. J. Am. Chem. Soc. 2011, 133, 11597–11604. [Google Scholar] [CrossRef]
- Goodman, R.P.; Heilemann, M.; Doose, S.; Erben, C.M.; Kapanidis, A.N.; Turberfield, A.J. Reconfigurable, braced, three-dimensional DNA nanostructures. Nat. Nanotechnol. 2008, 3, 93–96. [Google Scholar] [CrossRef]
- Goodman, R.P.; Turberfield, A.J. The single-step synthesis of a DNA tetrahedron. Chem. Commun. 2004, 4, 1372–1373. [Google Scholar] [CrossRef] [PubMed]
- Goodman, R.P.; Schaap, I.A.T.; Tardin, C.F.; Erben, C.M.; Berry, R.M.; Schmidt, C.F.; Turberfield, A.J. Chemistry: Rapid chiral assembly of rigid DNA building blocks for molecular nanofabrication. Science 2005, 310, 1661–1665. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Zhang, P.; Yue, L.; Willner, I. Triggered Interconversion of Dynamic Networks Composed of DNA-Tetrahedra Nanostructures. Nano Lett. 2019, 19, 7540–7547. [Google Scholar] [CrossRef]
- He, L.; Lu, D.; Liang, H.; Xie, S.; Zhang, X.; Liu, Q.; Yuan, Q.; Tan, W. mRNA-Initiated, Three-Dimensional DNA Amplifier Able to Function inside Living Cells. J. Am. Chem. Soc. 2018, 140, 258–263. [Google Scholar] [CrossRef]
- Walsh, A.S.; Yin, H.; Erben, C.M.; Wood, M.J.A.; Turberfield, A.J. DNA cage delivery to mammalian cells. ACS Nano 2011, 5, 5427–5432. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Pei, H.; Zhu, B.; Liang, L.; Wei, M.; He, Y.; Chen, N.; Li, D.; Huang, Q.; Fan, C. Self-assembled multivalent DNA nanostructures for noninvasive intracellular delivery of immunostimulatory CpG oligonucleotides. ACS Nano 2011, 5, 8783–8789. [Google Scholar] [CrossRef]
- Peng, P.; Du, Y.; Zheng, J.; Wang, H.; Li, T. Reconfigurable Bioinspired Framework Nucleic Acid Nanoplatform Dynamically Manipulated in Living Cells for Subcellular Imaging. Angew. Chem. Int. Ed. 2019, 58, 1648–1653. [Google Scholar] [CrossRef]
- Pei, H.; Liang, L.; Yao, G.; Li, J.; Huang, Q.; Fan, C. Reconfigurable three-dimensional DNA nanostructures for the construction of intracellular logic sensors. Angew. Chem. Int. Ed. 2012, 51, 9020–9024. [Google Scholar] [CrossRef]
- Zhou, Z.; Sohn, Y.S.; Nechushtai, R.; Willner, I. DNA Tetrahedra Modules as Versatile Optical Sensing Platforms for Multiplexed Analysis of miRNAs, Endonucleases, and Aptamer-Ligand Complexes. ACS Nano 2020, 14, 9021–9031. [Google Scholar] [CrossRef]
- Chen, W.H.; Luo, G.F.; Vázquez-González, M.; Cazelles, R.; Sohn, Y.S.; Nechushtai, R.; Mandel, Y.; Willner, I. Glucose-responsive metal-organic-framework nanoparticles act as “smart” sense-and-treat carriers. ACS Nano 2018, 12, 7538–7545. [Google Scholar] [CrossRef]
- Manzano, M.; Vallet-Regí, M. Mesoporous Silica Nanoparticles for Drug Delivery. Adv. Funct. Mater. 2020, 30, 1902634. [Google Scholar] [CrossRef]
- García-Fernández, A.; Aznar, E.; Martínez-Máñez, R.; Sancenón, F. New Advances in In Vivo Applications of Gated Mesoporous Silica as Drug Delivery Nanocarriers. Small 2020, 16, 1902242. [Google Scholar] [CrossRef]
- Tang, F.; Li, L.; Chen, D. Mesoporous silica nanoparticles: Synthesis, biocompatibility and drug delivery. Adv. Mater. 2012, 24, 1504–1534. [Google Scholar] [CrossRef]
- Wu, M.X.; Yang, Y.W. Metal–Organic Framework (MOF)-Based Drug/Cargo Delivery and Cancer Therapy. Adv. Mater. 2017, 29, 1606134. [Google Scholar] [CrossRef]
- Cai, W.; Chu, C.C.; Liu, G.; Wáng, Y.X.J. Metal-Organic Framework-Based Nanomedicine Platforms for Drug Delivery and Molecular Imaging. Small 2015, 11, 4806–4822. [Google Scholar] [CrossRef]
- Kong, F.Y.; Zhang, J.W.; Li, R.F.; Wang, Z.X.; Wang, W.J.; Wang, W. Unique roles of gold nanoparticles in drug delivery, targeting and imaging applications. Molecules 2017, 22, 1445. [Google Scholar] [CrossRef]
- Press, D. A new era of cancer treatment: Carbon nanotubes as drug delivery tools. Int. J. Nanomed. 2011, 6, 2963–2979. [Google Scholar]
- Liu, W.; Li, C.; Ren, Y.; Sun, X.; Pan, W.; Li, Y.; Wang, J.; Wang, W. Carbon dots: Surface engineering and applications. J. Mater. Chem. B 2016, 4, 5772–5788. [Google Scholar] [CrossRef]
- Deng, Y.; Ling, J.; Li, M.H. Physical stimuli-responsive liposomes and polymersomes as drug delivery vehicles based on phase transitions in the membrane. Nanoscale 2018, 10, 6781–6800. [Google Scholar] [CrossRef] [PubMed]
- Chandrawati, R.; Caruso, F. Biomimetic liposome- and polymersome-based multicompartmentalized assemblies. Langmuir 2012, 28, 13798–13807. [Google Scholar] [CrossRef]
- Deng, L.; Li, Q.; Al-Rehili, S.; Omar, H.; Almalik, A.; Alshamsan, A.; Zhang, J.; Khashab, N.M. Hybrid Iron Oxide-Graphene Oxide-Polysaccharides Microcapsule: A Micro-Matryoshka for On-Demand Drug Release and Antitumor Therapy in Vivo. ACS Appl. Mater. Interfaces 2016, 8, 6859–6868. [Google Scholar] [CrossRef] [PubMed]
- Xiao, W.; Zeng, X.; Lin, H.; Han, K.; Jia, H.Z.; Zhang, X.Z. Dual stimuli-responsive multi-drug delivery system for the individually controlled release of anti-cancer drugs. Chem. Commun. 2015, 51, 1475–1478. [Google Scholar] [CrossRef]
- Guo, J.; Ping, Y.; Ejima, H.; Alt, K.; Meissner, M.; Richardson, J.J.; Yan, Y.; Peter, K.; Von Elverfeldt, D.; Hagemeyer, C.E.; et al. Engineering multifunctional capsules through the assembly of metal-phenolic networks. Angew. Chem. Int. Ed. 2014, 53, 5546–5551. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Ju, X.J.; Zou, X.Y.; Xie, R.; Wang, W.; Liu, Y.M.; Chu, L.Y. Multi-stimuli-responsive microcapsules for adjustable controlled-release. Adv. Funct. Mater. 2014, 24, 3312–3323. [Google Scholar] [CrossRef]
- Mane, S.R.; Sathyan, A.; Shunmugam, R. Biomedical applications of pH-responsive amphiphilic polymer nanoassemblies. ACS Appl. Nano Mater. 2020, 3, 2104–21117. [Google Scholar] [CrossRef]
- Fu, X.; Hosta-Rigau, L.; Chandrawati, R.; Cui, J. Multi-Stimuli-Responsive Polymer Particles, Films, and Hydrogels for Drug Delivery. Chem 2018, 4, 2084–2107. [Google Scholar] [CrossRef]
- Kamaly, N.; Yameen, B.; Wu, J.; Farokhzad, O.C. Degradable controlled-release polymers and polymeric nanoparticles: Mechanisms of controlling drug release. Chem. Rev. 2016, 116, 2602–2663. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, K.; Liu, P.; Chen, M.; Zhong, Y.; Ye, Q.; Wei, M.Q.; Zhao, H.; Tang, Z. Encapsulation of Plasmid DNA by Nanoscale Metal–Organic Frameworks for Efficient Gene Transportation and Expression. Adv. Mater. 2019, 31, 1901570. [Google Scholar] [CrossRef]
- Zheng, H.; Zhang, Y.; Liu, L.; Wan, W.; Guo, P.; Nyström, A.M.; Zou, X. One-pot Synthesis of Metal-Organic Frameworks with Encapsulated Target Molecules and Their Applications for Controlled Drug Delivery. J. Am. Chem. Soc. 2016, 138, 962–968. [Google Scholar] [CrossRef]
- Min, H.; Wang, J.; Qi, Y.; Zhang, Y.; Han, X.; Xu, Y.; Xu, J.; Li, Y.; Chen, L.; Cheng, K.; et al. Biomimetic Metal–Organic Framework Nanoparticles for Cooperative Combination of Antiangiogenesis and Photodynamic Therapy for Enhanced Efficacy. Adv. Mater. 2019, 31, 1808200. [Google Scholar] [CrossRef] [PubMed]
- Lei, B.; Wang, M.; Jiang, Z.; Qi, W.; Su, R.; He, Z. Constructing Redox-Responsive Metal-Organic Framework Nanocarriers for Anticancer Drug Delivery. ACS Appl. Mater. Interfaces 2018, 10, 16698–16706. [Google Scholar] [CrossRef]
- Silva, J.Y.R.; Proenza, Y.G.; da Luz, L.L.; de Sousa Araújo, S.; Filho, M.A.G.; Junior, S.A.; Soares, T.A.; Longo, R.L. A thermo-responsive adsorbent-heater-thermometer nanomaterial for controlled drug release: (ZIF-8,EuxTby)@AuNP core-shell. Mater. Sci. Eng. C 2019, 102, 578–588. [Google Scholar] [CrossRef]
- Wu, M.X.; Gao, J.; Wang, F.; Yang, J.; Song, N.; Jin, X.; Mi, P.; Tian, J.; Luo, J.; Liang, F.; et al. Multistimuli Responsive Core–Shell Nanoplatform Constructed from Fe3O4@MOF Equipped with Pillar[6]arene Nanovalves. Small 2018, 14, 1704440. [Google Scholar] [CrossRef]
- Skirtach, A.G.; Dejugnat, C.; Braun, D.; Susha, A.S.; Rogach, A.L.; Parak, W.J.; Möhwald, H.; Sukhorukovt, G.B. The role of metal nanoparticles in remote release of encapsulated materials. Nano Lett. 2005, 5, 1371–1377. [Google Scholar] [CrossRef] [PubMed]
- Roth Stefaniak, K.; Epley, C.C.; Novak, J.J.; McAndrew, M.L.; Cornell, H.D.; Zhu, J.; McDaniel, D.K.; Davis, J.L.; Allen, I.C.; Morris, A.J.; et al. Photo-triggered release of 5-fluorouracil from a MOF drug delivery vehicle. Chem. Commun. 2018, 54, 7617–7620. [Google Scholar] [CrossRef]
- Collet, G.; Lathion, T.; Besnard, C.; Piguet, C.; Petoud, S. On-Demand Degradation of Metal-Organic Framework Based on Photocleavable Dianthracene-Based Ligand. J. Am. Chem. Soc. 2018, 140, 10820–10828. [Google Scholar] [CrossRef]
- Meng, X.; Gui, B.; Yuan, D.; Zeller, M.; Wang, C. Mechanized azobenzene-functionalized zirconium metal-organic framework for on-command cargo release. Sci. Adv. 2016, 2, e1600480. [Google Scholar] [CrossRef]
- Wang, X.; Chen, X.Z.; Alcântara, C.C.J.; Sevim, S.; Hoop, M.; Terzopoulou, A.; de Marco, C.; Hu, C.; de Mello, A.J.; Falcaro, P.; et al. MOFBOTS: Metal–Organic-Framework-Based Biomedical Microrobots. Adv. Mater. 2019, 31, 1901592. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Prouty, M.D.; Quo, Z.; Golub, V.O.; Kumar, C.S.S.R.; Lvov, Y.M. Magnetic switch of permeability for polyelectrolyte microcapsules embedded with Co@Aunanoparticles. Langmuir 2005, 21, 2042–2050. [Google Scholar] [CrossRef]
- Pavlov, A.M.; Saez, V.; Cobley, A.; Graves, J.; Sukhorukov, G.B.; Mason, T.J. Controlled protein release from microcapsules with composite shells using high frequency ultrasound—Potential for in vivo medical use. Soft Matter 2011, 7, 4341–4347. [Google Scholar] [CrossRef]
- Kolesnikova, T.A.; Gorin, D.A.; Fernandes, P.; Kessel, S.; Khomutov, G.B.; Fery, A.; Shchukin, D.G.; Möhwald, H. Nanocomposite microcontainers with high ultrasound sensitivity. Adv. Funct. Mater. 2010, 20, 1189–1195. [Google Scholar] [CrossRef]
- Shchukin, D.G.; Gorin, D.A.; Möhwald, H. Ultrasonically induced opening of polyelectrolyte microcontainers. Langmuir 2006, 22, 7400–7404. [Google Scholar] [CrossRef]
- Del Mercato, L.L.; Gonzalez, E.; Abbasi, A.Z.; Parak, W.J.; Puntes, V. Synthesis and evaluation of gold nanoparticle-modified polyelectrolyte capsules under microwave irradiation for remotely controlled release for cargo. J. Mater. Chem. 2011, 21, 11468–11471. [Google Scholar] [CrossRef]
- Chen, W.H.; Luo, G.F.; Sohn, Y.S.; Nechushtai, R.; Willner, I. miRNA-Specific Unlocking of Drug-Loaded Metal–Organic Framework Nanoparticles: Targeted Cytotoxicity toward Cancer Cells. Small 2019, 15, 1–10. [Google Scholar] [CrossRef]
- Fischer, A.; Lilienthal, S.; Vázquez-González, M.; Fadeev, M.; Sohn, Y.S.; Nechushtai, R.; Willner, I. Triggered Release of Loads from Microcapsule-in-Microcapsule Hydrogel Microcarriers: En-Route to an “artificial Pancreas”. J. Am. Chem. Soc. 2020, 142, 4223–4234. [Google Scholar] [CrossRef]
- Chen, C.; Pu, F.; Huang, Z.; Liu, Z.; Ren, J.; Qu, X. Stimuli-responsive controlled-release system using quadruplex DNA-capped silica nanocontainers. Nucleic Acids Res. 2011, 39, 1638–1644. [Google Scholar] [CrossRef]
- Chen, X.; Chen, T.; Ren, L.; Chen, G.; Gao, X.; Li, G.; Zhu, X. Triplex DNA Nanoswitch for pH-Sensitive Release of Multiple Cancer Drugs. ACS Nano 2019, 13, 7333–7734. [Google Scholar] [CrossRef] [PubMed]
- Liao, W.C.; Riutin, M.; Parak, W.J.; Willner, I. Programmed pH-Responsive Microcapsules for the Controlled Release of CdSe/ZnS Quantum Dots. ACS Nano 2016, 10, 8683–8689. [Google Scholar] [CrossRef] [PubMed]
- Kahn, J.S.; Freage, L.; Enkin, N.; Garcia, M.A.A.; Willner, I. Stimuli-Responsive DNA-Functionalized Metal–Organic Frameworks (MOFs). Adv. Mater. 2017, 29, 1602782. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, F.; Sohn, Y.S.; Nechushtai, R.; Willner, I. Gated Mesoporous SiO2 Nanoparticles Using K+ -Stabilized G-Quadruplexes. Adv. Funct. Mater. 2014, 24, 5662–5670. [Google Scholar] [CrossRef]
- Chen, W.H.; Luo, G.F.; Sohn, Y.S.; Nechushtai, R.; Willner, I. Enzyme-Driven Release of Loads from Nucleic Acid–Capped Metal–Organic Framework Nanoparticles. Adv. Funct. Mater. 2019, 29, 1805341. [Google Scholar] [CrossRef]
- Chen, W.H.; Yu, X.; Cecconello, A.; Sohn, Y.S.; Nechushtai, R.; Willner, I. Stimuli-responsive nucleic acid-functionalized metal-organic framework nanoparticles using pH- and metal-ion-dependent DNAzymes as locks. Chem. Sci. 2017, 8, 5769–5780. [Google Scholar] [CrossRef]
- Wang, C.; Vázquez-González, M.; Fadeev, M.; Sohn, Y.S.; Nechushtai, R.; Willner, I. Thermoplasmonic-Triggered Release of Loads from DNA-Modified Hydrogel Microcapsules Functionalized with Au Nanoparticles or Au Nanorods. Small 2020, 16, 2000880. [Google Scholar] [CrossRef]
- Juul, S.; Iacovelli, F.; Falconi, M.; Kragh, S.L.; Christensen, B.; Frøhlich, R.; Franch, O.; Kristoffersen, E.L.; Stougaard, M.; Leong, K.W.; et al. Temperature-controlled encapsulation and release of an active enzyme in the cavity of a self-assembled DNA nanocage. ACS Nano 2013, 7, 9724–9734. [Google Scholar] [CrossRef]
- He, D.; He, X.; Wang, K.; Cao, J.; Zhao, Y. A photon-fueled gate-like delivery system using i-motif DNA functionalized mesoporous silica nanoparticles. Adv. Funct. Mater. 2012, 22, 4704–4710. [Google Scholar] [CrossRef]
- Huang, F.; Liao, W.C.; Sohn, Y.S.; Nechushtai, R.; Lu, C.H.; Willner, I. Light-Responsive and pH-Responsive DNA Microcapsules for Controlled Release of Loads. J. Am. Chem. Soc. 2016, 138, 8936–8945. [Google Scholar] [CrossRef]
- Yuan, Q.; Zhang, Y.; Chen, T.; Lu, D.; Zhao, Z.; Zhang, X.; Li, Z.; Yan, C.H.; Tan, W. Photon-manipulated drug release from a mesoporous nanocontainer controlled by azobenzene-modified nucleic acid. ACS Nano 2012, 6, 6337–6344. [Google Scholar] [CrossRef]
- Takenaka, T.; Endo, M.; Suzuki, Y.; Yang, Y.; Emura, T.; Hidaka, K.; Kato, T.; Miyata, T.; Namba, K.; Sugiyama, H. Photoresponsive DNA nanocapsule having an open/close system for capture and release of nanomaterials. Chem. Eur. J. 2014, 20, 14951–14954. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, F.; Mochizuki, T.; Liang, X.; Asanuma, H.; Tanaka, S.; Suzuki, K.; Kitamura, S.I.; Nishikawa, A.; Ui-Tei, K.; Hagiya, M. Robust and photocontrollable DNA capsules using azobenzenes. Nano Lett. 2010, 10, 3560–3565. [Google Scholar] [CrossRef]
- Huang, F.; Duan, R.; Zhou, Z.; Vázquez-González, M.; Xia, F.; Willner, I. Near-infrared light-activated membrane fusion for cancer cell therapeutic applications. Chem. Sci. 2020, 11, 5592–5600. [Google Scholar] [CrossRef] [PubMed]
- Willner, I. Aptamer-Functionalized Micro- and Nanocarriers for Controlled Release. ACS Appl. Mater. Interfaces 2021. [Google Scholar] [CrossRef]
- Chen, W.H.; Yu, X.; Liao, W.C.; Sohn, Y.S.; Cecconello, A.; Kozell, A.; Nechushtai, R.; Willner, I. ATP-Responsive Aptamer-Based Metal–Organic Framework Nanoparticles (NMOFs) for the Controlled Release of Loads and Drugs. Adv. Funct. Mater. 2017, 27, 1702102. [Google Scholar] [CrossRef]
- Chen, W.H.; Yang Sung, S.; Fadeev, M.; Cecconello, A.; Nechushtai, R.; Willner, I. Targeted VEGF-triggered release of an anti-cancer drug from aptamer-functionalized metal-organic framework nanoparticles. Nanoscale 2018, 10, 4650–4657. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.H.; Karmi, O.; Willner, B.; Nechushtai, R.; Willner, I. Thrombin aptamer-modified metal–organic framework nanoparticles: Functional nanostructures for sensing thrombin and the triggered controlled release of anti-blood clotting drugs. Sensors 2019, 19, 5260. [Google Scholar] [CrossRef]
- Simon-Yarza, T.; Mielcarek, A.; Couvreur, P.; Serre, C. Nanoparticles of Metal-Organic Frameworks: On the Road to In Vivo Efficacy in Biomedicine. Adv. Mater. 2018, 30, 1707365. [Google Scholar] [CrossRef]
- McKinlay, A.C.; Morris, R.E.; Horcajada, P.; Férey, G.; Gref, R.; Couvreur, P.; Serre, C. BioMOFs: Metal-organic frameworks for biological and medical applications. Angew. Chem. Int. Ed. 2010, 49, 6260–6266. [Google Scholar] [CrossRef]
- Wen, J.; Yang, K.; Liu, F.; Li, H.; Xu, Y.; Sun, S. Diverse gatekeepers for mesoporous silica nanoparticle based drug delivery systems. Chem. Soc. Rev. 2017, 46, 6024–6045. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Balogh, D.; Wang, F.; Sung, S.Y.; Nechushtai, R.; Willner, I. Biocatalytic release of an anticancer drug from nucleic-acids-capped mesoporous SiO2 using DNA or molecular biomarkers as triggering stimuli. ACS Nano 2013, 7, 8455–8468. [Google Scholar] [CrossRef]
- Georgakilas, V.; Tiwari, J.N.; Kemp, K.C.; Perman, J.A.; Bourlinos, A.B.; Kim, K.S.; Zboril, R. Noncovalent Functionalization of Graphene and Graphene Oxide for Energy Materials, Biosensing, Catalytic, and Biomedical Applications. Chem. Rev. 2016, 116, 5464–5519. [Google Scholar] [CrossRef]
- Wu, Q.; Yang, L.; Wang, X.; Hu, Z. From Carbon-Based Nanotubes to Nanocages for Advanced Energy Conversion and Storage. Acc. Chem. Res. 2017, 50, 435–444. [Google Scholar] [CrossRef]
- Benzigar, M.R.; Talapaneni, S.N.; Joseph, S.; Ramadass, K.; Singh, G.; Scaranto, J.; Ravon, U.; Al-Bahily, K.; Vinu, A. Recent advances in functionalized micro and mesoporous carbon materials: Synthesis and applications. Chem. Soc. Rev. 2018, 47, 2680–2721. [Google Scholar] [CrossRef]
- Lim, S.Y.; Shen, W.; Gao, Z. Carbon quantum dots and their applications. Chem. Soc. Rev. 2015, 44, 362–381. [Google Scholar] [CrossRef]
- Wang, S.; Cazelles, R.; Liao, W.C.; Vázquez-González, M.; Zoabi, A.; Abu-Reziq, R.; Willner, I. Mimicking Horseradish Peroxidase and NADH Peroxidase by Heterogeneous Cu2+-Modified Graphene Oxide Nanoparticles. Nano Lett. 2017, 17, 2043–2048. [Google Scholar] [CrossRef] [PubMed]
- Posthuma-Trumpie, G.A.; Wichers, J.H.; Koets, M.; Berendsen, L.B.J.M.; Van Amerongen, A. Amorphous carbon nanoparticles: A versatile label for rapid diagnostic (immuno)assays. Anal. Bioanal. Chem. 2012, 402, 593–600. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, H.M.R.; Duarte, A.J.; Esteves da Silva, J.C.G. Optical fiber sensor for Hg(II) based on carbon dots. Biosens. Bioelectron. 2010, 26, 1302–1306. [Google Scholar] [CrossRef]
- Liu, C.; Zhang, P.; Zhai, X.; Tian, F.; Li, W.; Yang, J.; Liu, Y.; Wang, H.; Wang, W.; Liu, W. Nano-carrier for gene delivery and bioimaging based on carbon dots with PEI-passivation enhanced fluorescence. Biomaterials 2012, 33, 3604–3613. [Google Scholar] [CrossRef] [PubMed]
- Shoval, A.; Markus, A.; Zhou, Z.; Liu, X.; Cazelles, R.; Willner, I.; Mandel, Y. Anti-VEGF-Aptamer Modified C-Dots—A Hybrid Nanocomposite for Topical Treatment of Ocular Vascular Disorders. Small 2019, 15, 1902776. [Google Scholar] [CrossRef]
- Liao, W.C.; Lilienthal, S.; Kahn, J.S.; Riutin, M.; Sohn, Y.S.; Nechushtai, R.; Willner, I. pH-and ligand-induced release of loads from DNA-acrylamide hydrogel microcapsules. Chem. Sci. 2017, 8, 3362–3373. [Google Scholar] [CrossRef]
- Golub, E.; Albada, H.B.; Liao, W.C.; Biniuri, Y.; Willner, I. Nucleoapzymes: Hemin/G-Quadruplex DNAzyme-Aptamer Binding Site Conjugates with Superior Enzyme-like Catalytic Functions. J. Am. Chem. Soc. 2016, 138, 164–172. [Google Scholar] [CrossRef]
- Biniuri, Y.; Albada, B.; Willner, I. Probing ATP/ATP-Aptamer or ATP-Aptamer Mutant Complexes by Microscale Thermophoresis and Molecular Dynamics Simulations: Discovery of an ATP-Aptamer Sequence of Superior Binding Properties. J. Phys. Chem. B 2018, 122, 9102–9109. [Google Scholar] [CrossRef]
- Bauke Albada, H.; Golub, E.; Willner, I. Computational docking simulations of a DNA-aptamer for argininamide and related ligands. J. Comput. Aided. Mol. Des. 2015, 29, 643–654. [Google Scholar] [CrossRef]
- Caulfield, T.; Devkota, B. Motion of transfer RNA from the A/T state into the A-site using docking and simulations. Proteins Struct. Funct. Bioinform. 2012, 80, 2489–2500. [Google Scholar] [CrossRef] [PubMed]
- Krieger, E.; Darden, T.; Nabuurs, S.B.; Finkelstein, A.; Vriend, G. Making optimal use of empirical energy functions: Force-field parameterization in crystal space. Proteins Struct. Funct. Genet. 2004, 57, 678–683. [Google Scholar] [CrossRef] [PubMed]
- Biniuri, Y.; Shpilt, Z.; Albada, B.; Vázquez-González, M.; Wolff, M.; Hazan, C.; Golub, E.; Gelman, D.; Willner, I. A Bis-Zn2+-Pyridyl-Salen-Type Complex Conjugated to the ATP Aptamer: An ATPase-Mimicking Nucleoapzyme. ChemBioChem 2020, 21, 53–58. [Google Scholar] [CrossRef] [PubMed]
- Biniuri, Y.; Albada, B.; Wolff, M.; Golub, E.; Gelman, D.; Willner, I. Cu2+ or Fe3+ Terpyridine/Aptamer Conjugates: Nucleoapzymes Catalyzing the Oxidation of Dopamine to Aminochrome. ACS Catal. 2018, 8, 1802–1809. [Google Scholar] [CrossRef]
- Flanagan, M.L.; Arguello, A.E.; Colman, D.E.; Kim, J.; Krejci, J.N.; Liu, S.; Yao, Y.; Zhang, Y.; Gorin, D.J. A DNA-conjugated small molecule catalyst enzyme mimic for site-selective ester hydrolysis. Chem. Sci. 2018, 9, 2105–2112. [Google Scholar] [CrossRef]
- Wang, F.; Liu, X.; Willner, I. DNA switches: From principles to applications. Angew. Chem. Int. Ed. 2015, 54, 1098–1129. [Google Scholar] [CrossRef]
- Liu, X.; Lu, C.H.; Willner, I. Switchable reconfiguration of nucleic acid nanostructures by stimuli-responsive DNA machines. Acc. Chem. Res. 2014, 47, 1673–1680. [Google Scholar] [CrossRef]
- Wang, W.; Yang, Y.; Cheng, E.; Zhao, M.; Meng, H.; Liu, D.; Zhou, D. A pH-driven, reconfigurable DNA nanotriangle. Chem. Commun. 2009, 824–826. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Nishioka, H.; Takenaka, N.; Asanuma, H. A DNA nanomachine powered by light irradiation. ChemBioChem 2008, 9, 702–705. [Google Scholar] [CrossRef]
- Yurke, B.; Turber, A.J.; Mills, A.P., Jr.; Simmel, F.C.; Neumann, J.L. A DNA-fulled molecular machine made of DNA. Nature 2000, 406, 605–608. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.G.; Elbaz, J.; Willner, I. DNA machines: Bipedal walker and stepper. Nano Lett. 2011, 11, 304–309. [Google Scholar] [CrossRef]
- Yin, P.; Yan, H.; Daniell, X.G.; Turberfield, A.J.; Reif, J.H. A unidirectional DNA walker that moves autonomously along a track. Angew. Chem. Int. Ed. 2004, 43, 4906–4911. [Google Scholar] [CrossRef]
- Qi, X.J.; Lu, C.H.; Liu, X.; Shimron, S.; Yang, H.H.; Willner, I. Autonomous control of interfacial electron transfer and the activation of DNA machines by an oscillatory pH system. Nano Lett. 2013, 13, 4920–4924. [Google Scholar] [CrossRef]
- Wang, J.; Zhou, Z.; Yue, L.; Wang, S.; Willner, I. Switchable Triggered Interconversion and Reconfiguration of DNA Origami Dimers and Their Use for Programmed Catalysis. Nano Lett. 2018, 18, 2718–2724. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Lu, C.H.; Willner, I. Switchable catalytic DNA catenanes. Nano Lett. 2015, 15, 2099–2103. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.-H.; Qi, X.-J.; Cecconello, A.; Jester, S.-S.; Famulok, M.; Willner, I. Switchable Reconfiguration of an Interlocked DNA Olympiadane Nanostructure. Angew. Chem. Int. Ed. 2014, 126, 7629–7633. [Google Scholar] [CrossRef]
- Li, T.; Lohmann, F.; Famulok, M. Interlocked DNA nanostructures controlled by a reversible logic circuit. Nat. Commun. 2014, 5, 4940. [Google Scholar] [CrossRef][Green Version]
- Elbaz, J.; Cecconello, A.; Fan, Z.; Govorov, A.O.; Willner, I. Powering the programmed nanostructure and function of gold nanoparticles with catenated DNA machines. Nat. Commun. 2013, 4, 2000. [Google Scholar] [CrossRef] [PubMed]
- Ackermann, D.; Jester, S.S.; Famulok, M. Design strategy for DNA rotaxanes with a mechanically reinforced PX100 axle. Angew. Chem. Int. Ed. 2012, 51, 6771–6775. [Google Scholar] [CrossRef]
- Lohmann, F.; Ackermann, D.; Famulok, M. Reversible light switch for macrocycle mobility in a DNA rotaxane. J. Am. Chem. Soc. 2012, 134, 11884–11887. [Google Scholar] [CrossRef]
- Ackermann, D.; Schmidt, T.L.; Hannam, J.S.; Purohit, C.S.; Heckel, A.; Famulok, M. A double-stranded DNA rotaxane. Nat. Nanotechnol. 2010, 5, 436–442. [Google Scholar] [CrossRef]
- Douglas, S.M.; Bachelet, I.; Church, G.M. A logic-gated nanorobot for targeted transport of molecular payloads. Science 2012, 335, 831–834. [Google Scholar] [CrossRef]
- Wang, J.; Yue, L.; Li, Z.; Zhang, J.; Tian, H.; Willner, I. Active generation of nanoholes in DNA origami scaffolds for programmed catalysis in nanocavities. Nat. Commun. 2019, 10, 4963. [Google Scholar] [CrossRef] [PubMed]
- Tikhomirov, G.; Petersen, P.; Qian, L. Fractal assembly of micrometre-scale DNA origami arrays with arbitrary patterns. Nature 2017, 552, 67–71. [Google Scholar] [CrossRef] [PubMed]
- Gerasimova, Y.V.; Kolpashchikov, D.M. Towards a DNA Nanoprocessor: Reusable Tile-Integrated DNA Circuits. Angew. Chem. Int. Ed. 2016, 55, 10244–10247. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Huang, S.; Li, J.; Rui, K.; Bi, S.; Zhang, J.R.; Zhu, J.J. Evaluation of intracellular telomerase activity through cascade DNA logic gates. Chem. Sci. 2016, 8, 174–180. [Google Scholar] [CrossRef]
- Cecconello, A.; Besteiro, L.V.; Govorov, A.O.; Willner, I. Chiroplasmonic DNA-based nanostructures. Nat. Rev. Mater. 2017, 2, 17039. [Google Scholar] [CrossRef]
- Zhang, Q.; Jiang, Q.; Li, N.; Dai, L.; Liu, Q.; Song, L.; Wang, J.; Li, Y.; Tian, J.; Ding, B.; et al. DNA origami as an in vivo drug delivery vehicle for cancer therapy. ACS Nano 2014, 8, 6633–6643. [Google Scholar] [CrossRef]
- Zhao, Y.X.; Shaw, A.; Zeng, X.; Benson, E.; Nyström, A.M.; Högberg, B. DNA origami delivery system for cancer therapy with tunable release properties. ACS Nano 2012, 6, 8684–8691. [Google Scholar] [CrossRef]
- Elbaz, J.; Moshe, M.; Willner, I. Coherent activation of DNA tweezers: A “SET-RESET” logic system. Angew. Chem. Int. Ed. 2009, 48, 3834–3837. [Google Scholar] [CrossRef] [PubMed]
- Endo, M.; Sugiyama, H. Single-molecule imaging of dynamic motions of biomolecules in DNA origami nanostructures using high-speed atomic force microscopy. Acc. Chem. Res. 2014, 47, 1645–1653. [Google Scholar] [CrossRef]
- Rothemund, P.W.K. Folding DNA to create nanoscale shapes and patterns. Nature 2006, 440, 297–302. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Halverson, J.; Tian, Y.; Tkachenko, A.V.; Gang, O. Self-organized architectures from assorted DNA-framed nanoparticles. Nat. Chem. 2016, 8, 867–873. [Google Scholar] [CrossRef]
- Ding, B.; Deng, Z.; Yan, H.; Cabrini, S.; Zuckermann, R.N.; Bokor, J. Gold nanoparticle self-similar chain structure organized by DNA origami. J. Am. Chem. Soc. 2010, 132, 3248–3249. [Google Scholar] [CrossRef] [PubMed]
- Udomprasert, A.; Bongiovanni, M.N.; Sha, R.; Sherman, W.B.; Wang, T.; Arora, P.S.; Canary, J.W.; Gras, S.L.; Seeman, N.C. Amyloid fibrils nucleated and organized by DNA origami constructions. Nat. Nanotechnol. 2014, 9, 537–541. [Google Scholar] [CrossRef] [PubMed]
- Knudsen, J.B.; Liu, L.; Kodal, A.L.B.; Madsen, M.; Li, Q.; Song, J.; Woehrstein, J.B.; Wickham, S.F.J.; Strauss, M.T.; Schueder, F.; et al. Routing of individual polymers in designed patterns. Nat. Nanotechnol. 2015, 10, 892–898. [Google Scholar] [CrossRef]
- Wang, Z.G.; Liu, Q.; Ding, B. Shape-controlled nanofabrication of conducting polymer on planar DNA templates. Chem. Mater. 2014, 26, 3364–3367. [Google Scholar] [CrossRef]
- Kopperger, E.; List, J.; Madhira, S.; Rothfischer, F.; Lamb, D.C.; Simmel, F.C. A self-assembled nanoscale robotic arm controlled by electric fields. Science 2018, 301, 296–301. [Google Scholar] [CrossRef]
- Wickham, S.F.J.; Endo, M.; Katsuda, Y.; Hidaka, K.; Bath, J.; Sugiyama, H.; Turberfield, A.J. Direct observation of stepwise movement of a synthetic molecular transporter. Nat. Nanotechnol. 2011, 6, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Lund, K.; Manzo, A.J.; Dabby, N.; Michelotti, N.; Johnson-Buck, A.; Nangreave, J.; Taylor, S.; Pei, R.; Stojanovic, M.N.; Walter, N.G.; et al. Molecular robots guided by prescriptive landscapes. Nature 2010, 465, 206–209. [Google Scholar] [CrossRef] [PubMed]
- Omabegho, T.; Sha, R.; Seeman, N.C. with Coordinated Legs. Science 2009, 324, 67–71. [Google Scholar] [CrossRef]
- Wu, N.; Willner, I. Programmed dissociation of dimer and trimer origami structures by aptamer-ligand complexes. Nanoscale 2017, 9, 1416–1422. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhou, Z.; Li, Z.; Willner, I. Programmed catalysis within stimuli-responsive mechanically unlocked nanocavities in DNA origami tiles. Chem. Sci. 2020. [Google Scholar] [CrossRef]

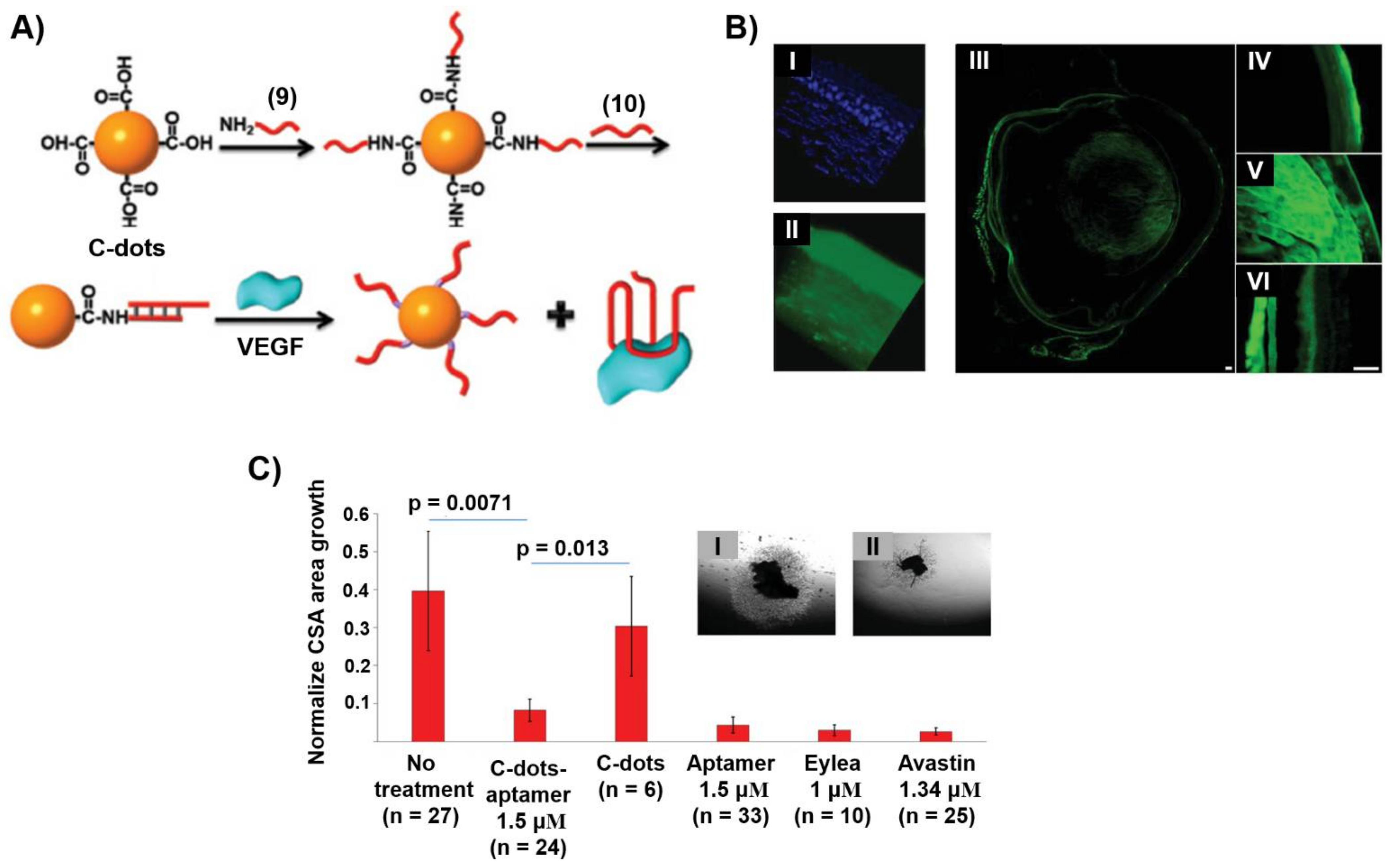
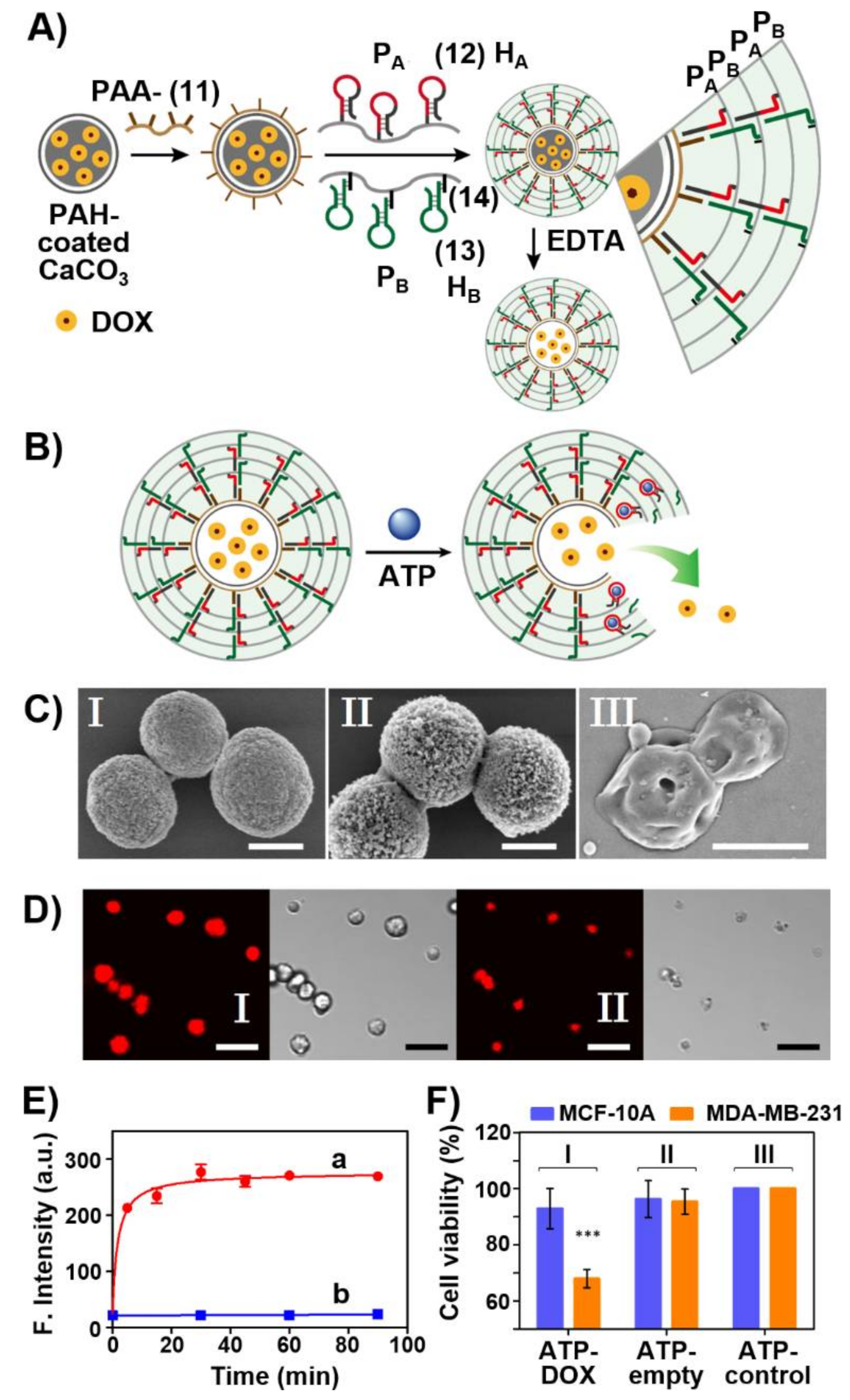
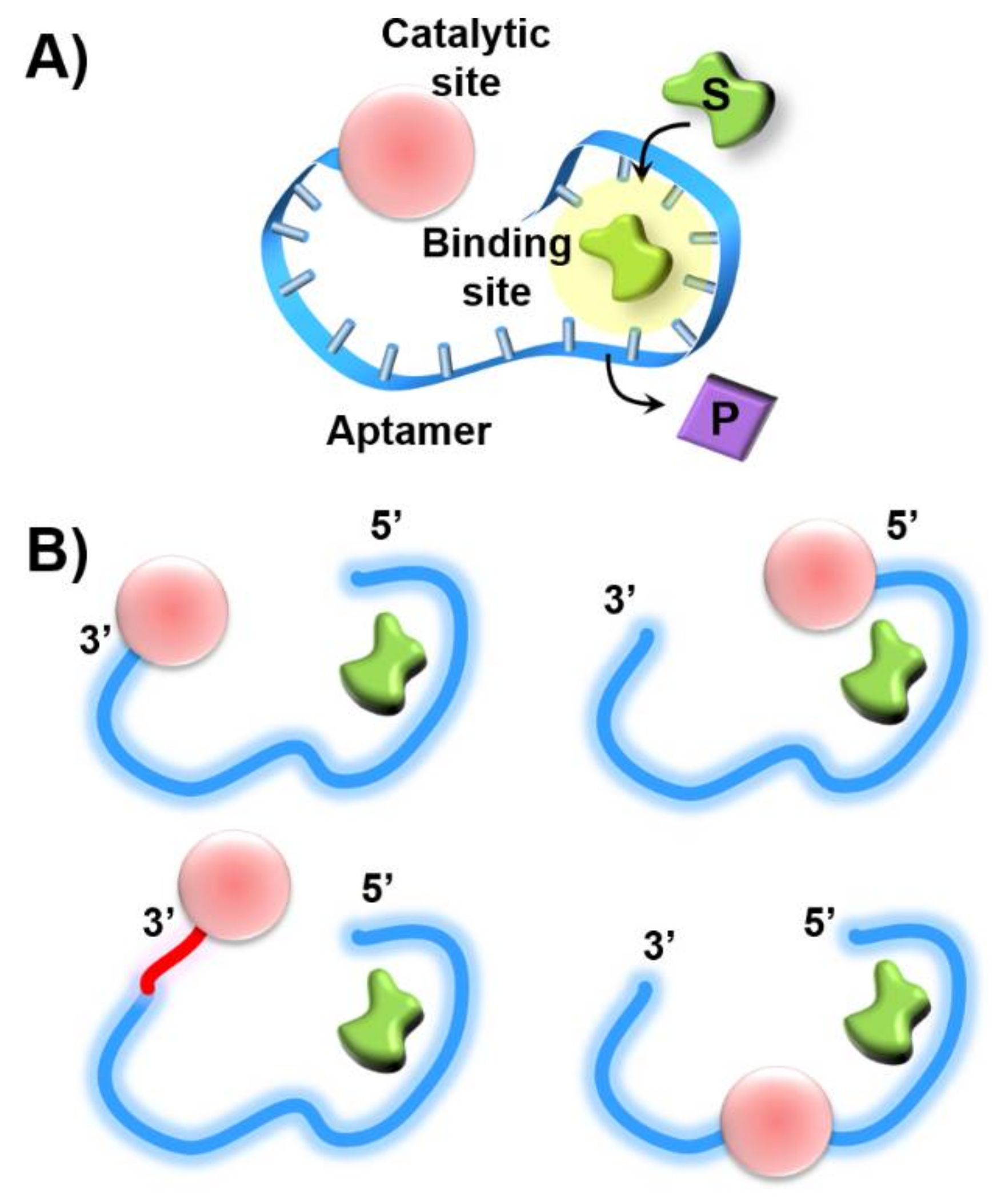
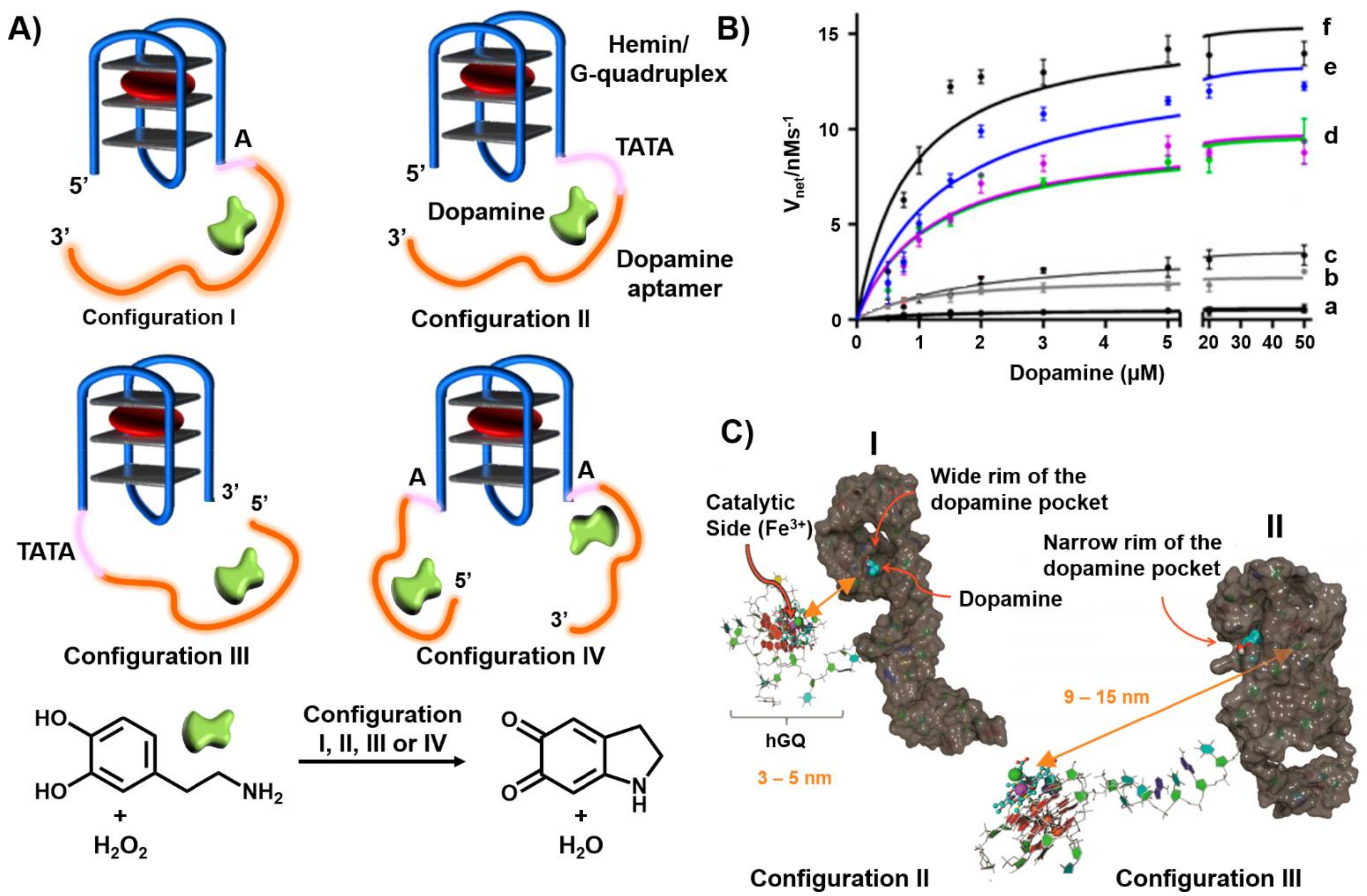
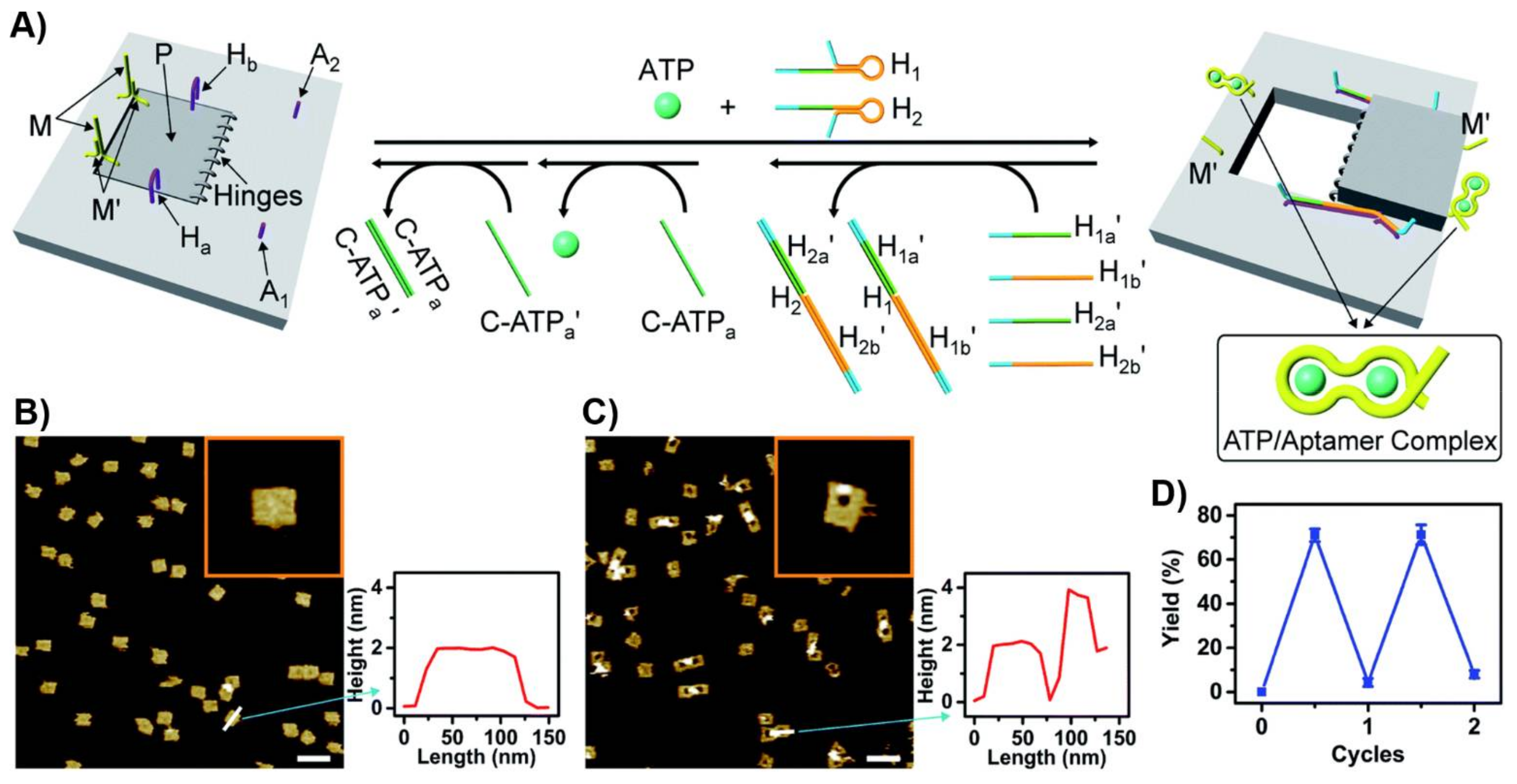
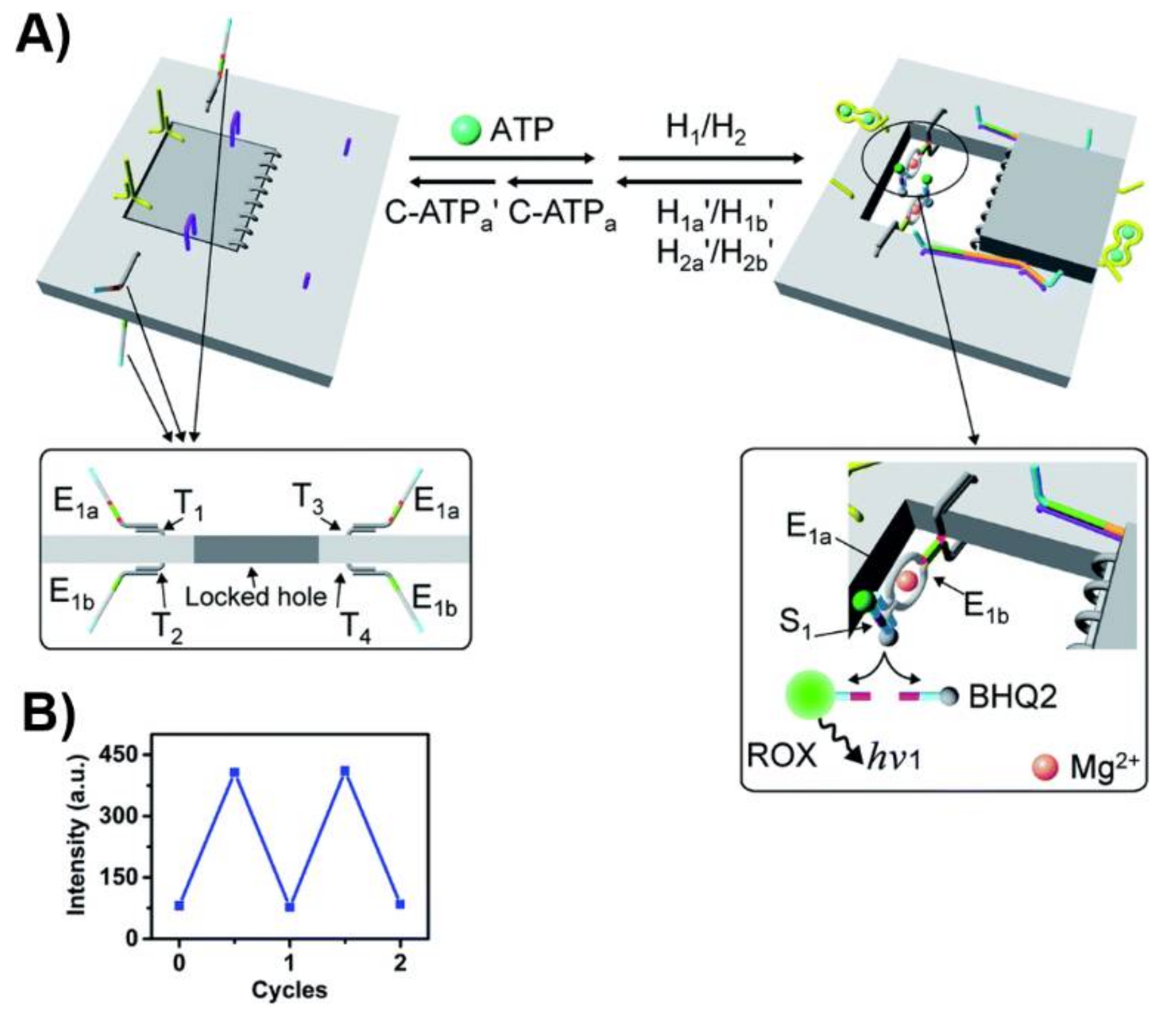
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vázquez-González, M.; Willner, I. Aptamer-Functionalized Hybrid Nanostructures for Sensing, Drug Delivery, Catalysis and Mechanical Applications. Int. J. Mol. Sci. 2021, 22, 1803. https://doi.org/10.3390/ijms22041803
Vázquez-González M, Willner I. Aptamer-Functionalized Hybrid Nanostructures for Sensing, Drug Delivery, Catalysis and Mechanical Applications. International Journal of Molecular Sciences. 2021; 22(4):1803. https://doi.org/10.3390/ijms22041803
Chicago/Turabian StyleVázquez-González, Margarita, and Itamar Willner. 2021. "Aptamer-Functionalized Hybrid Nanostructures for Sensing, Drug Delivery, Catalysis and Mechanical Applications" International Journal of Molecular Sciences 22, no. 4: 1803. https://doi.org/10.3390/ijms22041803
APA StyleVázquez-González, M., & Willner, I. (2021). Aptamer-Functionalized Hybrid Nanostructures for Sensing, Drug Delivery, Catalysis and Mechanical Applications. International Journal of Molecular Sciences, 22(4), 1803. https://doi.org/10.3390/ijms22041803




