Role of Exosomal miRNA in Bladder Cancer: A Promising Liquid Biopsy Biomarker
Abstract
1. Liquid Biopsy in Bladder Cancer
1.1. Current Landscape
1.2. Properties of Liquid Biopsy
1.3. Future Trends
2. Exosomal miRNAs in Bladder Cancer Diagnosis and Prognosis
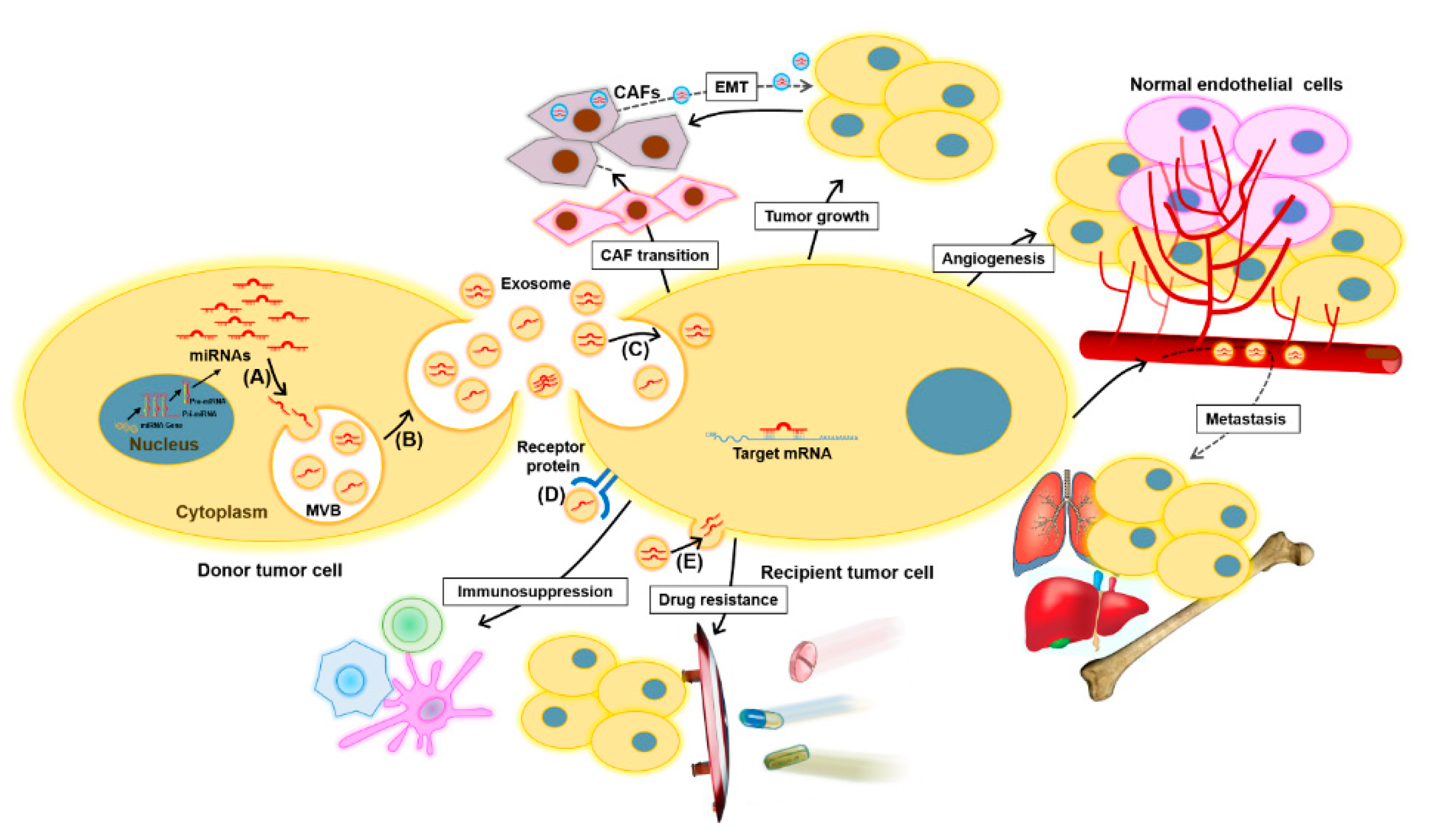
2.1. Exosomes and Exosomal miRNAs in Cancer
2.2. Exosomal miRNAs as Potential Biomarkers for Bladder Cancer
2.2.1. Diagnostic Markers
2.2.2. Prognostic Markers
2.2.3. Treatment Prediction Markers
3. Challenges Associated with the Use of Exosomal miRNAs as Biomarkers
4. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AUC | Area under the receiver-operator characteristics curve |
| BCa | Bladder cancer |
| CAF | Cancer-associated fibroblast |
| CAPP-Seq | Cancer personalized profiling by deep sequencing |
| CTC | Circulating tumor cells |
| ctDNA | Circulating cell-free tumor DNA |
| ddPCR | Digital droplet PCR |
| EMT | Epithelial-mesenchymal transition |
| EV | Extracellular vesicles |
| exomiR | Exosomal miRNAs |
| HD-CTC | High-definition CTC |
| NGS | Next generation sequencing |
| NMIBC | Non-muscle invasive bladder cancer |
| MIBC | Muscle invasive bladder cancer |
| MVB | Multivesicular body |
| RT-PCR | Reverse-transcription PCR |
References
- Cancer Statistics. Available online: https://www.cancer.gov/about-cancer/understanding/statistics (accessed on 25 September 2020).
- Su, H.; Jiang, H.; Tao, T.; Kang, X.; Zhang, X.; Kang, D.; Li, S.; Li, C.; Wang, H.; Yang, Z. Hope and challenge: Precision medicine in bladder cancer. Cancer Med. 2019, 8, 1806–1816. [Google Scholar] [CrossRef] [PubMed]
- Di Meo, A.; Bartlett, J.; Cheng, Y.; Pasic, M.D.; Yousef, G.M. Liquid biopsy: A step forward towards precision medicine in urologic malignancies. Mol. Cancer 2017, 16, 80. [Google Scholar] [CrossRef]
- Siravegna, G.; Marsoni, S.; Siena, S.; Bardelli, A. Integrating liquid biopsies into the management of cancer. Nat. Rev. Clin. Oncol. 2017, 14, 531–548. [Google Scholar] [CrossRef] [PubMed]
- Mandel, P. Les acides nucleiques du plasma sanguin chez 1 homme. CR Seances Soc. Biol. Fil. 1948, 142, 241–243. [Google Scholar]
- Swanton, C. Intratumor heterogeneity: Evolution through space and time. Cancer Res. 2012, 72, 4875–4882. [Google Scholar] [CrossRef]
- Gerlinger, M.; Rowan, A.J.; Horswell, S.; Larkin, J.; Endesfelder, D.; Gronroos, E.; Martinez, P.; Matthews, N.; Stewart, A.; Tarpey, P. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 2012, 366, 883–892. [Google Scholar] [CrossRef] [PubMed]
- Meeks, J.J.; Al-Ahmadie, H.; Faltas, B.M.; Taylor, J.A.; Flaig, T.W.; DeGraff, D.J.; Christensen, E.; Woolbright, B.L.; McConkey, D.J.; Dyrskjøt, L. Genomic heterogeneity in bladder cancer: Challenges and possible solutions to improve outcomes. Nat. Rev. Urol. 2020, 17, 259–270. [Google Scholar] [CrossRef]
- Ashworth, T. A case of cancer in which cells similar to those in the tumours were seen in the blood after death. Aust. Med. J. 1869, 14, 146. [Google Scholar]
- Guan, Y.; Xu, F.; Tian, J.; Chen, H.; Yang, C.; Huang, S.; Gao, K.; Wan, Z.; Li, M.; He, M. Pathology of circulating tumor cells and the available capture tools. Oncol. Rep. 2020, 43, 1355–1364. [Google Scholar] [CrossRef]
- Zhang, Z.; Fan, W.; Deng, Q.; Tang, S.; Wang, P.; Xu, P.; Wang, J.; Yu, M. The prognostic and diagnostic value of circulating tumor cells in bladder cancer and upper tract urothelial carcinoma: A meta-analysis of 30 published studies. Oncotarget 2017, 8, 59527–59538. [Google Scholar] [CrossRef]
- Lotan, Y.; O’Sullivan, P.; Raman, J.D.; Shariat, S.F.; Kavalieris, L.; Frampton, C.; Guilford, P.; Luxmanan, C.; Suttie, J.; Crist, H. Clinical comparison of noninvasive urine tests for ruling out recurrent urothelial carcinoma. Urol. Oncol. 2017, 531, e15–e22. [Google Scholar] [CrossRef]
- Lu, J.-J.; Kakehi, Y.; Takahashi, T.; Wu, X.-X.; Yuasa, T.; Yoshiki, T.; Okada, Y.; Terachi, T.; Ogawa, O. Detection of circulating cancer cells by reverse transcription-polymerase chain reaction for uroplakin II in peripheral blood of patients with urothelial cancer. Clin. Cancer Res. 2000, 6, 3166–3171. [Google Scholar] [PubMed]
- Flaig, T.W.; Wilson, S.; van Bokhoven, A.; Varella-Garcia, M.; Wolfe, P.; Maroni, P.; Genova, E.E.; Morales, D.; Lucia, M.S. Detection of circulating tumor cells in metastatic and clinically localized urothelial carcinoma. Urology 2011, 78, 863–867. [Google Scholar] [CrossRef] [PubMed]
- Busetto, G.M.; Ferro, M.; Del Giudice, F.; Antonini, G.; Chung, B.I.; Sperduti, I.; Giannarelli, D.; Lucarelli, G.; Borghesi, M.; Musi, G. The prognostic role of circulating tumor cells (CTC) in high-risk non–muscle-invasive bladder cancer. Clin. Genitourin. Cancer 2017, 15, e661–e666. [Google Scholar] [CrossRef] [PubMed]
- Naoe, M.; Ogawa, Y.; Morita, J.; Omori, K.; Takeshita, K.; Shichijyo, T.; Okumura, T.; Igarashi, A.; Yanaihara, A.; Iwamoto, S. Detection of circulating urothelial cancer cells in the blood using the CellSearch System. Cancer 2007, 109, 1439–1445. [Google Scholar] [CrossRef]
- Lodewijk, I.; Dueñas, M.; Rubio, C.; Munera-Maravilla, E.; Segovia, C.; Bernardini, A.; Teijeira, A.; Paramio, J.M.; Suárez-Cabrera, C. Liquid biopsy biomarkers in bladder cancer: A current need for patient diagnosis and monitoring. Int. J. Mol. Sci. 2018, 19, 2514. [Google Scholar] [CrossRef]
- Bettegowda, C.; Sausen, M.; Leary, R.J.; Kinde, I.; Wang, Y.; Agrawal, N.; Bartlett, B.R.; Wang, H.; Luber, B.; Alani, R.M. Detection of circulating tumor DNA in early-and late-stage human malignancies. Sci. Transl. Med. 2014, 6, 224ra24. [Google Scholar] [CrossRef]
- Wan, J.C.; Massie, C.; Garcia-Corbacho, J.; Mouliere, F.; Brenton, J.D.; Caldas, C.; Pacey, S.; Baird, R.; Rosenfeld, N. Liquid biopsies come of age: Towards implementation of circulating tumour DNA. Nat. Rev. Cancer 2017, 17, 223–238. [Google Scholar] [CrossRef] [PubMed]
- Francis, G.; Stein, S. Circulating cell-free tumour DNA in the management of cancer. Int. J. Mol. Sci. 2015, 16, 14122–14142. [Google Scholar] [CrossRef]
- Birkenkamp-Demtröder, K.; Nordentoft, I.; Christensen, E.; Høyer, S.; Reinert, T.; Vang, S.; Borre, M.; Agerbæk, M.; Jensen, J.B.; Ørntoft, T.F. Genomic alterations in liquid biopsies from patients with bladder cancer. Eur. Urol. 2016, 70, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Christensen, E.; Birkenkamp-Demtröder, K.; Nordentoft, I.; Høyer, S.; Van Der Keur, K.; Van Kessel, K.; Zwarthoff, E.; Agerbæk, M.; Ørntoft, T.F.; Jensen, J.B. Liquid biopsy analysis of FGFR3 and PIK3CA hotspot mutations for disease surveillance in bladder cancer. Eur. Urol. 2017, 71, 961–969. [Google Scholar] [CrossRef]
- Vandekerkhove, G.; Todenhöfer, T.; Annala, M.; Struss, W.J.; Wong, A.; Beja, K.; Ritch, E.; Brahmbhatt, S.; Volik, S.V.; Hennenlotter, J. Circulating tumor DNA reveals clinically actionable somatic genome of metastatic bladder cancer. Clin. Cancer Res. 2017, 23, 6487–6497. [Google Scholar] [CrossRef]
- Patel, K.; Van Der Vos, K.; Smith, C.G.; Mouliere, F.; Tsui, D.; Morris, J.; Chandrananda, D.; Marass, F.; Van Den Broek, D.; Neal, D. Association of plasma and urinary mutant DNA with clinical outcomes in muscle invasive bladder cancer. Sci. Rep. 2017, 7, 5554. [Google Scholar] [CrossRef] [PubMed]
- Brohawn, P.Z.; Higgs, B.W.; Kuziora, M.; Englert, J.; Ranade, K. Early reduction in circulating tumor DNA (ctDNA) and survival in gastric cancer patients (pts) treated with durvalumab (D), tremelimumab (T), or durvalumab in combination with tremelimumab (D+ T). J. Clin. Oncol. 2018, 36, e15027. [Google Scholar] [CrossRef]
- Cheng, M.L.; Shady, M.; Cipolla, C.K.; Funt, S.; Arcila, M.E.; Al-Ahmadie, H.; Rosenberg, J.E.; Bajorin, D.F.; Berger, M.F.; Tsui, D. Comparison of somatic mutation profiles from cell free DNA (cfDNA) versus tissue in metastatic urothelial carcinoma (mUC). J. Clin. Oncol. 2017, 35, 4533. [Google Scholar] [CrossRef]
- Hegemann, M.; Stenzl, A.; Bedke, J.; Chi, K.N.; Black, P.C.; Todenhöfer, T. Liquid biopsy: Ready to guide therapy in advanced prostate cancer? BJU Int. 2016, 118, 855–863. [Google Scholar] [CrossRef]
- Alix-Panabières, C.; Pantel, K. Clinical applications of circulating tumor cells and circulating tumor DNA as liquid biopsy. Cancer Discov. 2016, 6, 479–491. [Google Scholar] [CrossRef] [PubMed]
- Thind, A.; Wilson, C. Exosomal miRNAs as cancer biomarkers and therapeutic targets. J. Extracell. Vesicles 2016, 5, 31292. [Google Scholar] [CrossRef] [PubMed]
- Mathieu, M.; Martin-Jaular, L.; Lavieu, G.; Thery, C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell Biol. 2019, 21, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Berrondo, C.; Flax, J.; Kucherov, V.; Siebert, A.; Osinski, T.; Rosenberg, A.; Fucile, C.; Richheimer, S.; Beckham, C.J. Expression of the long non-coding RNA HOTAIR correlates with disease progression in bladder cancer and is contained in bladder cancer patient urinary exosomes. PLoS ONE 2016, 11, e0147236. [Google Scholar] [CrossRef] [PubMed]
- Silvers, C.R.; Miyamoto, H.; Messing, E.M.; Netto, G.J.; Lee, Y.-F. Characterization of urinary extracellular vesicle proteins in muscle-invasive bladder cancer. Oncotarget 2017, 8, 91199. [Google Scholar] [CrossRef] [PubMed]
- Baumgart, S.; Hölters, S.; Ohlmann, C.-H.; Bohle, R.; Stöckle, M.; Ostenfeld, M.S.; Dyrskjøt, L.; Junker, K.; Heinzelmann, J. Exosomes of invasive urothelial carcinoma cells are characterized by a specific miRNA expression signature. Oncotarget 2017, 8, 58278–91208. [Google Scholar] [CrossRef] [PubMed]
- Beckham, C.J.; Olsen, J.; Yin, P.-N.; Wu, C.-H.; Ting, H.-J.; Hagen, F.K.; Scosyrev, E.; Messing, E.M.; Lee, Y.-F. Bladder cancer exosomes contain EDIL-3/Del1 and facilitate cancer progression. J. Urol. 2014, 192, 583–592. [Google Scholar] [CrossRef]
- Armstrong, D.A.; Green, B.B.; Seigne, J.D.; Schned, A.R.; Marsit, C.J. MicroRNA molecular profiling from matched tumor and bio-fluids in bladder cancer. Mol. Cancer 2015, 14, 194. [Google Scholar] [CrossRef] [PubMed]
- Piao, X.M.; Jeong, P.; Kim, Y.H.; Byun, Y.J.; Xu, Y.; Kang, H.W.; Ha, Y.S.; Kim, W.T.; Lee, J.Y.; Woo, S.H. Urinary cell-free microRNA biomarker could discriminate bladder cancer from benign hematuria. Int. J. Cancer 2019, 144, 380–388. [Google Scholar] [CrossRef]
- Harouaka, R.; Kang, Z.; Zheng, S.-Y.; Cao, L. Circulating tumor cells: Advances in isolation and analysis, and challenges for clinical applications. Pharm. Ther. 2014, 141, 209–221. [Google Scholar] [CrossRef] [PubMed]
- Hillig, T.; Nygaard, A.B.; Nekiunaite, L.; Klingelhöfer, J.; Sölétormos, G. In vitro validation of an ultra-sensitive scanning fluorescence microscope for analysis of Circulating Tumor Cells. APMIS 2014, 122, 545–551. [Google Scholar] [CrossRef]
- Diaz, L.A., Jr.; Bardelli, A. Liquid biopsies: Genotyping circulating tumor DNA. J. Clin. Oncol. 2014, 32, 579–586. [Google Scholar] [CrossRef]
- Olsson, E.; Winter, C.; George, A.; Chen, Y.; Howlin, J.; Tang, M.H.E.; Dahlgren, M.; Schulz, R.; Grabau, D.; van Westen, D. Serial monitoring of circulating tumor DNA in patients with primary breast cancer for detection of occult metastatic disease. Embo Mol. Med. 2015, 7, 1034–1047. [Google Scholar] [CrossRef]
- Li, M.; Diehl, F.; Dressman, D.; Vogelstein, B.; Kinzler, K.W. BEAMing up for detection and quantification of rare sequence variants. Nat. Methods 2006, 3, 95–97. [Google Scholar] [CrossRef]
- Newman, A.M.; Bratman, S.V.; To, J.; Wynne, J.F.; Eclov, N.C.; Modlin, L.A.; Liu, C.L.; Neal, J.W.; Wakelee, H.A.; Merritt, R.E. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat. Med. 2014, 20, 548–554. [Google Scholar] [CrossRef]
- Todenhöfer, T.; Struss, W.J.; Seiler, R.; Wyatt, A.W.; Black, P.C. Liquid biopsy-analysis of circulating tumor DNA (ctDNA) in bladder cancer. Bladder Cancer 2018, 4, 19–29. [Google Scholar] [CrossRef] [PubMed]
- Bellingham, S.A.; Shambrook, M.; Hill, A.F. Quantitative analysis of exosomal miRNA via qPCR and digital PCR, in Exosomes and Microvesicles. Methods Mol. Biol. 2017, 1545, 55–70. [Google Scholar] [CrossRef]
- Liu, X.; Yuan, W.; Yang, L.; Li, J.; Cai, J. miRNA profiling of exosomes from spontaneous hypertensive rats using next-generation sequencing. J. Cardiovasc Transl. Res. 2019, 12, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Salimu, J.; Webber, J.; Gurney, M.; Al-Taei, S.; Clayton, A.; Tabi, Z. Dominant immunosuppression of dendritic cell function by prostate-cancer-derived exosomes. J. Extracell. Vesicles 2017, 6, 1368823. [Google Scholar] [CrossRef] [PubMed]
- Pisitkun, T.; Johnstone, R.; Knepper, M.A. Discovery of urinary biomarkers. Mol. Cell Proteom. 2006, 5, 1760–1771. [Google Scholar] [CrossRef] [PubMed]
- Gámez-Valero, A.; Lozano-Ramos, S.I.; Bancu, I.; Lauzurica-Valdemoros, R.; Borràs, F.E. Urinary extracellular vesicles as source of biomarkers in kidney diseases. Front. Immunol. 2015, 6, 6. [Google Scholar] [CrossRef]
- Iyer, M.; Niknafs, Y.; Malik, R.; Malik, R.; Singhal, U.; Sahu, A.; Hosono, Y.; Barrette, T.R.; Prensner, J.R.; Evans, J.R.; et al. The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 2015, 47, 199–208. [Google Scholar] [CrossRef]
- Jeppesen, D.K.; Fenix, A.M.; Franklin, J.L.; Higginbotham, J.N.; Zhang, Q.; Zimmerman, L.J.; Liebler, D.C.; Ping, J.; Liu, Q.; Evans, R.; et al. Reassessment of exosome composition. Cell 2019, 177, 428–445.e18. [Google Scholar] [CrossRef]
- Yokoi, A.; Villar-Prados, A.; Oliphint, P.A.; Zhang, J.; Song, X.; De Hoff, P.; Morey, R.; Liu, J.; Roszik, J.; Clise-Dwyer, K.; et al. Mechanisms of nuclear content loading to exosomes. Sci. Adv. 2019, 5, eaax8849. [Google Scholar] [CrossRef]
- Weber, J.A.; Baxter, D.H.; Zhang, S.; Huang, D.Y.; How Huang, K.; Jen Lee, M.; Galas, D.J.; Wang, K. The microRNA spectrum in 12 body fluids. Clin. Chem. 2010, 56, 1733–1741. [Google Scholar] [CrossRef]
- Yun, S.J.; Jeong, P.; Kim, W.-T.; Kim, T.H.; Lee, Y.-S.; Song, P.H.; Choi, Y.-H.; Kim, I.Y.; Moon, S.-K.; Kim, W.-J. Cell-free microRNAs in urine as diagnostic and prognostic biomarkers of bladder cancer. Int. J. Oncol. 2012, 41, 1871–1878. [Google Scholar] [CrossRef]
- Minciacchi, V.R.; Freeman, M.R.; Di Vizio, D. Extracellular vesicles in cancer: Exosomes, microvesicles and the emerging role of large oncosomes. Semi. Cell Dev. Biol. 2015, 40, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Tomasetti, M.; Lee, W.; Santarelli, L.; Neuzil, J. Exosome-derived microRNAs in cancer metabolism: Possible implications in cancer diagnostics and therapy. Exp. Mol. Med. 2017, 49, e285. [Google Scholar] [CrossRef]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef] [PubMed]
- Xi, Y.; Nakajima, G.; Gavin, E.; Morris, C.G.; Kudo, K.; Hayashi, K.; Ju, J. Systematic analysis of microRNA expression of RNA extracted from fresh frozen and formalin-fixed paraffin-embedded samples. RNA 2007, 13, 1668–1674. [Google Scholar] [CrossRef]
- Rabinowits, G.; Gerçel-Taylor, C.; Day, J.M.; Taylor, D.D.; Kloecker, G.H. Exosomal microRNA: A diagnostic marker for lung cancer. Clin. Lung Cancer 2009, 10, 42–46. [Google Scholar] [CrossRef]
- Taylor, D.D.; Gercel-Taylor, C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008, 110, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Andre, F.; Schartz, N.E.; Movassagh, M.; Flament, C.; Pautier, P.; Morice, P.; Pomel, C.; Lhomme, C.; Escudier, B.; Le Chevalier, T. Malignant effusions and immunogenic tumour-derived exosomes. Lancet 2002, 360, 295–305. [Google Scholar] [CrossRef]
- Valenti, R.; Huber, V.; Filipazzi, P.; Pilla, L.; Sovena, G.; Villa, A.; Corbelli, A.; Fais, S.; Parmiani, G.; Rivoltini, L. Human tumor-released microvesicles promote the differentiation of myeloid cells with transforming growth factor-β–mediated suppressive activity on T lymphocytes. Cancer Res. 2006, 66, 9290–9298. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Xu, K.; Zheng, X.; Chen, T.; Wang, J.; Song, Y.; Shao, Y.; Zheng, S. Application of exosomes as liquid biopsy in clinical diagnosis. Signal. Transduct. Target. Ther. 2020, 5, 144. [Google Scholar] [CrossRef]
- Magy-Bertrand, N. Actualités sur les amyloses. La Rev. De Médecine Interne 2016, 37, 529–535. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Yu, F.; Ding, H.; Wang, Y.; Li, P.; Wang, K. Emerging function and clinical values of exosomal MicroRNAs in cancer. Mol. Ther. Nucleic Acids 2019, 16, 791–804. [Google Scholar] [CrossRef] [PubMed]
- Rahbarghazi, R.; Jabbari, N.; Sani, N.A.; Asghari, R.; Salimi, L.; Kalashani, S.A.; Feghhi, M.; Etemadi, T.; Akbariazar, E.; Mahmoudi, M. Tumor-derived extracellular vesicles: Reliable tools for Cancer diagnosis and clinical applications. Cell Commun. Signal. 2019, 17, 73. [Google Scholar] [CrossRef]
- Liu, J.; Sun, H.; Wang, X.; Yu, Q.; Li, S.; Yu, X.; Gong, W. Increased exosomal microRNA-21 and microRNA-146a levels in the cervicovaginal lavage specimens of patients with cervical cancer. Int. J. Mol. Sci. 2014, 15, 758–773. [Google Scholar] [CrossRef] [PubMed]
- Cui, S.; Cheng, Z.; Qin, W.; Jiang, L. Exosomes as a liquid biopsy for lung cancer. Lung Cancer 2018, 116, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Hsu, Y.; Hung, J.; Chang, W.; Lin, Y.; Pan, Y.; Tsai, P.; Wu, C.; Kuo, P. Hypoxic lung cancer-secreted exosomal miR-23a increased angiogenesis and vascular permeability by targeting prolyl hydroxylase and tight junction protein ZO-1. Oncogene 2017, 36, 4929–4942. [Google Scholar] [CrossRef] [PubMed]
- Bao, L.; You, B.; Shi, S.; Shan, Y.; Zhang, Q.; Yue, H.; Zhang, J.; Zhang, W.; Shi, Y.; Liu, Y. Metastasis-associated miR-23a from nasopharyngeal carcinoma-derived exosomes mediates angiogenesis by repressing a novel target gene TSGA10. Oncogene 2018, 37, 2873–2889. [Google Scholar] [CrossRef]
- Sruthi, T.; Edatt, L.; Raji, G.R.; Kunhiraman, H.; Shankar, S.S.; Shankar, V.; Ramachandran, V.; Poyyakkara, A.; Kumar, S.V. Horizontal transfer of miR-23a from hypoxic tumor cell colonies can induce angiogenesis. J. Cell Physiol. 2018, 233, 3498–3514. [Google Scholar] [CrossRef]
- Lin, X.-J.; Fang, J.-H.; Yang, X.-J.; Zhang, C.; Yuan, Y.; Zheng, L.; Zhuang, S.-M. Hepatocellular carcinoma cell-secreted exosomal microRNA-210 promotes angiogenesis in vitro and in vivo. Mol. Ther. Nucleic Acids 2018, 11, 243–252. [Google Scholar] [CrossRef]
- Fang, J.H.; Zhang, Z.J.; Shang, L.R.; Luo, Y.W.; Lin, Y.F.; Yuan, Y.; Zhuang, S.M. Hepatoma cell-secreted exosomal microRNA-103 increases vascular permeability and promotes metastasis by targeting junction proteins. Hepatology 2018, 68, 1459–1475. [Google Scholar] [CrossRef]
- Liu, Y.; Luo, F.; Wang, B.; Li, H.; Xu, Y.; Liu, X.; Shi, L.; Lu, X.; Xu, W.; Lu, L. STAT3-regulated exosomal miR-21 promotes angiogenesis and is involved in neoplastic processes of transformed human bronchial epithelial cells. Cancer Lett. 2016, 370, 125–135. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.-F.; Zhang, X.-W.; Hua, R.-X.; Du, Y.-Q.; Huang, M.-Z.; Liu, Y.; Cheng, Y.F.; Guo, W.-J. Mel-18 negatively regulates stem cell-like properties through downregulation of miR-21 in gastric cancer. Oncotarget 2016, 7, 63352–63361. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yasui, T.; Yanagida, T.; Ito, S.; Konakade, Y.; Takeshita, D.; Naganawa, T.; Nagashima, K.; Shimada, T.; Kaji, N.; Nakamura, Y.; et al. Unveiling massive numbers of cancer-related urinary-microRNA candidates via nanowires. Sci. Adv. 2017, 3, e1701133. [Google Scholar] [CrossRef]
- Andreu, Z.; Oshiro, R.O.; Redruello, A.; López-Martín, S.; Gutiérrez-Vázquez, C.; Morato, E.; Marina, A.I.; Gómez, C.O.; Yáñez-Mó, M. Extracellular vesicles as a source for non-invasive biomarkers in bladder cancer progression. Eur. J. Pharm. Sci. 2017, 98, 70–79. [Google Scholar] [CrossRef]
- Matsuzaki, K.; Fujita, K.; Jingushi, K.; Kawashima, A.; Ujike, T.; Nagahara, A.; Ueda, Y.; Tanigawa, G.; Yoshioka, I.; Ueda, K.; et al. MiR-21-5p in urinary extracellular vesicles is a novel biomarker of urothelial carcinoma. Oncotarget 2017, 8, 24668–24678. [Google Scholar] [CrossRef]
- Long, J.D.; Sullivan, T.B.; Humphrey, J.; Logvinenko, T.; Summerhayes, K.A.; Kozinn, S.; Harty, N.; Summerhayes, I.C.; Libertino, J.A.; Holway, A.H.; et al. A non-invasive miRNA based assay to detect bladder cancer in cell-free urine. Am. J. Transl. Res. 2015, 7, 2500–2509. [Google Scholar]
- Pardini, B.; Cordero, F.; Naccarati, A.; Viberti, C.; Birolo, G.; Oderda, M.; Di Gaetano, C.; Arigoni, M.; Martina, F.; Calogero, R.A.; et al. microRNA profiles in urine by next-generation sequencing can stratify bladder cancer subtypes. Oncotarget 2018, 9, 20658–20669. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Du, L.; Wang, L.; Li, J.; Liu, Y.; Zheng, G.; Qu, A.; Zhang, X.; Pan, H.; Yang, Y.; et al. Serum microRNA expression signatures identified from genome-wide microRNA profiling serve as novel noninvasive biomarkers for diagnosis and recurrence of bladder cancer. Int. J. Cancer 2015, 136, 854–862. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Jiang, X.; Duan, W.; Wang, R.; Wang, L.; Zheng, G.; Yan, K.; Wang, L.; Li, J.; Zhang, X.; et al. Cell-free microRNA expression signatures in urine serve as novel noninvasive biomarkers for diagnosis and recurrence prediction of bladder cancer. Oncotarget 2017, 8, 40832–40842. [Google Scholar] [CrossRef]
- Urquidi, V.; Netherton, M.; Gomes-Giacoia, E.; Serie, D.J.; Eckel-Passow, J.; Rosser, C.J.; Goodison, S. A microRNA biomarker panel for the non-invasive detection of bladder cancer. Oncotarget 2016, 7, 86290–86299. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.Z.; Lau, K.M.; Chan, E.S.; Wang, G.; Szeto, C.C.; Wong, K.; Choy, R.K.; Ng, C.F. Cell-free urinary microRNA-99a and microRNA-125b are diagnostic markers for the non-invasive screening of bladder cancer. PLoS ONE 2014, 9, e100793. [Google Scholar] [CrossRef]
- Baumgart, S.; Meschkat, P.; Edelmann, P.; Hartmann, A.; Bohle, R.; Pryalukhin, A.; Heinzelmann, J.; Stöckle, M.; Junker, K. Invasion-associated miRNAs as possible diagnostic biomarkers of muscle invasive bladder cancer in tumor tissues and urinary exosomes. J. Urol. 2018, 199, e1038. [Google Scholar] [CrossRef][Green Version]
- Kim, S.M.; Kang, H.W.; Kim, W.T.; Kim, Y.J.; Yun, S.J.; Lee, S.C.; Kim, W.J. Cell-Free microRNA-214 From Urine as a Biomarker for Non-Muscle-Invasive Bladder Cancer. Korean J. Urol. 2013, 54, 791–796. [Google Scholar] [CrossRef] [PubMed]
- Sapre, N.; Macintyre, G.; Clarkson, M.; Naeem, H.; Cmero, M.; Kowalczyk, A.; Anderson, P.D.; Costello, A.J.; Corcoran, N.M.; Hovens, C.M. A urinary microRNA signature can predict the presence of bladder urothelial carcinoma in patients undergoing surveillance. Br. J. Cancer 2016, 114, 454–462. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Du, L.; Duan, W.; Wang, R.; Yan, K.; Wang, L.; Li, J.; Zheng, G.; Zhang, X.; Yang, Y.; et al. Serum microRNA expression signatures as novel noninvasive biomarkers for prediction and prognosis of muscle-invasive bladder cancer. Oncotarget 2016, 7, 36733–36742. [Google Scholar] [CrossRef]
- Santos, P.; Almeida, F. Role of Exosomal miRNAs and the Tumor Microenvironment in Drug Resistance. Cells 2020, 9, 1450. [Google Scholar] [CrossRef]
- Li, X.J.; Ren, Z.J.; Tang, J.H.; Yu, Q. Exosomal MicroRNA MiR-1246 promotes cell proliferation, invasion and drug resistance by targeting CCNG2 in breast cancer. Cell Physiol. Biochem. 2017, 44, 1741–1748. [Google Scholar] [CrossRef]
- Bayraktar, R.; Van Roosbroeck, K. miR-155 in cancer drug resistance and as target for miRNA-based therapeutics. Cancer Metastasis Rev. 2018, 37, 33–44. [Google Scholar] [CrossRef]
- Santos, J.C.; da Silva Lima, N.; Sarian, L.O.; Matheu, A.; Ribeiro, M.L.; Derchain, S.F.M. Exosome-mediated breast cancer chemoresistance via miR-155 transfer. Sci. Rep. 2018, 8, 829. [Google Scholar] [CrossRef]
- Patel, G.K.; Khan, M.A.; Bhardwaj, A.; Srivastava, S.K.; Zubair, H.; Patton, M.C.; Singh, S.; Singh, A.P. Exosomes confer chemoresistance to pancreatic cancer cells by promoting ROS detoxification and miR-155-mediated suppression of key gemcitabine-metabolising enzyme, DCK. Br. J. Cancer 2017, 116, 609–619. [Google Scholar] [CrossRef]
- Liu, T.; Zhang, X.; Du, L.; Wang, Y.; Liu, X.; Tian, H.; Wang, L.; Li, P.; Zhao, Y.; Duan, W. Exosome-transmitted miR-128-3p increase chemosensitivity of oxaliplatin-resistant colorectal cancer. Mol. Cancer 2019, 18, 43. [Google Scholar] [CrossRef]
- Wang, B.; Zhang, Y.; Ye, M.; Wu, J.; Ma, L.; Chen, H. Cisplatin-resistant MDA-MB-231 cell-derived exosomes increase the resistance of recipient cells in an exosomal miR-423-5p-dependent manner. Curr. Drug Metab. 2019, 20, 804–814. [Google Scholar] [CrossRef]
- Fanous, H.; Sullivan, T.; Rieger-Christ, K. Distinct exosomal miRNA profiles in chemoresistant bladder carcinoma cell lines. J. Urol. 2017, 197, e1179–e1180. [Google Scholar] [CrossRef][Green Version]
- Li, K.; Rodosthenous, R.S.; Kashanchi, F.; Gingeras, T.; Gould, S.J.; Kuo, L.S.; Kurre, P.; Lee, H.; Leonard, J.N.; Liu, H. Advances, challenges, and opportunities in extracellular RNA biology: Insights from the NIH exRNA Strategic Workshop. JCI Insight 2018, 3, e98942. [Google Scholar] [CrossRef] [PubMed]
- Witwer, K.W.; Buzás, E.I.; Bemis, L.T.; Bora, A.; Lässer, C.; Lötvall, J.; Nolte- ‘t Hoen, E.N.; Piper, M.G.; Sivaraman, S.; Skog, J. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J. Extracell. Vesicles 2013, 2, 20360. [Google Scholar] [CrossRef] [PubMed]
- Van Deun, J.; Mestdagh, P.; Agostinis, P.; Akay, Ö.; Anand, S.; Anckaert, J.; Martinez, Z.A.; Baetens, T.; Beghein, E.; Bertier, L. EV-TRACK: Transparent reporting and centralizing knowledge in extracellular vesicle research. Nat. Methods 2017, 14, 228–232. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Yuan, Y.; Cho, J.-H.; McClarty, S.; Baxter, D.; Galas, D.J. Comparing the MicroRNA spectrum between serum and plasma. PLoS ONE 2012, 7, e41561. [Google Scholar] [CrossRef] [PubMed]
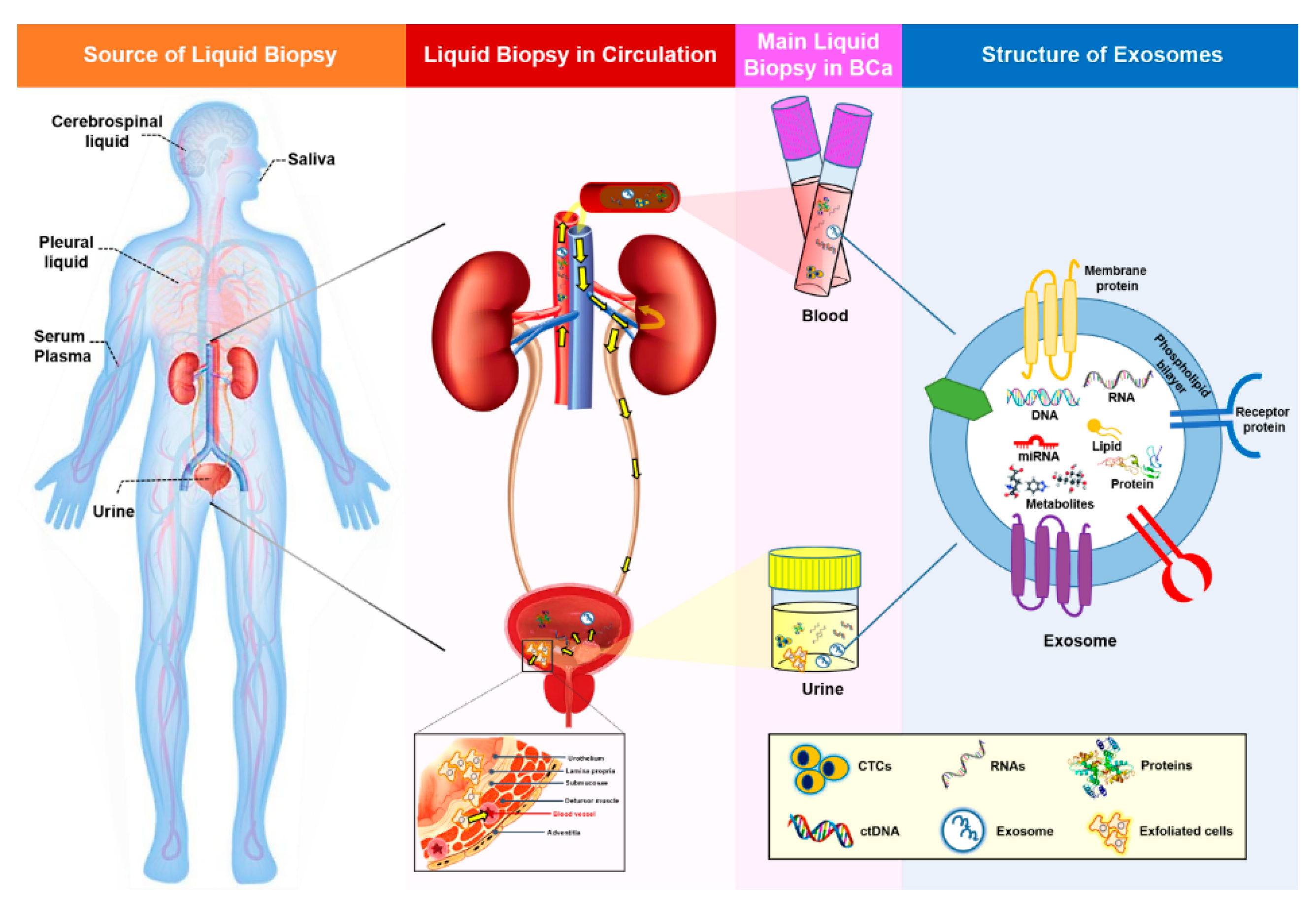
| Type of Liquid Biopsy Sample | Origin | Detection Methods | Advantages | Disadvantages | Applications in BCa | References |
|---|---|---|---|---|---|---|
| CTC | Primary tumors; Metastatic sites. | Microfilters; Immunocytochemistry; Immunomagnetic assays; CellSearch System; CytoTrack; Epic CTC Platform; HD-CTC; RT-PCR. | High specificity; Non-invasive analysis; Represent genomic information from the tumor or metastatic sites in real time; Can observe response to therapeutic treatments. | Low sensitivity to monitor tumors, particularly early stage tumors. | Valuable prognosis marker associated with disease-free survival in metastatic BCa; Useful for deciding therapeutic approaches, since it is strongly correlated to tumor stage, histological grade, and metastasis. | [9,11,12,13,14,15,16,37,38] |
| ctDNA | Released from viable tumors by activating secretion, apoptosis, or necrosis; Destruction of CTCs. | PCR; Pyrosequencing; RT-PCR; NGS; ddPCR; BEAMing technology; CAPP-Seq. | High sensitivity; Non-invasive analysis; Stability in variable degradation microenvironments; Can be used to detect mutations related to carcinogenesis. | Require a specific method to distinguish tumor origin DNA from healthy origin DNA, since both release ctDNA; Low yield in plasma; Low sensitivity to monitor tumors, particularly early stage tumors. | Genomic alterations such as specific mutations, deletions, and methylation variations detected in ctDNA from BCa urine and plasma are correlated to disease recurrence and progression; Alterations in ctDNAs with differential sensitivity to therapeutic agents could be used as markers of therapy response in metastatic patients. | [12,13,15,17,21,22,23,24,25,26,39,40,41,42,43] |
| Exosome | Primary tumors; Cells in various body fluids | Real-time PCR; ddPCR; NGS; Microarray; Western blotting; ELISA. | Non-invasive analysis; High stability in variable degradation microenvironments; Better sensitivity and specificity than CTC and ctDNA in various bio-specimens. | Challenging protocols to analyze genomic materials in exosomes | Urinary exosomal miRNAs show potential for BCa detection; Long non-coding RNAs and proteins in urinary exosomes are proposed to be enriched in BCa. | [28,31,32,33,34,35,36,44,45,46] |
| Markers | Biological Source | Regulation | Clinical Significance | Reference |
|---|---|---|---|---|
| miR-1285-3p, miR-142-3p, miR-16-1-3p, miR-195-3p, miR-196b-5p, miR-23b-3p, miR-28-5p, miR-299-3p, miR-3155a, miR-3162-5p, miR-3678-3p, miR-4283, miR-4295, miR-4311, miR-4531, miR-492, miR-5096, miR-513b-5p, miR-5187-5p, miR-601, miR-619-5p, miR-92a-2-5p | Urine | Up | Diagnosis | [75] |
| miR-375, miR-146a | Urine | Up | Diagnosis | [76] |
| miR-155-5p, miR-15a-5p, miR-21-5p, miR-132-3p, miR-31-5p (especially miR-21-5p) | Urine | Up | Diagnosis | [77] |
| Four-miRNA panel: mir-21, miR-93, miR-200c, and miR-940 | Urine | Up | Diagnosis | [78] |
| Three-miRNA panel: miR-30a-5p, let-7c-5p and miR-486-5p | Urine | Up | Diagnosis | [79] |
| Six-miRNA panel: miR-152, miR-148b-3p, miR-3187-3p, miR-15b-5p, miR-27a-3p and miR-30a-5p | Serum | Up | Diagnosis | [80] |
| Seven-miRNA panel: miR-7-5p, miR-22-3p, miR-29a-3p, miR-126-5p, miR-200a-3p, miR-375 and miR-423-5p | Urine | Up | Diagnosis | [81] |
| Twenty-five-miRNA panel: miR-652, miR-199a-3p, miR-140-5p, miR-93, miR-142-5p, miR-1305, miR-30a, miR-224, miR-96, miR-766, miR-223, miR-99b, miR-140-3p, let-7b, miR-141, miR-191, miR-146b-5p, miR-491-5p, miR-339-3p, miR-200c, miR-106b, miR-143, miR-429, miR-222 and miR-200a | Urine | Up | Diagnosis | [82] |
| Ratio of miR-6124/miR-4511 | Urine | Up | Diagnosis | [36] |
| miR-30a-3p, miR-99a-5p, miR-137-3p | Cells | Up | Discrimination of MIBC from NMIBC | [84] |
| miR-141-3p, miR-205-5p | Cells | Down | Discrimination of MIBC from NMIBC | [84] |
| miR-99a, miR-125b | Urine | Down | Diagnosis | [83] |
| miR-152 | Serum | Up | Prediction of NMIBC recurrence | [80] |
| miR-22-3p, miR-200a-3p | Urine | Up | Prediction of NMIBC recurrence | [81] |
| Six-miRNA panel: miR16, miR200c, miR205, miR21, miR221 and miR34a | Urine | Up | Prediction of NMIBC recurrence | [86] |
| Four-miRNA panel: miR-422a-3p, miR-486-3p, miR-103a-3p and miR-27a-3p | Serum | Up | Prediction of MIBC survival | [87] |
| miR-214 | Urine | Down | Prediction of NMIBC recurrence | [85] |
| miR-21-5p | Cells | Up | Prediction of response to chemotherapy | [95] |
| miR-Let-7i-3p | Cells | Down | Prediction of response to chemotherapy | [95] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piao, X.-M.; Cha, E.-J.; Yun, S.J.; Kim, W.-J. Role of Exosomal miRNA in Bladder Cancer: A Promising Liquid Biopsy Biomarker. Int. J. Mol. Sci. 2021, 22, 1713. https://doi.org/10.3390/ijms22041713
Piao X-M, Cha E-J, Yun SJ, Kim W-J. Role of Exosomal miRNA in Bladder Cancer: A Promising Liquid Biopsy Biomarker. International Journal of Molecular Sciences. 2021; 22(4):1713. https://doi.org/10.3390/ijms22041713
Chicago/Turabian StylePiao, Xuan-Mei, Eun-Jong Cha, Seok Joong Yun, and Wun-Jae Kim. 2021. "Role of Exosomal miRNA in Bladder Cancer: A Promising Liquid Biopsy Biomarker" International Journal of Molecular Sciences 22, no. 4: 1713. https://doi.org/10.3390/ijms22041713
APA StylePiao, X.-M., Cha, E.-J., Yun, S. J., & Kim, W.-J. (2021). Role of Exosomal miRNA in Bladder Cancer: A Promising Liquid Biopsy Biomarker. International Journal of Molecular Sciences, 22(4), 1713. https://doi.org/10.3390/ijms22041713






