Origin and Fates of TERT Gene Copies in Polyploid Plants
Abstract
1. Introduction
2. Results
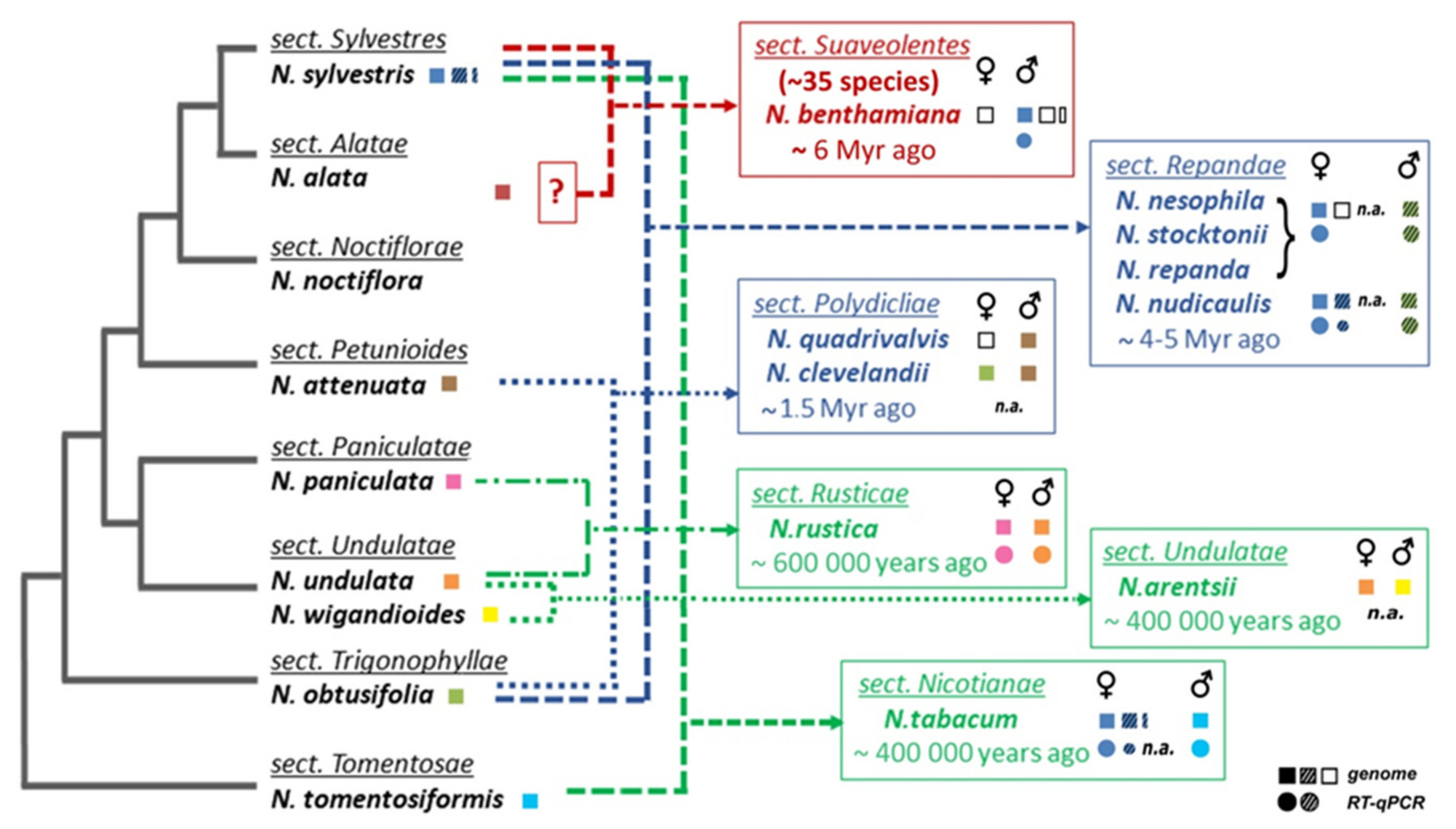
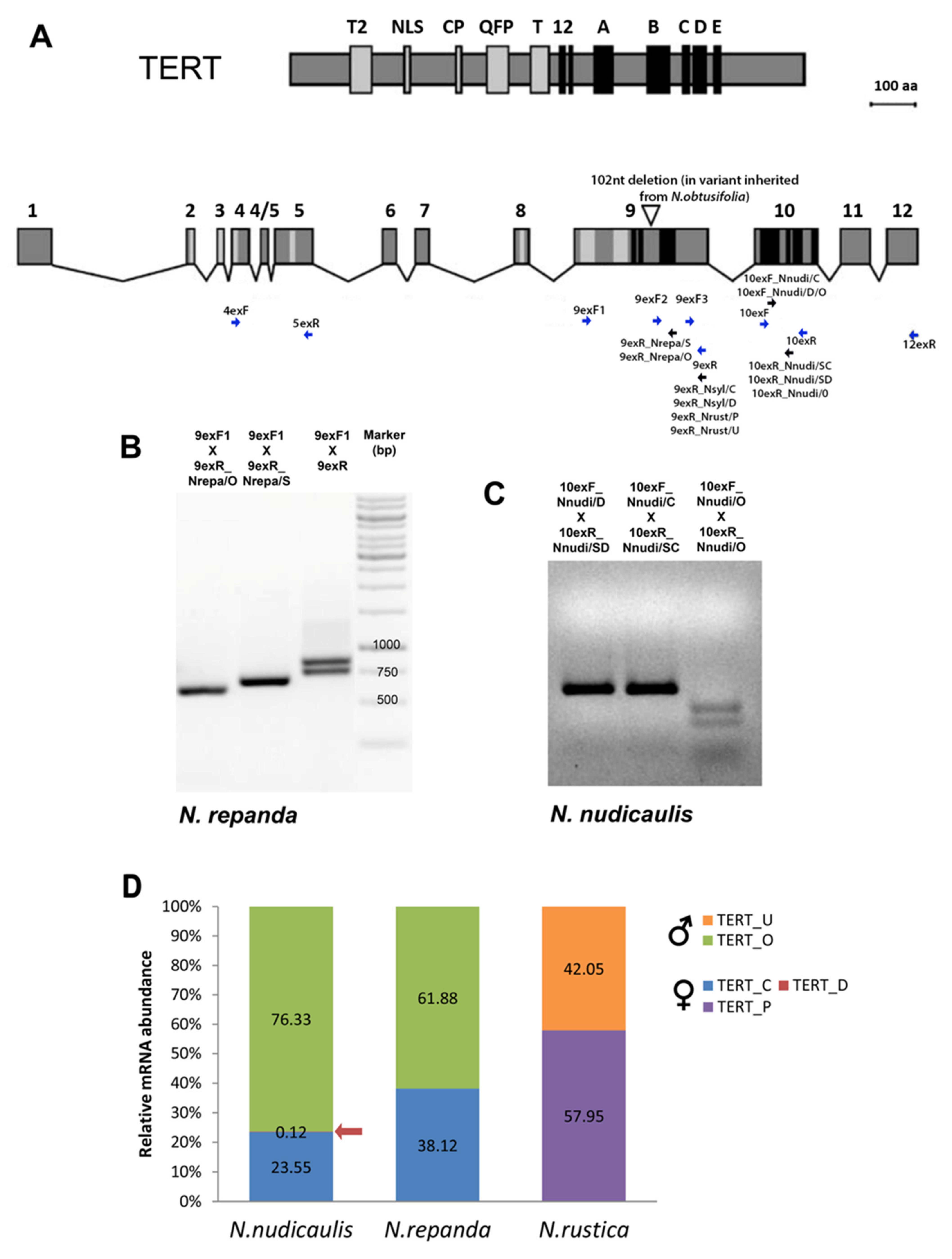
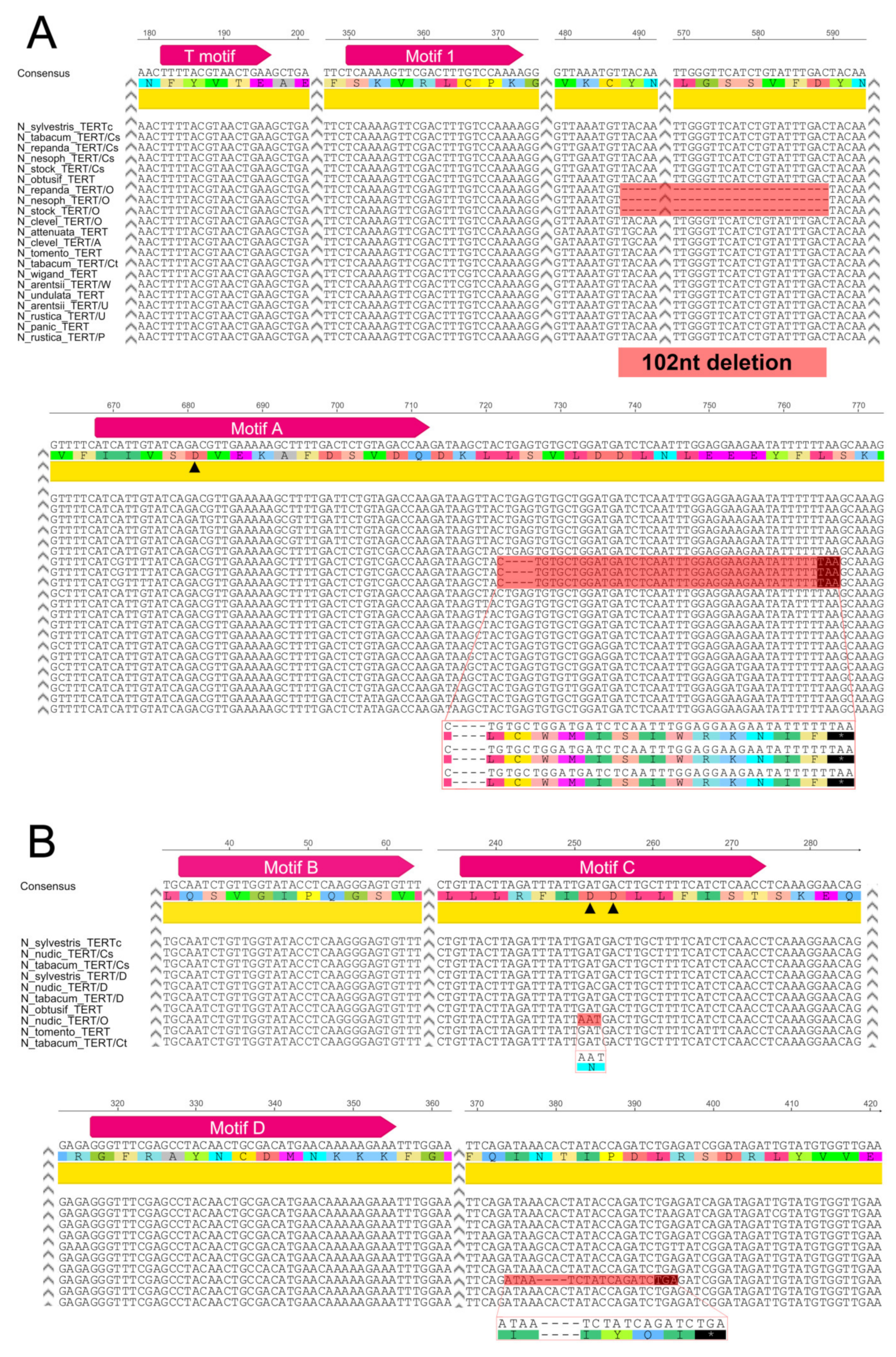
2.1. Number of TERT Variants in Nicotiana Polyploids as a Case Study
2.2. Origin of TERT Genes in Polyploids with the Ancestral N. sylvestris Donor Genome
2.2.1. Suaveolentes
2.2.2. Repandae
2.2.3. Nicotianae
2.3. Origin of the TERT Gene in Polyploid Sections Polydicliae, Rusticae and Undulatae
2.3.1. Polydicliae
2.3.2. Undulatae and Rusticae
2.4. Expression of TERT Variants in Nicotiana Polyploids
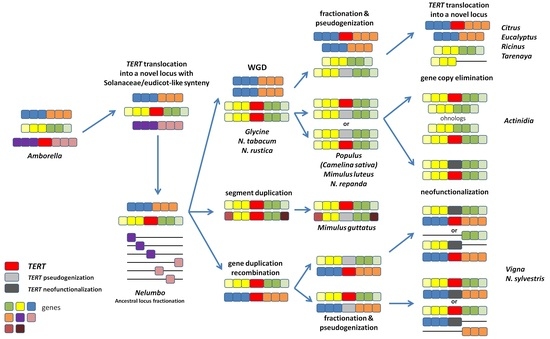
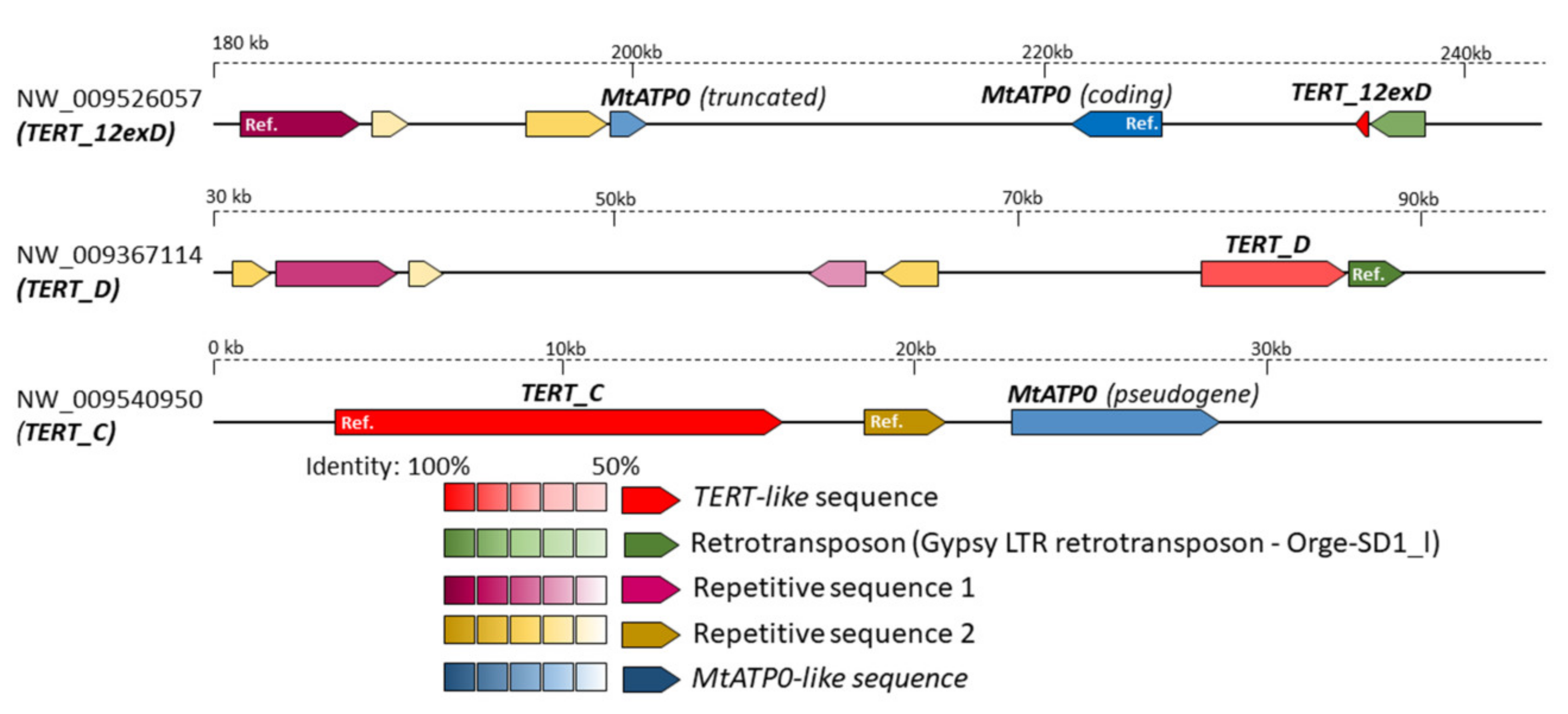
2.5. In Silico Analysis of N. sylvestris Genome Assembly Illustrates a Possible Evolutionary Scenario and the Origin of Subsequent Multiple TERT Loci
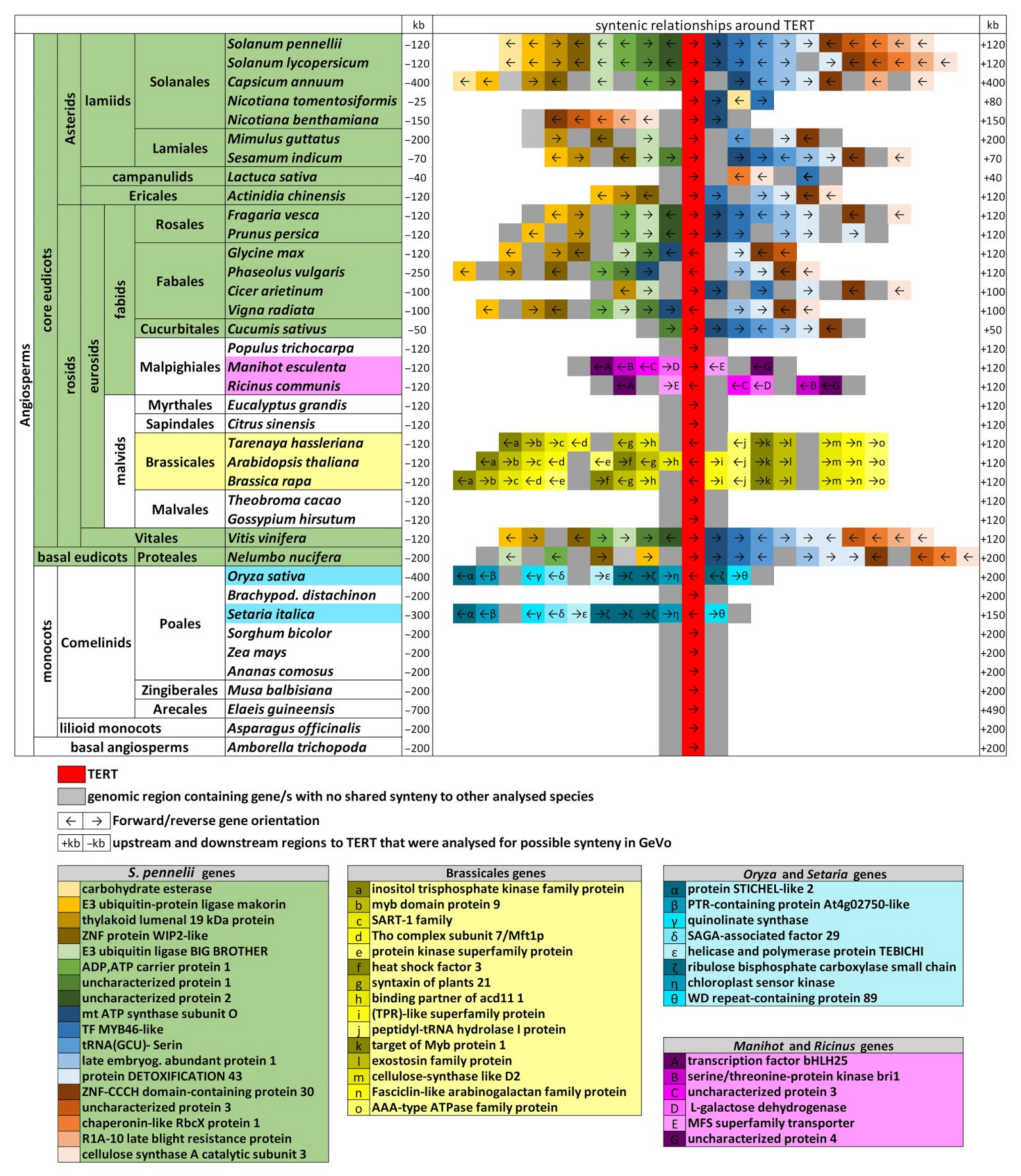
2.6. Genomic TERT Loci Analysis Defines Ancestral Synteny within Flowering Plants
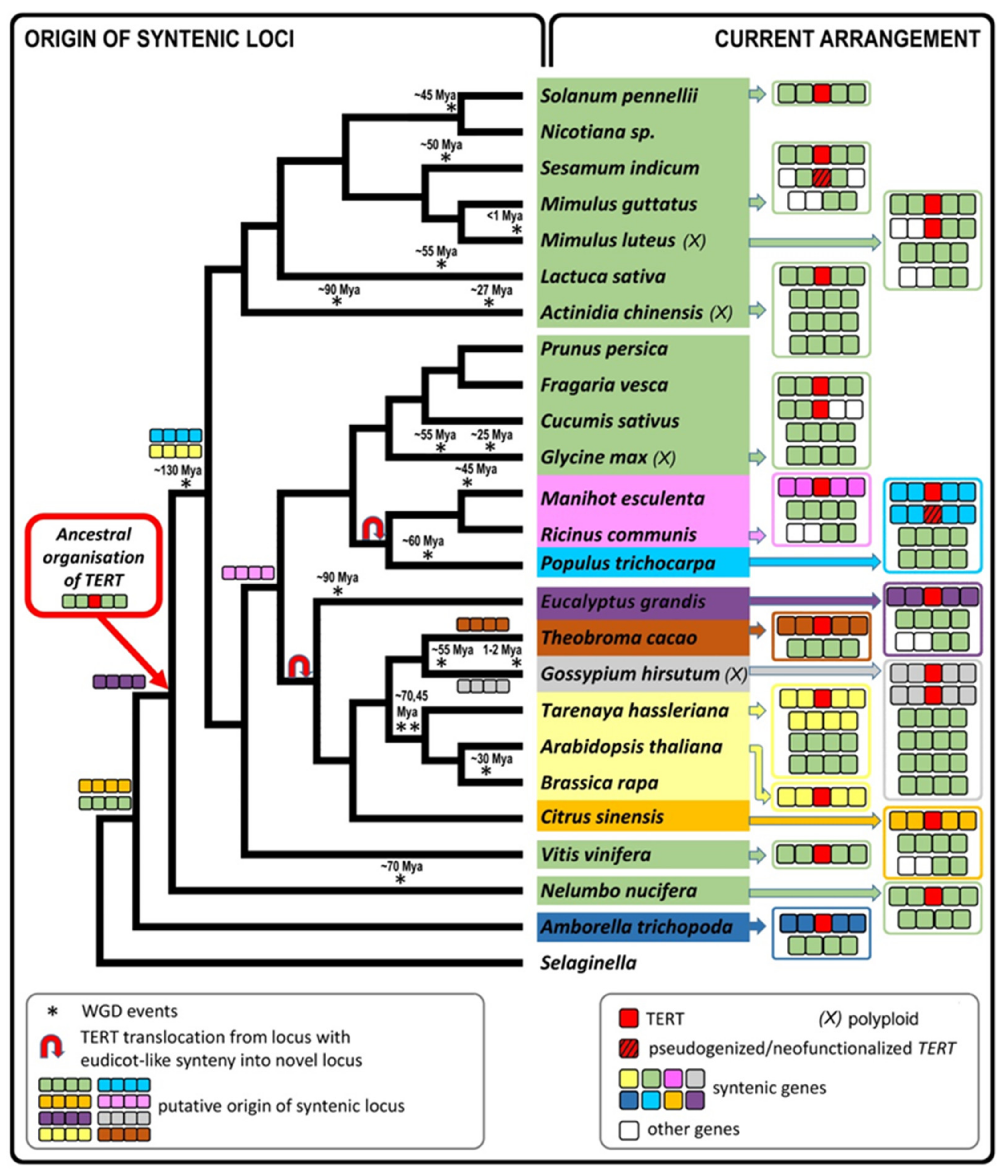
2.7. The Occurrence of TERT Homologs in Model Species Illustrates a Possible Origin of TERT Variants
3. Discussion
4. Materials and Methods
4.1. Isolation of Plant Material, Genomic DNA and RNA
4.2. PCR Amplification of TERT Sequence Variants
4.3. Quantitative PCR and RT–qPCR
4.4. In Silico Identification of TERT Variants in Nicotiana Species
4.5. Analysis of Gene Synteny of the TERT Locus within Angiosperms
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BLASTn | Basic local alignment search tool, nucleotide to nucleotide |
| LTR | Long terminal repeat |
| MtATPO | Mitochondrial ATP synthase subunit delta |
| NGS | Next-generation sequencing |
| qPCR | Quantitative PCR |
| SNP | Single nucleotide polymorphism |
| SRA | Short read archive |
| TERT | Telomerase reverse transcriptase |
| WGD | Whole-genome duplication |
References
- Ohno, S. Evolution by Gene Duplication; Springer: New York, NY, USA, 1970. [Google Scholar]
- Soltis, D.E.; Visger, C.J.; Marchant, D.B.; Soltis, P.S. Polyploidy: Pitfalls and paths to a paradigm. Am. J. Bot. 2016, 103, 1146–1166. [Google Scholar] [CrossRef]
- Lohaus, R.; Van de Peer, Y. Of dups and dinos: Evolution at the K/Pg boundary. Curr. Opin. Plant Biol. 2016, 30, 62–69. [Google Scholar] [CrossRef]
- Barker, M.S.; Husband, B.C.; Pires, J.C. Spreading Winge and flying high: The evolutionary importance of polyploidy after a century of study. Am. J. Bot. 2016, 103, 1139–1145. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Wickett, N.J.; Ayyampalayam, S.; Chanderbali, A.S.; Landherr, L.; Ralph, P.E.; Tomsho, L.P.; Hu, Y.; Liang, H.; Soltis, P.S.; et al. Ancestral polyploidy in seed plants and angiosperms. Nature 2011, 473, 97–100. [Google Scholar] [CrossRef]
- Jiao, Y.; Leebens-Mack, J.; Ayyampalayam, S.; Bowers, J.E.; McKain, M.R.; McNeal, J.; Rolf, M.; Ruzicka, D.R.; Wafula, E.; Wickett, N.J.; et al. A genome triplication associated with early diversification of the core eudicots. Genome Biol. 2012, 13, R3. [Google Scholar] [CrossRef] [PubMed]
- Murat, F.; Armero, A.; Pont, C.; Klopp, C.; Salse, J. Reconstructing the genome of the most recent common ancestor of flowering plants. Nat. Genet. 2017, 49, 490–496. [Google Scholar] [CrossRef] [PubMed]
- Flagel, L.; Udall, J.; Nettleton, D.; Wendel, J. Duplicate gene expression in allopolyploid Gossypium reveals two temporally distinct phases of expression evolution. BMC Biol. 2008, 6, 16. [Google Scholar] [CrossRef] [PubMed]
- Parisod, C.; Mhiri, C.; Lim, K.Y.; Clarkson, J.J.; Chase, M.W.; Leitch, A.R.; Grandbastien, M.A. Differential dynamics of transposable elements during long-term diploidization of Nicotiana section Repandae (Solanaceae) allopolyploid genomes. PLoS ONE 2012, 7, e50352. [Google Scholar] [CrossRef]
- Birchler, J.A.; Veitia, R.A. The gene balance hypothesis: From classical genetics to modern genomics. Plant Cell 2007, 19, 395–402. [Google Scholar] [CrossRef] [PubMed]
- McClintock, B. The fusion of broken chromosome ends of sister half-chromatids following chromatid breakage at meiotic anaphases. Mo. Agric. Exp. Stn. Res. Bull. 1938, 290, 1–48. [Google Scholar]
- Blackburn, E.H.; Gall, J.G. Tandemly Repeated Sequence at Termini of Extrachromosomal Ribosomal-Rna Genes in Tetrahymena. J. Mol. Biol. 1978, 120, 33–53. [Google Scholar] [CrossRef]
- Greider, C.W.; Blackburn, E.H. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 1985, 43, 405–413. [Google Scholar] [CrossRef]
- Peska, V.; Garcia, S. Origin, Diversity, and Evolution of Telomere Sequences in Plants. Front. Plant Sci. 2020, 11, 117. [Google Scholar] [CrossRef]
- Sykorova, E.; Lim, K.Y.; Kunicka, Z.; Chase, M.W.; Bennett, M.D.; Fajkus, J.; Leitch, A.R. Telomere variability in the monocotyledonous plant order Asparagales. Proc. R. Soc. Lond. B Biol. Sci. 2003, 270, 1893–1904. [Google Scholar] [CrossRef]
- Fajkus, P.; Peska, V.; Sitova, Z.; Fulneckova, J.; Dvorackova, M.; Gogela, R.; Sykorova, E.; Hapala, J.; Fajkus, J. Allium telomeres unmasked: The unusual telomeric sequence (CTCGGTTATGGG)n is synthesized by telomerase. Plant J. 2016, 85, 337–347. [Google Scholar] [CrossRef]
- Sykorova, E.; Lim, K.Y.; Chase, M.W.; Knapp, S.; Leitch, I.J.; Leitch, A.R.; Fajkus, J. The absence of Arabidopsis-type telomeres in Cestrum and closely related genera Vestia and Sessea (Solanaceae): First evidence from eudicots. Plant J. 2003, 34, 283–291. [Google Scholar] [CrossRef]
- Peska, V.; Fajkus, P.; Fojtova, M.; Dvorackova, M.; Hapala, J.; Dvoracek, V.; Polanska, P.; Leitch, A.R.; Sykorova, E.; Fajkus, J. Characterisation of an unusual telomere motif (TTTTTTAGGG)n in the plant Cestrum elegans (Solanaceae), a species with a large genome. Plant J. 2015, 82, 644–654. [Google Scholar] [CrossRef]
- Peska, V.; Sitova, Z.; Fajkus, P.; Fajkus, J. BAL31-NGS approach for identification of telomeres de novo in large genomes. Methods 2017, 114, 16–27. [Google Scholar] [CrossRef] [PubMed]
- Fajkus, J.; Sykorova, E.; Leitch, A.R. Telomeres in evolution and evolution of telomeres. Chromosome Res. 2005, 13, 469–479. [Google Scholar] [CrossRef] [PubMed]
- Fajkus, P.; Peska, V.; Zavodnik, M.; Fojtova, M.; Fulneckova, J.; Dobias, S.; Kilar, A.; Dvorackova, M.; Zachova, D.; Necasova, I.; et al. Telomerase RNAs in land plants. Nucleic Acids Res. 2019, 47, 9842–9856. [Google Scholar] [CrossRef]
- Peska, V.; Matl, M.; Mandakova, T.; Vitales, D.; Fajkus, P.; Fajkus, J.; Garcia, S. Human-like telomeres in Zostera marina reveal a mode of transition from the plant to the human telomeric sequences. J. Exp. Bot. 2020, 71, 5786–5793. [Google Scholar] [CrossRef] [PubMed]
- Sykorova, E.; Fajkus, J. Structure-Function relationships in telomerase genes. Biol. Cell 2009, 101, 375–392. [Google Scholar] [CrossRef]
- Belfort, M.; Curcio, M.J.; Lue, N.F. Telomerase and retrotransposons: Reverse transcriptases that shaped genomes. Proc. Natl. Acad. Sci. USA 2011, 108, 20304–20310. [Google Scholar] [CrossRef]
- Fitzgerald, M.S.; Riha, K.; Gao, F.; Ren, S.; McKnight, T.D.; Shippen, D.E. Disruption of the telomerase catalytic subunit gene from Arabidopsis inactivates telomerase and leads to a slow loss of telomeric DNA. Proc. Natl. Acad. Sci. USA 1999, 96, 14813–14818. [Google Scholar] [CrossRef] [PubMed]
- Oguchi, K.; Liu, H.; Tamura, K.; Takahashi, H. Molecular cloning and characterization of AtTERT, a telomerase reverse transcriptase homolog in Arabidopsis thaliana. FEBS Lett. 1999, 457, 465–469. [Google Scholar] [CrossRef]
- Harrington, L.; Zhou, W.; McPhail, T.; Oulton, R.; Yeung, D.S.; Mar, V.; Bass, M.B.; Robinson, M.O. Human telomerase contains evolutionarily conserved catalytic and structural subunits. Genes Dev. 1997, 11, 3109–3115. [Google Scholar] [CrossRef] [PubMed]
- Lingner, J.; Hughes, T.R.; Shevchenko, A.; Mann, M.; Lundblad, V.; Cech, T.R. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science 1997, 276, 561–567. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, T.M.; Morin, G.B.; Chapman, K.B.; Weinrich, S.L.; Andrews, W.H.; Lingner, J.; Harley, C.B.; Cech, T.R. Telomerase catalytic subunit homologs from fission yeast and human. Science 1997, 277, 955–959. [Google Scholar] [CrossRef]
- Sykorova, E.; Fulneckova, J.; Mokros, P.; Fajkus, J.; Fojtova, M.; Peska, V. Three TERT genes in Nicotiana tabacum. Chromosome Res. 2012, 20, 381–394. [Google Scholar] [CrossRef]
- Clarkson, J.J.; Dodsworth, S.; Chase, M.W. Time-Calibrated phylogenetic trees establish a lag between polyploidisation and diversification in Nicotiana (Solanaceae). Plant Syst. Evol. 2017, 303, 1001–1012. [Google Scholar] [CrossRef]
- Leitch, I.J.; Hanson, L.; Lim, K.Y.; Kovarik, A.; Chase, M.W.; Clarkson, J.J.; Leitch, A.R. The ups and downs of genome size evolution in polyploid species of Nicotiana (Solanaceae). Ann. Bot. 2008, 101, 805–814. [Google Scholar] [CrossRef]
- Clarkson, J.J.; Lim, K.Y.; Kovarik, A.; Chase, M.W.; Knapp, S.; Leitch, A.R. Long-Term genome diploidization in allopolyploid Nicotiana section Repandae (Solanaceae). New Phytol. 2005, 168, 241–252. [Google Scholar] [CrossRef] [PubMed]
- Knapp, S.; Lughadha, E.N.; Paton, A. Taxonomic inflation, species concepts and global species lists. Trends Ecol. Evol. 2005, 20, 7–8. [Google Scholar] [CrossRef] [PubMed]
- Kelly, L.J.; Leitch, A.R.; Clarkson, J.J.; Knapp, S.; Chase, M.W. Reconstructing the complex evolutionary origin of wild allopolyploid tobaccos (Nicotiana section suaveolentes). Evolution 2013, 67, 80–94. [Google Scholar] [CrossRef] [PubMed]
- Jureckova, J.F.; Sykorova, E.; Hafidh, S.; Honys, D.; Fajkus, J.; Fojtova, M. Tissue-Specific expression of telomerase reverse transcriptase gene variants in Nicotiana tabacum. Planta 2017, 245, 549–561. [Google Scholar] [CrossRef]
- Pfaffl, M.W. Quantification strategies in real-time PCR. In A-Z of Quantitative PCR; Bustin, S.A., Ed.; International University Line: La Jolla, CA, USA, 2004; pp. 87–112. [Google Scholar]
- Clarkson, J.J.; Knapp, S.; Garcia, V.F.; Olmstead, R.G.; Leitch, A.R.; Chase, M.W. Phylogenetic relationships in Nicotiana (Solanaceae) inferred from multiple plastid DNA regions. Mol. Phylogenet. Evol. 2004, 33, 75–90. [Google Scholar] [CrossRef]
- Bombarely, A.; Rosli, H.G.; Vrebalov, J.; Moffett, P.; Mueller, L.A.; Martin, G.B. A draft genome sequence of Nicotiana benthamiana to enhance molecular plant-microbe biology research. Mol. Plant Microbe Interact. 2012, 25, 1523–1530. [Google Scholar] [CrossRef]
- Lim, K.Y.; Matyasek, R.; Kovarik, A.; Leitch, A.R. Genome evolution in allotetraploid Nicotiana. Biol. J. Linn. Soc. 2004, 82, 599–606. [Google Scholar] [CrossRef]
- Renny-Byfield, S.; Chester, M.; Kovarik, A.; Le Comber, S.C.; Grandbastien, M.A.; Deloger, M.; Nichols, R.A.; Macas, J.; Novak, P.; Chase, M.W.; et al. Next generation sequencing reveals genome downsizing in allotetraploid Nicotiana tabacum, predominantly through the elimination of paternally derived repetitive DNAs. Mol. Biol. Evol. 2011, 28, 2843–2854. [Google Scholar] [CrossRef] [PubMed]
- Chase, M.W.; Knapp, S.; Cox, A.V.; Clarkson, J.J.; Butsko, Y.; Joseph, J.; Savolainen, V.; Parokonny, A.S. Molecular systematics, GISH and the origin of hybrid taxa in Nicotiana (Solanaceae). Ann. Bot. 2003, 92, 107–127. [Google Scholar] [CrossRef]
- Kohany, O.; Gentles, A.J.; Hankus, L.; Jurka, J. Annotation, submission and screening of repetitive elements in Repbase: RepbaseSubmitter and Censor. BMC Bioinform. 2006, 7, 474. [Google Scholar] [CrossRef]
- Gordenin, D.A.; Lobachev, K.S.; Degtyareva, N.P.; Malkova, A.L.; Perkins, E.; Resnick, M.A. Inverted DNA repeats: A source of eukaryotic genomic instability. Mol. Cell. Biol. 1993, 13, 5315–5322. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Bomhoff, M.D.; Briones, E.; Zhang, L.; Schnable, J.C.; Lyons, E. SynFind: Compiling Syntenic Regions across Any Set of Genomes on Demand. Genome Biol. Evol. 2015, 7, 3286–3298. [Google Scholar] [CrossRef] [PubMed]
- He, Z.C.; Li, J.Q.; Cai, Q.; Wang, Q. The cytology of Actinidia, Saurauia and Clematoclethra (Actinidiaceae). Bot. J. Linn. Soc. 2005, 147, 369–374. [Google Scholar] [CrossRef]
- Shi, T.; Huang, H.W.; Barker, M.S. Ancient genome duplications during the evolution of kiwifruit (Actinidia) and related Ericales. Ann. Bot. 2010, 106, 497–504. [Google Scholar] [CrossRef]
- Schmutz, J.; Cannon, S.B.; Schlueter, J.; Ma, J.; Mitros, T.; Nelson, W.; Hyten, D.L.; Song, Q.; Thelen, J.J.; Cheng, J.; et al. Genome sequence of the palaeopolyploid soybean. Nature 2010, 463, 178–183. [Google Scholar] [CrossRef]
- Zhang, T.; Hu, Y.; Jiang, W.; Fang, L.; Guan, X.; Chen, J.; Zhang, J.; Saski, C.A.; Scheffler, B.E.; Stelly, D.M.; et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 2015, 33, 531–537. [Google Scholar] [CrossRef]
- Xie, M.; Podlevsky, J.D.; Qi, X.; Bley, C.J.; Chen, J.J. A novel motif in telomerase reverse transcriptase regulates telomere repeat addition rate and processivity. Nucleic Acids Res. 2010, 38, 1982–1996. [Google Scholar] [CrossRef] [PubMed]
- Madlung, A.; Tyagi, A.P.; Watson, B.; Jiang, H.; Kagochi, T.; Doerge, R.W.; Martienssen, R.; Comai, L. Genomic changes in synthetic Arabidopsis polyploids. Plant J. 2005, 41, 221–230. [Google Scholar] [CrossRef]
- Byng, J.W.; Chase, M.W.; Christenhusz, M.J.M.; Fay, M.F.; Judd, W.S.; Mabberley, D.J.; Sennikov, A.N.; Soltis, D.E.; Soltis, P.S.; Stevens, P.F.; et al. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc. 2016, 181, 1–20. [Google Scholar] [CrossRef]
- Huang, S.; Ding, J.; Deng, D.; Tang, W.; Sun, H.; Liu, D.; Zhang, L.; Niu, X.; Zhang, X.; Meng, M.; et al. Draft genome of the kiwifruit Actinidia chinensis. Nat. Commun. 2013, 4, 2640. [Google Scholar] [CrossRef] [PubMed]
- Dellaporta, S.L.; Wood, J.; Hicks, J.B. A plant DNA minipreparation: Version II. Plant Mol. Biol. Report. 1983, 1, 19–21. [Google Scholar] [CrossRef]
- Fajkus, J.; Dvorackova, M.; Sykorova, E. Analysis of telomeres and telomerase. Methods Mol. Biol. 2008, 463, 267–296. [Google Scholar] [CrossRef] [PubMed]

| Allopolyploids | GeneBank Accessions | Sequence Similarity [%] | Analyzed Region 1 | |||
|---|---|---|---|---|---|---|
| Maternal Parent | Paternal Parent | |||||
| SUAVEOLENTES | N. alata | N. noctiflora2 | N. syl. C var. | N. syl. D var. | ||
| N. benthamiana | NbS000104 | 96.3 | n.a. | 97.5 | n.a. | exon 4 to 5 |
| 27g0116.1 | n.a. | 96.1 | 97.6 | 93.4 | exons 10, 11, 12 | |
| REPANDAE | N. syl. C var. | N. syl. D var. | N. obtusifolia | |||
| N. repanda | MG242402 1 | 95.9 | 91.6 | 97.4 | exon 9 | |
| MG242403 1 | 97.9 | 92.4 | 96.4 | exon 9 | ||
| N. stocktonii | MG242407 1 | 95.6 | 91.7 | 97.6 | exon 9 | |
| MG242408 1 | 98.6 | 93.1 | 97.0 | exon 9 | ||
| N. nesophila | MG242405 1 | 95.2 | 91.6 | 97.0 | exon 9 | |
| MG242406 1 | 98.5 | 92.9 | 96.9 | exon 9 | ||
| N. nudicaulis | MG242409 1 | 98.6 | 94.3 | 94.4 | exon 10 to 12 | |
| MG545647 1 | 92.8 | 94.8 | 91.6 | exon 10 to 12 | ||
| MG242410 1 | 94.2 | 93.3 | 96.3 | exon 10 to 12 | ||
| POLYDICLIAE | N. obtusifolia | N. attenuata | ||||
| MG242422 1 | 94.3 | 99.3 | exon 4 to 5 | |||
| N. clevelandii | var1 2 | 97.3 | 98.9 | exon 9 2 | ||
| var2 2 | 99.2 | 97.3 | exon 9 2 | |||
| N. quadrivalvis | MG242423 1 | 94.9 | 98.6 | exon 4 to 5 | ||
| ARENTSII | N. undulata | N. wigandiodes | ||||
| N. arentsii | MG242418 1 | 99.5 | 98.4 | exon 9 | ||
| MG242419 1 | 98.8 | 99.8 | exon 9 | |||
| RUSTICA | N. paniculata | N. undulata | ||||
| N. rustica | MG242413 1 | 100.0 | 98.2 | exon 9 | ||
| MG242414 1 | 98.2 | 99.8 | exon 9 | |||
| Species/Genome Dataset Accession | Total No. of TERT Reads | Expected Genome Coverage (Depth) | No. of Detected TERT Variants | Read Counts Corresponding to Known TERT Variants | Ratio of TERT Variants in Genome | ||
|---|---|---|---|---|---|---|---|
| N. tabacum SRX338107 | 1259 | 35× | 3 | NtTERT_Cs | NtTERT_D | NtTERT_Ct | 1:1:1 |
| 425 | 424 | 410 | |||||
| N. sylvestris ERX248848 | 644 | 26× | 2 | NsTERT_C | NsTERT_D | 1:1 | |
| 332 | 312 | ||||||
| N. tomentosiformis ERX248865 | 203 | 15× | 1 | NtomTERT | - | ||
| 203 | |||||||
| N. benthamiana (raw data from [39]) | 286 | 20× | 1 | NbenTERT | - | ||
| 286 | |||||||
| N. sylvestris Accession | Ct (±SD) | ∆Ct (C-C1/2×) | ∆Ct (C-D) | C:D Ratio | ||
|---|---|---|---|---|---|---|
| NsTERT_C | NsTERT_C 1/2× (Control) | NsTERT_D | ||||
| A04750326 | 16.41 (±0.036) | 16.91 (±0.085) | 16.41 (±0.065) | −0.5 | 0.00 | 1:1 |
| 934750005 | 16.65 (±0.052) | 17.12 (±0.043) | 16.59 (±0.049) | −0.47 | 0.06 | 1:1 |
| ITB626 | 17.65 (±0.067) | 18.12 (±0.051) | 17.66 (±0.035) | −0.47 | −0.01 | 1:1 |
| TW136 | 17.25 (±0.02) | 17.68 (±0.015) | 17.13 (±0.043) | −0.43 | 0.12 | 1:1 |
| Ducrettet 101-268 | 17.31 (±0.023) | 17.95 (±0.063) | 17.26 (±0.08) | −0.64 | 0.05 | 1:1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fajkus, P.; Peška, V.; Fajkus, J.; Sýkorová, E. Origin and Fates of TERT Gene Copies in Polyploid Plants. Int. J. Mol. Sci. 2021, 22, 1783. https://doi.org/10.3390/ijms22041783
Fajkus P, Peška V, Fajkus J, Sýkorová E. Origin and Fates of TERT Gene Copies in Polyploid Plants. International Journal of Molecular Sciences. 2021; 22(4):1783. https://doi.org/10.3390/ijms22041783
Chicago/Turabian StyleFajkus, Petr, Vratislav Peška, Jiří Fajkus, and Eva Sýkorová. 2021. "Origin and Fates of TERT Gene Copies in Polyploid Plants" International Journal of Molecular Sciences 22, no. 4: 1783. https://doi.org/10.3390/ijms22041783
APA StyleFajkus, P., Peška, V., Fajkus, J., & Sýkorová, E. (2021). Origin and Fates of TERT Gene Copies in Polyploid Plants. International Journal of Molecular Sciences, 22(4), 1783. https://doi.org/10.3390/ijms22041783