Genome-Wide Identification and Expression Analysis of OsbZIP09 Target Genes in Rice Reveal Its Mechanism of Controlling Seed Germination
Abstract
1. Introduction
2. Results
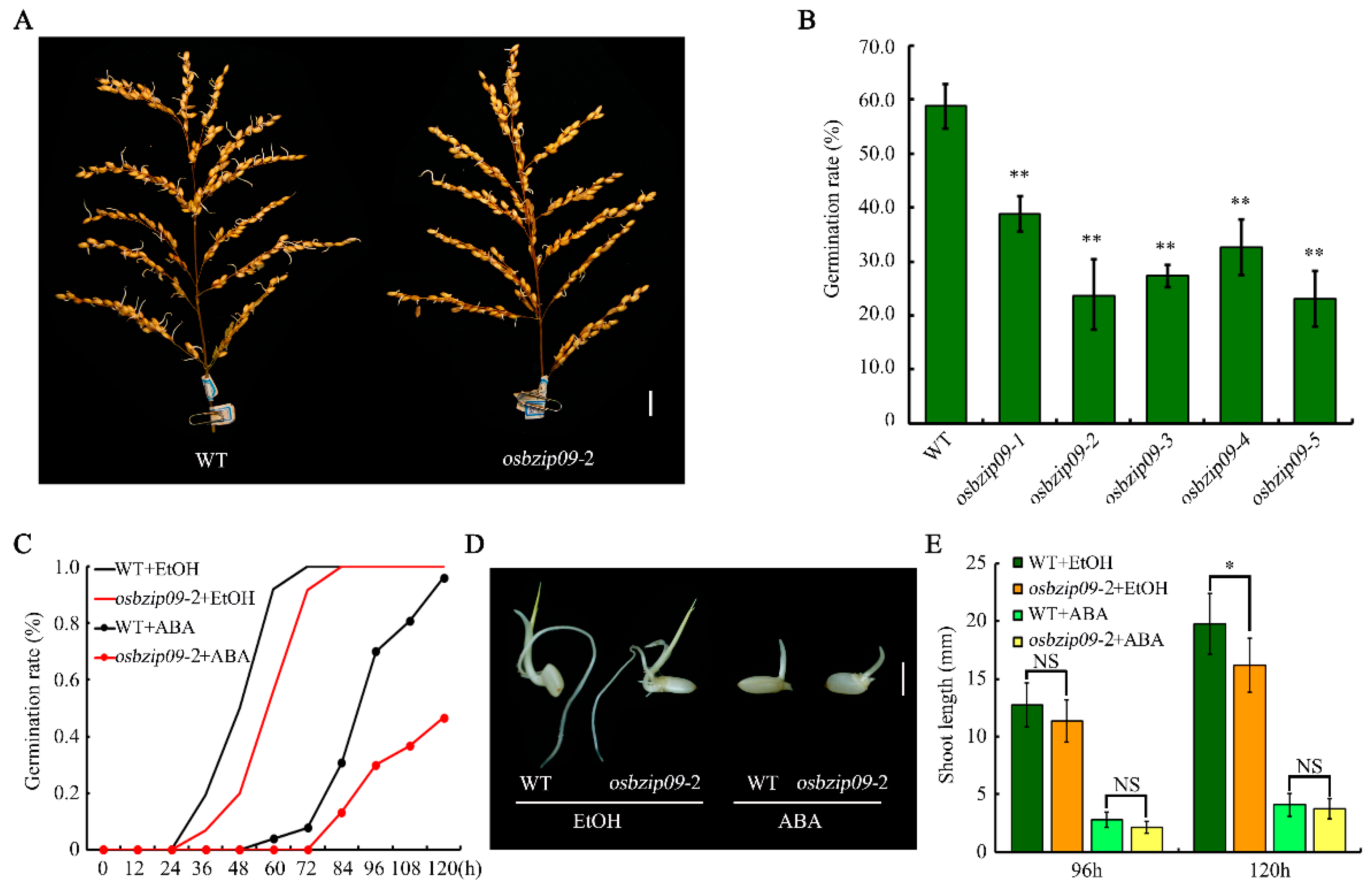
2.1. Mutation of OsbZIP09 Inhibits PHS in Rice and Enhances Rice Sensitivity to ABA during Seed Germination
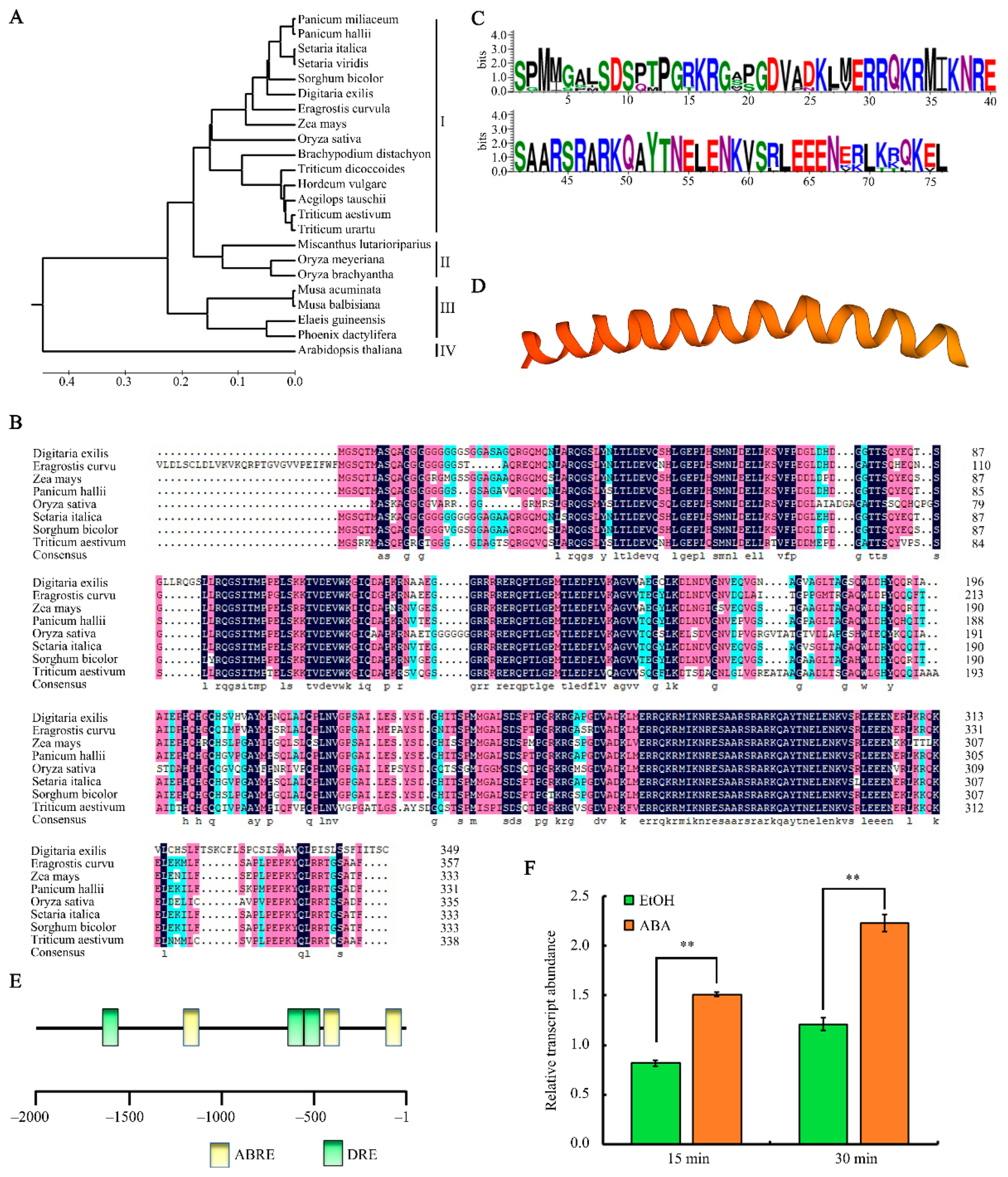
2.2. Analysis of the Phylogenetic Relationships, Protein Sequence and Structure, and Gene Expression of bZIP09
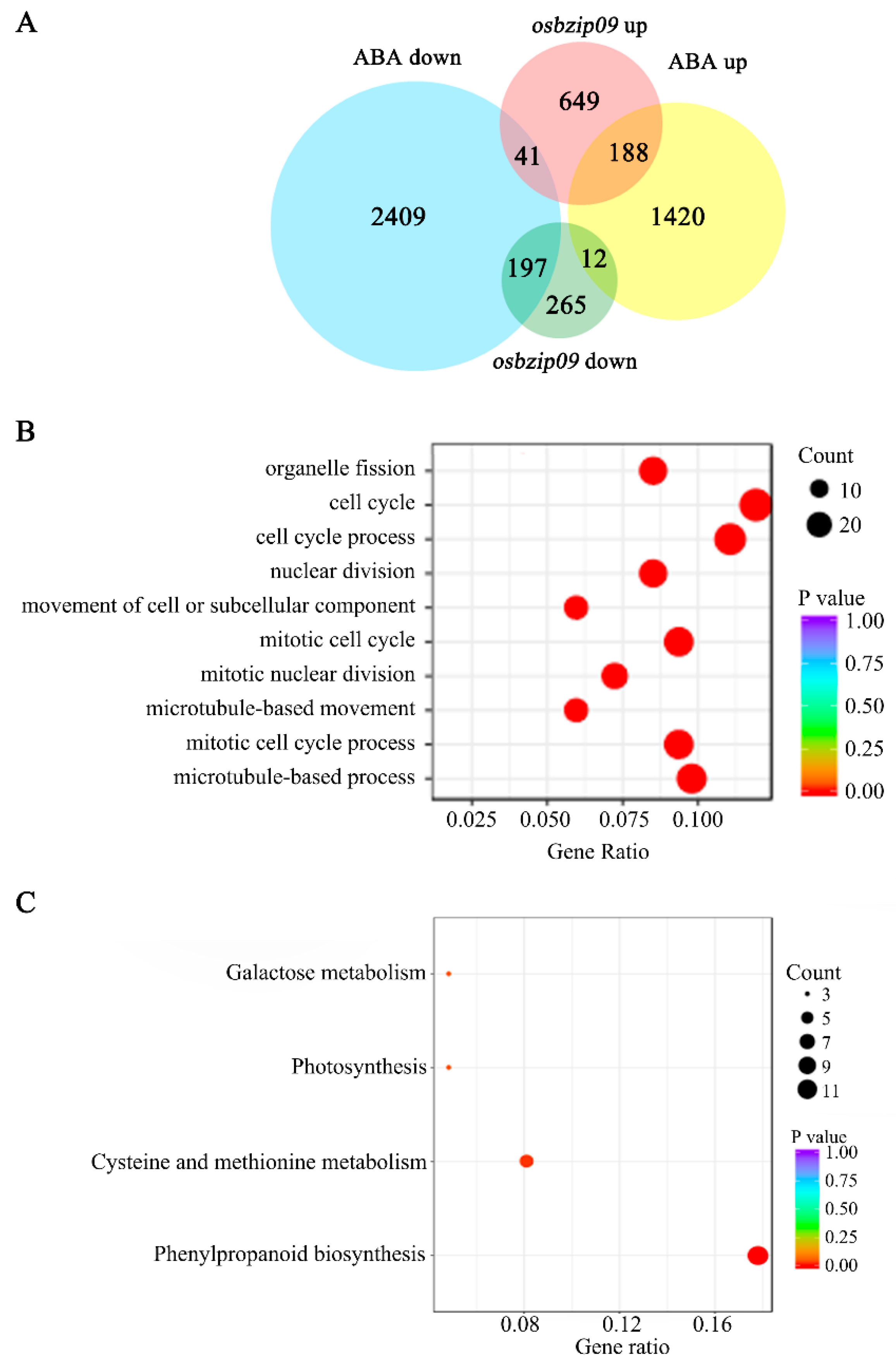
2.3. Identification of Genes Co-Regulated by ABA and OsbZIP09 Via RNA-Seq
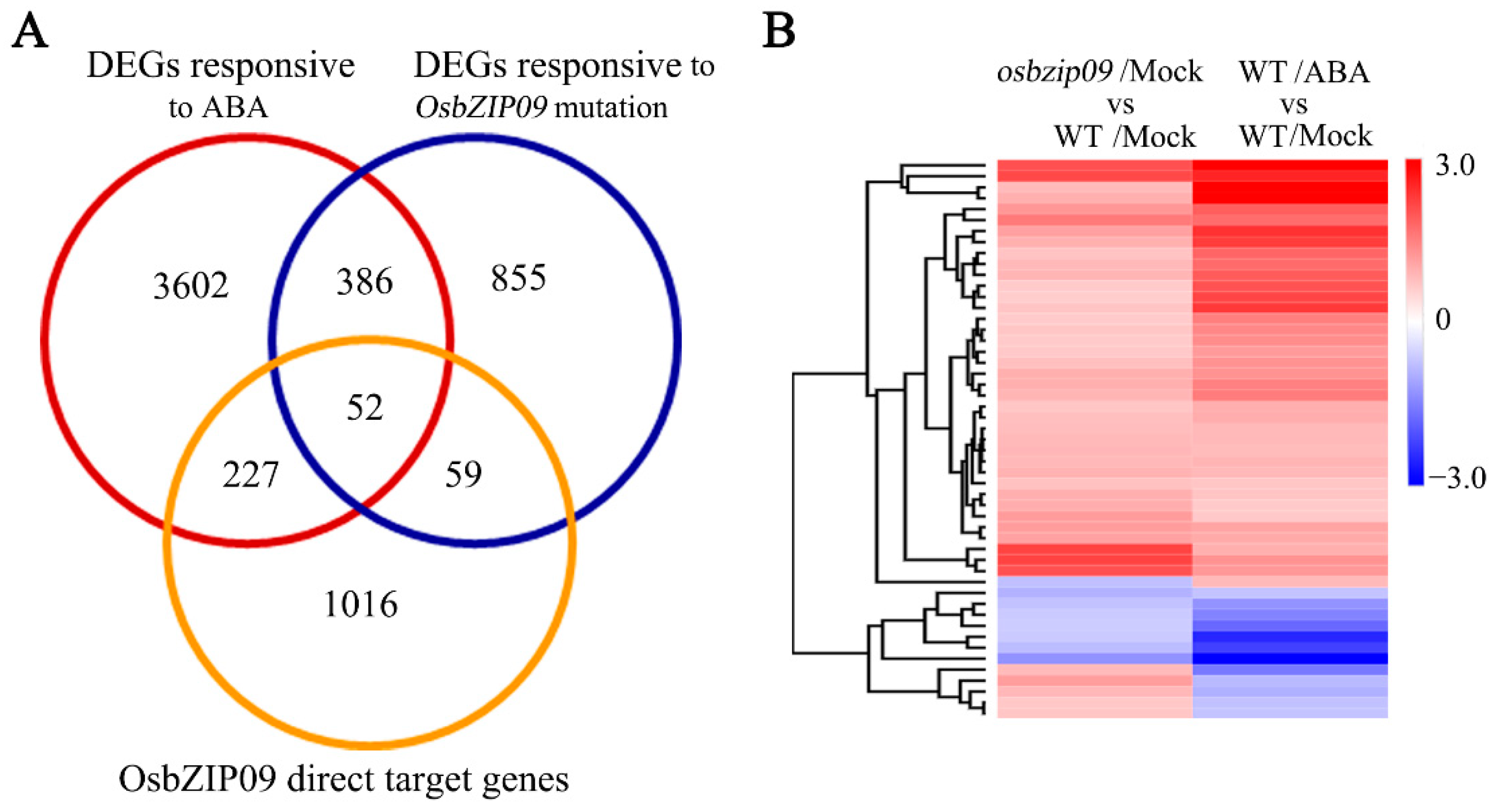
2.4. DAP-Seq Identification of the Genes Directly Targeted by OsbZIP09
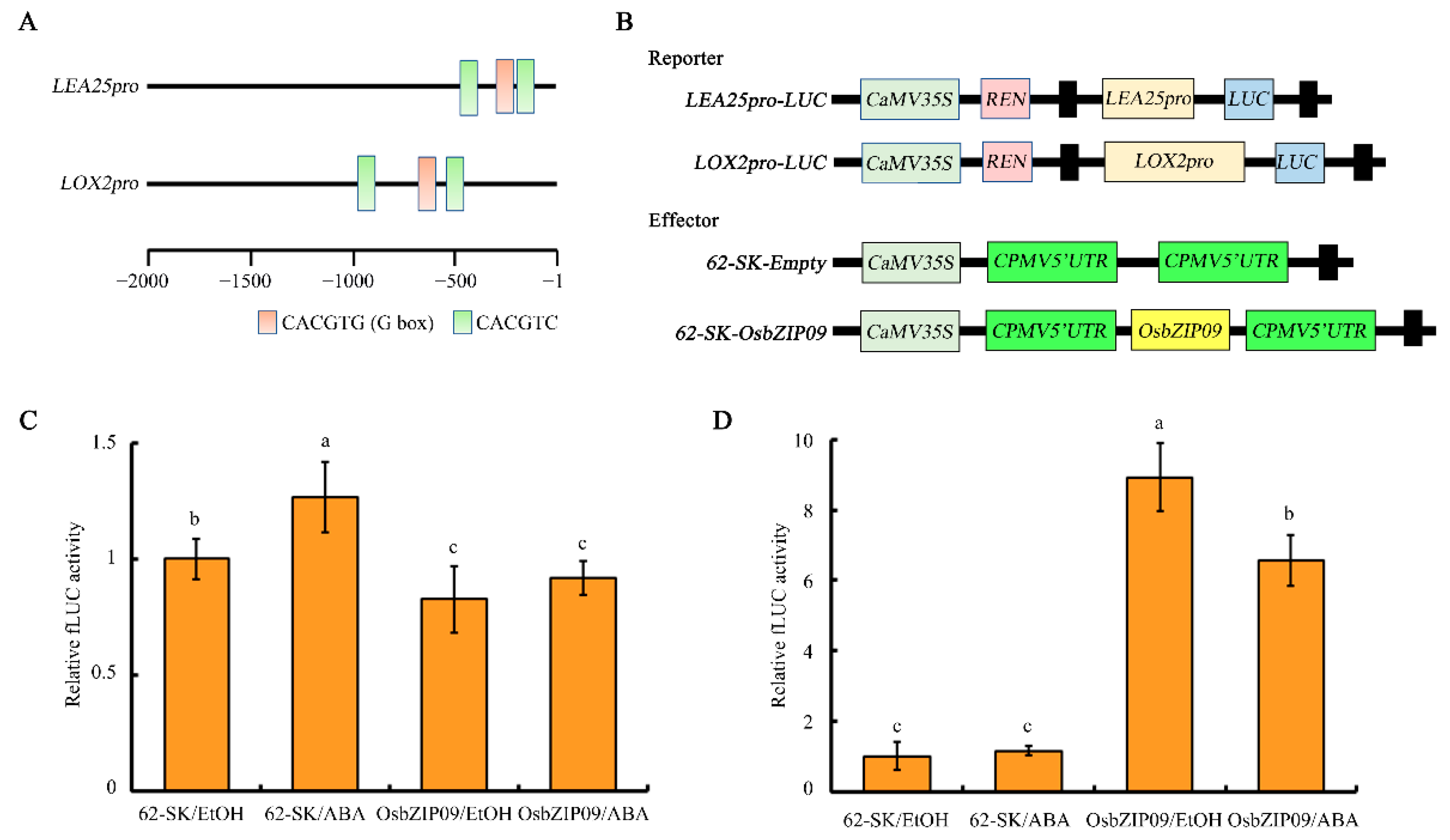
2.5. Identification and Analysis of the Core Direct Target Genes of OsbZIP09
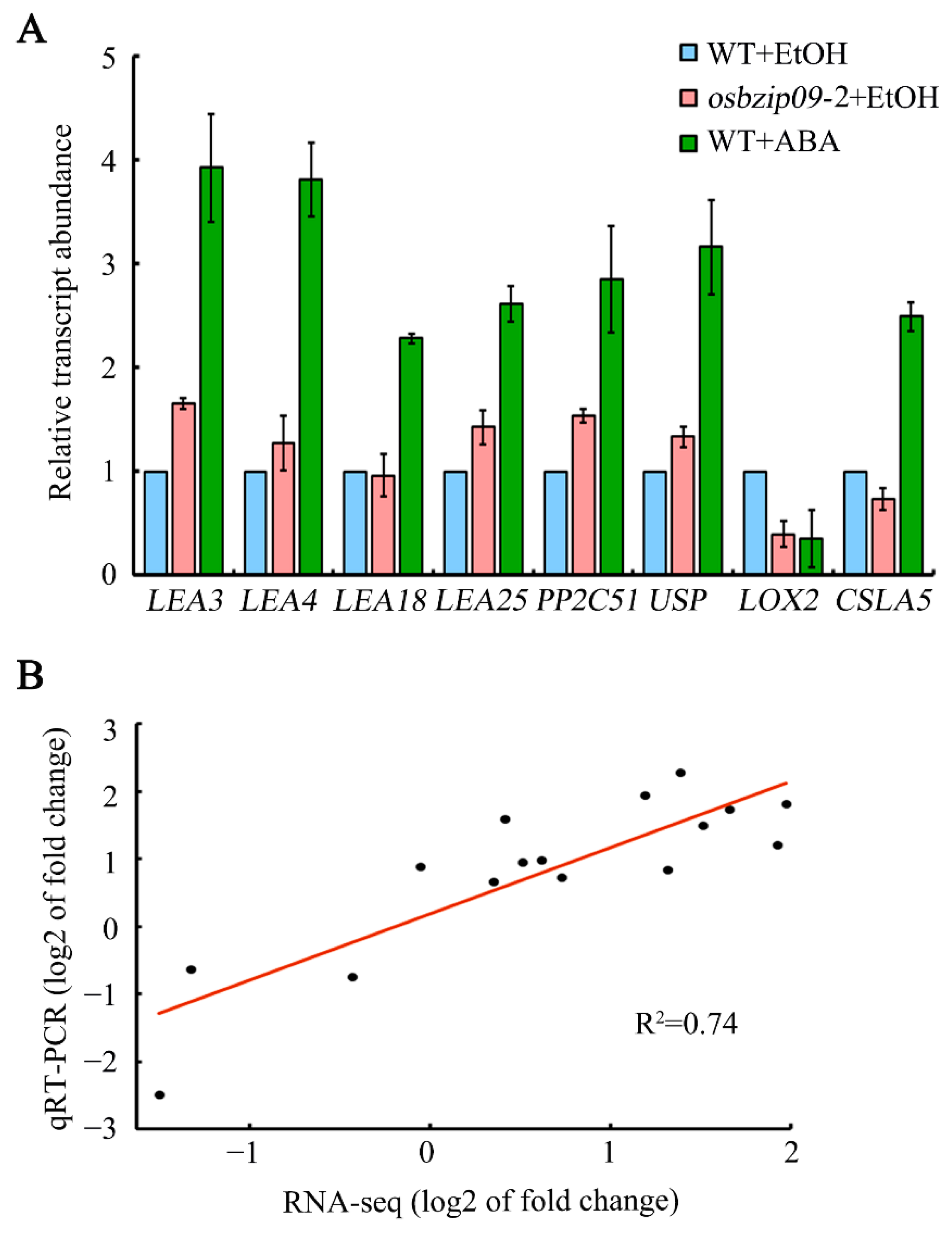
2.6. Expression Analysis and Validation of the Representative Target Genes of OsbZIP09
2.7. Expression Analysis of the Representative Target Genes Regulated by OsbZIP09 and ABA
3. Discussion
4. Materials and Methods
4.1. Generation of Osbzip09 Mutants Using CRISPR/Cas9
4.2. Rice Growth Conditions
4.3. Seed Germination Analysis
4.4. Phylogenetic Assay, Sequence Alignment, and Domain Analysis of bZIP09
4.5. RNA Extraction and qRT-PCR Analysis
4.6. RNA-Seq Analysis
4.7. DNA Affinity Purification Sequencing (DAP-Seq) Sampling
4.8. DAP-Seq Data Analysis
4.9. Promoter Analysis
4.10. Dual-Luciferase Reporter Assay
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ABA | abscisic acid |
| ABI5 | ABA-INSENSITIVE 5 |
| ABFs | ABRE-binding factors |
| ABRE | ABA-responsive element |
| AOC | Allene Oxide Cyclase |
| AREBs | ABA-responsive element binding proteins |
| AWPM-19 | ABA-induced Wheat Plasma Membrane Polypeptide-19 |
| ANOVA | analysis of variance |
| bZIP | basic leucine zipper |
| ChIP-seq | Chromatin immunoprecipitation sequencing |
| CNX | cofactor for nitrate reductase and xanthine dehydrogenase |
| CSLA5 | cellulose synthase-like A5 |
| DAP-seq | DNA affinity purification sequencing |
| DOG1 | DELAY OF GERMINATION 1 |
| DRE | drought-responsive element |
| HAI | hours after imbibition |
| DEGs | differentially expressed genes |
| DLT | DWARF AND LOW-TILLERING |
| GA | gibberellin |
| GAP | GTPase activating protein |
| GO | gene ontology |
| IPA1 | Ideal Plant Architecture 1 |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| LEA | Late Embryogenesis Abundant |
| LOX2 | lipoxygenase 2 |
| MFT2 | MOTHER OF FT AND TFL 2 |
| NCBI | National Center for Biotechnology Information |
| PHS | preharvest sprouting |
| PLA3 | PLASTOCHRON3 |
| PP2Cs | protein phosphatases 2C |
| PUFAs | polyunsaturated fatty acids |
| qRT-PCR | quantitative real-time polymerase chain reaction |
| QTLs | quantitative trait loci |
| RCAR1 | regulatory component of ABA receptor 1 |
| ROS | reactive oxygen species |
| RNA-Seq | RNA sequencing |
| SD | standard deviation |
| SnRK2 | sucrose nonfermenting 1-related protein kinase 2 |
| TF | transcription factor |
| TSS | transcription start site |
| TTS | transcription termination site |
| USP | universal stress protein |
References
- Shu, K.; Liu, X.D.; Xie, Q.; He, Z.H. Two faces of one seed: Hormonal regulation of dormancy and germination. Mol. Plant 2016, 9, 34–45. [Google Scholar] [CrossRef]
- Finkelstein, R.; Reeves, W.; Ariizumi, T.; Steber, C. Molecular aspects of seed dormancy. Annu. Rev. Plant Biol. 2008, 59, 387–415. [Google Scholar] [CrossRef]
- Wang, J.; Deng, Q.; Li, Y.; Yu, Y.; Liu, X.; Han, Y.; Luo, X.; Wu, X.; Ju, L.; Sun, J.; et al. Transcription factors Rc and OsVP 1 coordinately regulate preharvest sprouting tolerance in red pericarp rice. J. Agric. Food Chem. 2020, 68, 14748–14757. [Google Scholar] [CrossRef]
- Suzuki, Y.; Miura, K.; Shigemune, A.; Sasahara, H.; Ohta, H.; Uehara, Y.; Ishikawa, T.; Hamada, S.; Shirasawa, K. Marker-assisted breeding of a LOX-3-null rice line with improved storability and resistance to preharvest sprouting. Theor. Appl. Genet. 2015, 128, 1421–1430. [Google Scholar] [CrossRef]
- Liu, X.; Wang, J.; Yu, Y.; Kong, L.; Liu, Y.; Liu, Z.; Li, H.; Wei, P.; Liu, M.; Zhou, H.; et al. Identification and characterization of the rice pre-harvest sprouting mutants involved in molybdenum cofactor biosynthesis. New Phytol. 2019, 222, 275–285. [Google Scholar] [CrossRef] [PubMed]
- Nonogaki, H. Seed germination and dormancy: The classic story, new puzzles, and evolution. J. Integr. Plant Biol. 2019, 61, 541–563. [Google Scholar] [CrossRef]
- Yan, A.; Chen, Z. The pivotal role of abscisic acid signaling during transition from seed maturation to germination. Plant Cell Rep. 2017, 36, 689–703. [Google Scholar] [CrossRef] [PubMed]
- Née, G.; Xiang, Y.; Soppe, W.J. The release of dormancy, a wake–up call for seeds to germinate. Curr. Opin. Plant Biol. 2017, 35, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Andújar, C.; Ordiz, M.I.; Huang, Z.; Nonogaki, M.; Beachy, R.N.; Nonogaki, H. Induction of 9-cis-epoxycarotenoid dioxygenase in Arabidopsis thaliana seeds enhances seed dormancy. Proc. Natl. Acad. Sci. USA 2011, 108, 17225–17229. [Google Scholar] [CrossRef]
- Nonogaki, M.; Sall, K.; Nambara, E.; Nonogaki, H. Amplification of ABA biosynthesis and signaling through a positive feedback mechanism in seeds. Plant J. 2014, 78, 527–539. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Li, G.J.; Bressan, R.A.; Song, C.P.; Zhu, J.K.; Zhao, Y. Abscisic acid dynamics, signaling, and functions in plants. J. Integr. Plant Biol. 2020, 62, 25–54. [Google Scholar] [CrossRef] [PubMed]
- Miyakawa, T.; Fujita, Y.; Yamaguchi–Shinozaki, K.; Tanokura, M. Structure and function of abscisic acid receptors. Trends Plant Sci. 2013, 18, 259–266. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Nie, K.; Zhou, H.; Yan, X.; Zhan, Q.; Zheng, Y.; Song, C.P. ABI5 modulates seed germination via feedback regulation of the expression of the PYR/PYL/RCAR ABA receptor genes. New Phytol. 2020, 228, 596–608. [Google Scholar] [CrossRef]
- Deppmann, C.D.; Alvania, R.S.; Taparowsky, E.J. Cross-species annotation of basic leucine zipper factor interactions: Insight into the evolution of closed interaction networks. Mol. Biol. Evol. 2006, 23, 1480–1492. [Google Scholar] [CrossRef]
- Corrêa, L.G.G.; Riaño-Pachón, D.M.; Schrago, C.G.; dos Santos, R.V.; Mueller-Roeber, B.; Vincentz, M. The role of bZIP transcription factors in green plant evolution: Adaptive features emerging from four founder genes. PLoS ONE 2008, 3, e2944. [Google Scholar] [CrossRef]
- Dröge-Laser, W.; Snoek, B.L.; Snel, B.; Weiste, C. The Arabidopsis bZIP transcription factor family-an update. Curr. Opin. Plant Biol. 2018, 45, 36–49. [Google Scholar] [CrossRef]
- Nijhawan, A.; Jain, M.; Tyagi, A.K.; Khurana, J.P. Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant Physiol. 2008, 146, 333–350. [Google Scholar] [CrossRef]
- Liu, X.; Chu, Z. Genome-wide evolutionary characterization and analysis of bZIP transcription factors and their expression profiles in response to multiple abiotic stresses in Brachypodium distachyon. BMC Genom. 2015, 16, 227. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Zou, H.F.; Wei, W.; Hao, Y.J.; Tian, A.G.; Huang, J.; Liu, Y.F.; Zhang, J.S.; Chen, S.Y. Soybean GmbZIP44 GmbZIP62 and GmbZIP78 genes function as negative regulator of ABA signaling and confer salt and freezing tolerance in transgenic Arabidopsis. Planta 2008, 228, 225–240. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Xu, D.; Jia, L.; Huang, X.; Ma, G.; Wang, S.; Zhu, M.; Zhang, A.; Guan, M.; Lu, K.; et al. Genome-wide identification and structural analysis of bZIP transcription factor genes in Brassica napus. Genes 2017, 8, 288. [Google Scholar] [CrossRef]
- Johnson, D.S.; Mortazavi, A.; Myers, R.M.; Wold, B. Genome-wide mapping of in vivo protein-DNA interactions. Science 2007, 316, 1497–1502. [Google Scholar] [CrossRef]
- O’Malley, R.C.; Huang, S.C.; Song, L.; Lewsey, M.G.; Bartlett, A.; Nery, J.R.; Galli, M.; Gallavotti, A.; Ecker, J.R. Cistrome and epicistrome features shape the regulatory DNA landscape. Cell 2016, 165, 1280–1292. [Google Scholar] [CrossRef] [PubMed]
- Muro-Villanueva, F.; Mao, X.; Chapple, C. Linking phenylpropanoid metabolism, lignin deposition, and plant growth inhibition. Curr. Opin. Biotechnol. 2019, 56, 202–208. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, T.; Meyer, C.A.; Eeckhoute, J.; Johnson, D.S.; Bernstein, B.E.; Nusbaum, C.; Myers, R.M.; Brown, M.; Li, W.; et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008, 9, R137. [Google Scholar] [CrossRef]
- Li, Q.F.; Zhou, Y.; Xiong, M.; Ren, X.Y.; Han, L.; Wang, J.D.; Zhang, C.Q.; Fan, X.L.; Liu, Q.Q. Gibberellin recovers seed germination in rice with impaired brassinosteroid signalling. Plant Sci. 2020, 293, 110435. [Google Scholar] [CrossRef]
- Huang, J.; Cai, M.; Long, Q.; Liu, L.; Lin, Q.; Jiang, L.; Chen, S.; Wan, J. OsLOX2, a rice type I lipoxygenase, confers opposite effects on seed germination and longevity. Transgenic Res. 2014, 23, 643–655. [Google Scholar] [CrossRef]
- Fang, J.; Chai, C.; Qian, Q.; Li, C.; Tang, J.; Sun, L.; Huang, Z.; Guo, X.; Sun, C.; Liu, M.; et al. Mutations of genes in synthesis of the carotenoid precursors of ABA lead to pre-harvest sprouting and photo-oxidation in rice. Plant J. 2008, 54, 177–189. [Google Scholar] [CrossRef]
- Shu, K.; Meng, Y.J.; Shuai, H.W.; Liu, W.G.; Du, J.B.; Liu, L.; Yang, W.Y. Dormancy and germination: How does the crop seed decide? Plant Biol. 2015, 17, 1104–1112. [Google Scholar] [CrossRef]
- Yang, L.; Liu, S.; Lin, R. The role of light in regulating seed dormancy and germination. J. Integr. Plant Biol. 2020, 62, 1310–1326. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Zhu, L.; Xu, Y.; Zeng, D.; Wu, P.; Qian, Q. QTL analysis of seed dormancy in rice. Euphytica 2004, 140, 155–162. [Google Scholar] [CrossRef]
- Gao, F.Y.; Ren, G.J.; Lu, X.J.; Sun, S.X.; Li, H.J.; Gao, Y.M.; Luo, H.; Yan, W.G.; Zhang, Y.Z. QTL analysis for resistance to preharvest sprouting in rice (Oryza sativa). Plant Breed. 2008, 127, 268–273. [Google Scholar] [CrossRef]
- Ye, H.; Beighley, D.H.; Feng, J.; Gu, X.Y. Genetic and physiological characterization of two clusters of quantitative trait loci associated with seed dormancy and plant height in rice. G3 2013, 3, 323–331. [Google Scholar] [CrossRef]
- Lee, G.A.; Jeon, Y.A.; Lee, H.S.; Hyun, D.Y.; Lee, J.R.; Lee, M.C.; Lee, S.Y.; Ma, K.H.; Koh, H.J. New genetic loci associated with preharvest sprouting and its evaluation based on the model equation in rice. Front. Plant Sci. 2017, 8, 1393. [Google Scholar] [CrossRef] [PubMed]
- Cheon, K.S.; Won, Y.J.; Jeong, Y.M.; Lee, Y.Y.; Kang, D.Y.; Oh, J.; Oh, H.; Kim, S.L.; Kim, N.; Lee, E.; et al. QTL mapping for pre-harvest sprouting resistance in japonica rice varieties utilizing genome re-sequencing. Mol. Genet. Genom. 2020, 295, 1129–1140. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, G.K.; Yamazaki, M.; Kobayashi, M.; Hirochika, R.; Miyao, A.; Hirochika, H. Screening of the rice viviparous mutants generated by endogenous retrotransposon Tos17 insertion. Tagging of a zeaxanthin epoxidase gene and a novel OsTATC gene. Plant Physiol. 2001, 125, 1248–1257. [Google Scholar] [CrossRef]
- Kawakatsu, T.; Taramino, G.; Itoh, J.; Allen, J.; Sato, Y.; Hong, S.K.; Yule, R.; Nagasawa, N.; Kojima, M.; Kusaba, M.; et al. PLASTOCHRON3/GOLIATH encodes a glutamate carboxypeptidase required for proper development in rice. Plant J. 2009, 58, 1028–1040. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Xu, F.; Fang, J.; Gao, S.; Tang, J.; Fang, S.; Wang, H.; Tong, H.; Zhang, F.; Chu, J.; et al. Endosperm sugar accumulation caused by mutation of PHS8/ISA1 leads to pre-harvest sprouting in rice. Plant J. 2018, 95, 545–556. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Tang, J.; Gao, S.; Cheng, X.; Du, L.; Chu, C. Control of rice pre-harvest sprouting by glutaredoxin-mediated abscisic acid signaling. Plant J. 2019, 100, 1036–1051. [Google Scholar] [CrossRef]
- Miao, C.; Wang, Z.; Zhang, L.; Yao, J.; Hua, K.; Liu, X.; Shi, H.; Zhu, J.K. The grain yield modulator miR156 regulates seed dormancy through the gibberellin pathway in rice. Nat. Commun. 2019, 10, 3822. [Google Scholar] [CrossRef]
- Bi, C.; Ma, Y.; Wu, Z.; Yu, Y.T.; Liang, S.; Lu, K.; Wang, X.F. Arabidopsis ABI5 plays a role in regulating ROS homeostasis by activating CATALASE 1 transcription in seed germination. Plant Mol. Biol. 2017, 94, 197–213. [Google Scholar] [CrossRef]
- Bai, M.; Sun, J.; Liu, J.; Ren, H.; Wang, K.; Wang, Y.; Wang, C.; Dehesh, K. The B-box protein BBX19 suppresses seed germination via induction of ABI5. Plant J. 2019, 99, 1192–1202. [Google Scholar] [CrossRef] [PubMed]
- Zou, M.; Guan, Y.; Ren, H.; Zhang, F.; Chen, F. A bZIP transcription factor, OsABI5, is involved in rice fertility and stress tolerance. Plant Mol. Biol. 2008, 66, 675–683. [Google Scholar] [CrossRef] [PubMed]
- Ju, L.; Jing, Y.; Shi, P.; Liu, J.; Chen, J.; Yan, J.; Chu, J.; Chen, K.M.; Sun, J. JAZ proteins modulate seed germination through interaction with ABI5 in bread wheat and Arabidopsis. New Phytol. 2019, 223, 246–260. [Google Scholar] [CrossRef]
- Utsugi, S.; Ashikawa, I.; Nakamura, S.; Shibasaka, M. TaABI5, a wheat homolog of Arabidopsis thaliana ABA insensitive 5, controls seed germination. J. Plant Res. 2020, 133, 245–256. [Google Scholar] [CrossRef] [PubMed]
- Collin, A.; Daszkowska-Golec, A.; Kurowska, M.; Szarejko, I. Barley ABI5 (Abscisic Acid INSENSITIVE 5) is involved in abscisic acid-dependent drought response. Front. Plant Sci. 2020, 11, 1138. [Google Scholar] [CrossRef] [PubMed]
- Cantoro, R.; Crocco, C.D.; Benech-Arnold, R.L.; Rodríguez, M.V. In vitro binding of Sorghum bicolor transcription factors ABI4 and ABI5 to a conserved region of a GA 2-OXIDASE promoter: Possible role of this interaction in the expression of seed dormancy. J. Exp. Bot. 2013, 64, 5721–5735. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.A.; Cho, J.I.; Han, M.; Ahn, C.H.; Jeon, J.S.; An, G.; Park, P.B. The ABRE–binding bZIP transcription factor OsABF2 is a positive regulator of abiotic stress and ABA signaling in rice. J. Plant. Physiol. 2010, 167, 1512–1520. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Wang, G.; Wu, H.; Fan, X.; Liang, L.; Zhao, H.; Li, S.; Hu, Y.; Liu, H.; Ayaad, M.; et al. OsMFT2 is involved in the regulation of ABA signaling mediated seed germination through interacting with OsbZIP23/66/72 in rice. Plant J. 2020, 103, 532–546. [Google Scholar] [CrossRef]
- Wang, Y.; Hou, Y.; Qiu, J.; Wang, H.; Wang, S.; Tang, L.; Tong, X.; Zhang, J. Abscisic acid promotes jasmonic acid biosynthesis via a ‘SAPK10–bZIP72–AOC’ pathway to synergistically inhibit seed germination in rice (Oryza sativa). New Phytol. 2020, 228, 1336–1353. [Google Scholar] [CrossRef]
- Wang, Q.; Lin, Q.; Wu, T.; Duan, E.; Huang, Y.; Yang, C.; Mou, C.; Lan, J.; Zhou, C.; Xie, K.; et al. OsDOG1L–3 regulates seed dormancy through the abscisic acid pathway in rice. Plant Sci. 2020, 298, 110570. [Google Scholar] [CrossRef]
- Wise, M.J.; Tunnacliffe, A. POPP the question: What do LEA proteins do? Trends Plant Sci. 2004, 9, 13–17. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Li, X. Identification and phylogenetic analysis of late embryogenesis abundant proteins family in tomato (Solanum lycopersicum). Planta 2015, 241, 757–772. [Google Scholar] [CrossRef]
- Liu, Y.; Song, Q.; Li, D.; Yang, X.; Li, D. Multifunctional roles of plant dehydrins in response to environmental stresses. Front. Plant Sci. 2017, 8, 1018. [Google Scholar]
- Yang, W.; Zhang, L.; Lv, H.; Li, H.; Zhang, Y.; Xu, Y.; Yu, J. The K-segments of wheat dehydrin WZY2 are essential for its protective functions under temperature stress. Front. Plant Sci. 2015, 6, 406. [Google Scholar] [CrossRef]
- Hara, M.; Monna, S.; Murata, T.; Nakano, T.; Amano, S.; Nachbar, M.; Wätzig, H. The Arabidopsis KS-type dehydrin recovers lactate dehydrogenase activity inhibited by copper with the contribution of his residues. Plant Sci. 2016, 245, 135–142. [Google Scholar] [CrossRef]
- Wang, X.S.; Zhu, H.B.; Jin, G.L.; Liu, H.L.; Wu, W.R.; Zhu, J. Genome-scale identification and analysis of LEA genes in rice (Oryza sativa L.). Plant Sci. 2007, 172, 414–420. [Google Scholar] [CrossRef]
- Xiao, B.; Huang, Y.; Tang, N.; Xiong, L. Over-expression of a LEA gene in rice improves drought resistance under the field conditions. Theor. Appl. Genet. 2007, 115, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Zhang, M.; Jia, J.; Zhao, X.; Huang, X.; Ji, E.; Ni, L.; Jiang, M. An atypical late embryogenesis abundant protein OsLEA5 plays a positive role in ABA-induced antioxidant defense in oryza sativa L. Plant Cell Physiol. 2018, 59, 916–929. [Google Scholar] [CrossRef]
- Hu, T.; Liu, Y.; Zhu, S.; Qin, J.; Li, W.; Zhou, N. Overexpression of OsLea14-A improves the tolerance of rice and increases Hg accumulation under diverse stresses. Environ. Sci. Pollut. Res. Int. 2019, 26, 10537–10551. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Jia, J.; Zhao, X.; Zhang, M.; Huang, X.; Ji, E.; Ni, L.; Jiang, M. The ascorbate peroxidase APX1 is a direct target of a zinc finger transcription factor ZFP36 and a late embryogenesis abundant protein OsLEA5 interacts with ZFP36 to co-regulate OsAPX1 in seed germination in rice. Biochem. Biophys. Res. Commun. 2018, 495, 339–345. [Google Scholar] [CrossRef]
- Koike, M.; Takezawa, D.; Arakawa, K.; Yoshida, S. Accumulation of 19-kDa plasma membrane polypeptide during induction of freezing tolerance in wheat suspensioncultured cells by abscisic acid. Plant Cell Physiol. 1997, 38, 707–716. [Google Scholar] [CrossRef]
- Ranford, J.C.; Bryce, J.H.; Morris, P.C. PM19, a barley (Hordeum vulgare L.) gene encoding a putative plasma membrane protein, is expressed during embryo development and dormancy. J. Exp. Bot. 2002, 53, 147–148. [Google Scholar]
- Chen, H.; Lan, H.; Huang, P.; Zhang, Y.; Yuan, X.; Huang, X.; Huang, J.; Zhang, H. Characterization of OsPM19L1 encoding an AWPM-19-like family protein that is dramatically induced by osmotic stress in rice. Genet. Mol. Res. 2015, 14, 11994–12005. [Google Scholar] [CrossRef]
- Barrero, J.M.; Cavanagh, C.; Verbyla, K.L.; Tibbits, J.F.; Verbyla, A.P.; Huang, B.E.; Rosewarne, G.M.; Stephen, S.; Wang, P.; Whan, A.; et al. Transcriptomic analysis of wheat nearisogenic lines identifies PM19-A1 and A2 as candidates for a major dormancy QTL. Genome Biol. 2015, 16, 93. [Google Scholar] [CrossRef] [PubMed]
- Yao, L.; Cheng, X.; Gu, Z.; Huang, W.; Li, S.; Wang, L.; Wang, Y.F.; Xu, P.; Ma, H.; Ge, X. The AWPM-19 family protein OsPM1 mediates abscisic acid influx and drought response in rice. Plant Cell 2018, 30, 1258–1276. [Google Scholar] [CrossRef] [PubMed]
- Tong, H.; Liu, L.; Jin, Y.; Du, L.; Yin, Y.; Qian, Q.; Zhu, L.; Chu, C. Dwarf and low-tillering acts as a direct downstream target of a GSK3/SHAGGY-like kinase to mediate brassinosteroid responses in rice. Plant Cell 2012, 24, 2562–2577. [Google Scholar] [CrossRef] [PubMed]
- Hirano, K.; Yoshida, H.; Aya, K.; Kawamura, M.; Hayashi, M.; Hobo, T.; Sato-Izawa, K.; Kitano, H.; Ueguchi-Tanaka, M.; Matsuoka, M. Small organ size 1 and small organ size 2/dwarf and low-tillering form a complex to integrate auxin and brassinosteroid signaling in rice. Mol. Plant 2017, 10, 590–604. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Liu, Q.; Shen, Y.; Hua, Y.F.; Wang, J.J.; Lin, J.R.; Wu, M.G.; Sun, T.T.; Cheng, Z.K.; Mercier, R.; et al. Clonal seeds from hybrid rice by simultaneous genome engineering of meiosis and fertilization genes. Nat. Biotechnol. 2019, 37, 283–286. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.F.; Xiong, M.; Xu, P.; Huang, L.C.; Zhang, C.Q.; Liu, Q.Q. Dissection of brassinosteroid-regulated proteins in rice embryos during germination by quantitative proteomics. Sci. Rep. 2016, 6, 34583. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.Y.; Wu, Y.; Guo, J.P.; Du, B.; Chen, R.Z.; Zhu, L.L.; He, G.C. A rice lectin receptor-like kinase that is involved in innate immune responses also contributes to seed germination. Plant J. 2013, 76, 687–698. [Google Scholar] [CrossRef]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Bartlett, A.; O’Malley, R.C.; Huang, S.C.; Galli, M.; Nery, J.R.; Gallavotti, A.; Ecker, J.R. Mapping genome-wide transcription-factor binding sites using DAP-seq. Nat. Protoc. 2017, 12, 1659–1672. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Machanick, P.; Bailey, T.L. MEME-ChIP: Motif analysis of large DNA datasets. Bioinformatics 2011, 27, 1696–1697. [Google Scholar] [CrossRef]
- Hellens, R.P.; Allan, A.C.; Friel, E.N.; Bolitho, K.; Grafton, K.; Templeton, M.D.; Karunairetnam, S.; Gleave, A.P.; Laing, W.A. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 2005, 1, 13. [Google Scholar] [CrossRef]
- Li, Q.F.; He, J.X. BZR1 interacts with HY5 to mediate brassinosteroid- and light-regulated cotyledon opening in Arabidopsis in darkness. Mol. Plant 2016, 9, 113–125. [Google Scholar] [CrossRef]

| Gene ID | Transcriptome Analysis Log2(Fold Change) 1 | DAP-Seq Analysis | Description | ||
|---|---|---|---|---|---|
| osbzip09-2/WT | ABA/Mock (WT) | −Log10 (p Value) | Distance to TSS (bp) 2 | ||
| Os10g0173000 | 2.05 | 3.94 | 14.30 | 1616 | expressed protein |
| Os08g0529300 | 2.13 | 2.52 | 11.59 | 95 | F-box protein 463 |
| Os11g0154900 | 0.95 | 1.30 | 17.17 | 114 | OsZIP-2a; OsbZIP80 |
| Os03g0386000 | 0.82 | 3.04 | 12.68 | 1374 | SRWD5 |
| Os03g0820500 | 1.18 | 1.09 | 9.65 | 373 | OsADF3 |
| Os02g0716800 | 2.20 | 0.96 | 6.91 | 1543 | PTR2 |
| Os05g0542500 | 0.68 | 1.97 | 12.53 | 552 | OsLEA3 |
| Os01g0702500 | 0.68 | 1.42 | 10.54 | 3 | OsLEA22 |
| Os05g0563900 | 0.87 | 0.81 | 10.25 | 1498 | OsGRX17 |
| Os09g0325700 | 0.63 | 1.53 | 11.80 | 37 | OsPP108; SIPP2C1 |
| Os02g0740500 | 0.85 | 0.90 | 9.65 | 1299 | expressed protein |
| Os04g0617050 | 0.79 | 0.84 | 9.20 | 110 | OsSAUR21 |
| Os12g0478200 | 1.12 | 1.02 | 7.73 | 1383 | GRAM protein |
| Os05g0198400 | 0.97 | 0.71 | 11.26 | 52 | OsZIP7a |
| Os10g0524300 | 1.24 | 1.89 | 7.66 | 1330 | EMBRYO SAC 1 |
| Os09g0484800 | 0.84 | 0.85 | 9.25 | 101 | pirin, putative |
| Os10g0159033 | 2.20 | 1.29 | 12.19 | 135 | expressed protein |
| Os03g0723400 | 0.64 | 2.04 | 9.57 | 6 | expressed protein |
| Os10g0542100 | 1.13 | 2.38 | 8.48 | 2 | OsMT-II-1a |
| Os06g0110200 | 0.66 | 1.20 | 8.45 | 62 | OsLEA4 |
| Os04g0385600 | 0.77 | 0.99 | 10.06 | 522 | OsFBO3 |
| Os09g0127700 | 0.64 | 1.36 | 9.25 | 18 | expressed protein |
| Os03g0170900 | 0.99 | 0.63 | 10.24 | 1082 | OsSUT1 |
| Os11g0454200 | 0.95 | 3.05 | 8.23 | 97 | OsLEA28 |
| Os11g0451700 | 0.95 | 2.27 | 8.77 | 118 | OsLEA25, Rab17 |
| Os08g0150700 | 0.93 | 0.85 | 9.88 | 881 | OsFbox407 |
| Os05g0572700 | 0.97 | 1.49 | 12.68 | 745 | OsPP2C51 |
| Os05g0524100 | 0.77 | 1.30 | 7.79 | 234 | expressed protein |
| Os07g0422100 | 0.61 | 2.17 | 6.17 | 247 | OsPM19L2 |
| Os06g0697200 | 0.74 | 0.68 | 7.53 | 1321 | OsRH35B |
| Os10g0177200 | 0.71 | 2.29 | 12.03 | 856 | OsDSR-1 |
| Os04g0610600 | 0.88 | 1.93 | 5.95 | 12 | OsLEA18 |
| Os01g0135700 | 0.68 | 0.98 | 8.55 | 155 | OsCML16 |
| Os02g0718600 | 0.88 | 1.55 | 13.89 | 388 | expressed protein |
| Os01g0849600 | 1.58 | 1.73 | 6.90 | 70 | USP protein |
| Os09g0502500 | 1.19 | 0.66 | 8.12 | 162 | alcohol dehydrogenase |
| Os12g0510750 | 0.85 | 1.74 | 8.45 | 162 | expressed protein |
| Os06g0261300 | 2.05 | 1.22 | 4.79 | 486 | expressed protein |
| Gene ID | Transcriptome Analysis Log2(Fold Change) 1 | DAP-Seq Analysis | Description | ||
|---|---|---|---|---|---|
| osbzip09-2/WT | ABA/Mock(WT) | −Log10 (p Value) | Distance To TSS 2 | ||
| Os06g0127800 | −0.58 | −1.84 | 24.06 | 220 | DLT; OsGRAS-32 |
| Os04g0678700 | −0.63 | −1.47 | 15.45 | 111 | OsPORA |
| Os06g0112100 | −1.30 | −3.33 | 15.80 | 673 | Nucleoside phosphorylase |
| Os02g0769200 | −0.73 | −1.28 | 7.64 | 435 | LYP5, Os-LYP5 |
| Os10g0104900 | −0.80 | −2.22 | 7.29 | 701 | OsCMT3a |
| Os03g0306800 | −0.93 | −0.72 | 6.91 | 329 | Calvin-cycle protein CP12 |
| Os05g0406800 | −0.61 | −1.78 | 5.47 | 141 | receptor-like protein kinase |
| Os03g0738600 | −0.64 | −2.49 | 6.91 | 209 | OsLOX2 |
| Gene ID | Transcriptome Analysis Log2(Fold Change)1 | DAP-Seq Analysis | Description | ||
|---|---|---|---|---|---|
| osbzip09-2/WT | ABA/Mock (WT) | −Log10(p Value) | Distance To TSS (bp) 2 | ||
| Os08g0278900 | 0.67 | −0.74 | 12.61 | 80 | MIR domain protein |
| Os03g0835150 | 0.82 | −1.59 | 7.48 | 568 | expressed protein |
| Os02g0226200 | 1.17 | −0.83 | 8.95 | 260 | IB hydrolase |
| Os04g0678400 | 0.86 | −0.94 | 6.91 | 1566 | OsDof-17; OsDof18 |
| Os03g0377700 | −0.75 | 0.84 | 8.07 | 1375 | CSLA5-cellulose synthase |
| Os02g0115900 | 0.70 | −0.72 | 5.95 | 22 | OsBip1; BiP3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, C.-C.; Wang, C.-X.; Lu, C.-Y.; Wang, J.-D.; Zhou, Y.; Xiong, M.; Zhang, C.-Q.; Liu, Q.-Q.; Li, Q.-F. Genome-Wide Identification and Expression Analysis of OsbZIP09 Target Genes in Rice Reveal Its Mechanism of Controlling Seed Germination. Int. J. Mol. Sci. 2021, 22, 1661. https://doi.org/10.3390/ijms22041661
Zhu C-C, Wang C-X, Lu C-Y, Wang J-D, Zhou Y, Xiong M, Zhang C-Q, Liu Q-Q, Li Q-F. Genome-Wide Identification and Expression Analysis of OsbZIP09 Target Genes in Rice Reveal Its Mechanism of Controlling Seed Germination. International Journal of Molecular Sciences. 2021; 22(4):1661. https://doi.org/10.3390/ijms22041661
Chicago/Turabian StyleZhu, Cheng-Chao, Chu-Xin Wang, Chen-Ya Lu, Jin-Dong Wang, Yu Zhou, Min Xiong, Chang-Quan Zhang, Qiao-Quan Liu, and Qian-Feng Li. 2021. "Genome-Wide Identification and Expression Analysis of OsbZIP09 Target Genes in Rice Reveal Its Mechanism of Controlling Seed Germination" International Journal of Molecular Sciences 22, no. 4: 1661. https://doi.org/10.3390/ijms22041661
APA StyleZhu, C.-C., Wang, C.-X., Lu, C.-Y., Wang, J.-D., Zhou, Y., Xiong, M., Zhang, C.-Q., Liu, Q.-Q., & Li, Q.-F. (2021). Genome-Wide Identification and Expression Analysis of OsbZIP09 Target Genes in Rice Reveal Its Mechanism of Controlling Seed Germination. International Journal of Molecular Sciences, 22(4), 1661. https://doi.org/10.3390/ijms22041661






