Genome-Wide Identification of the B-BOX Genes that Respond to Multiple Ripening Related Signals in Sweet Cherry Fruit
Abstract
1. Introduction
2. Results
2.1. Identification of PavBBX Genes in Prunus avium
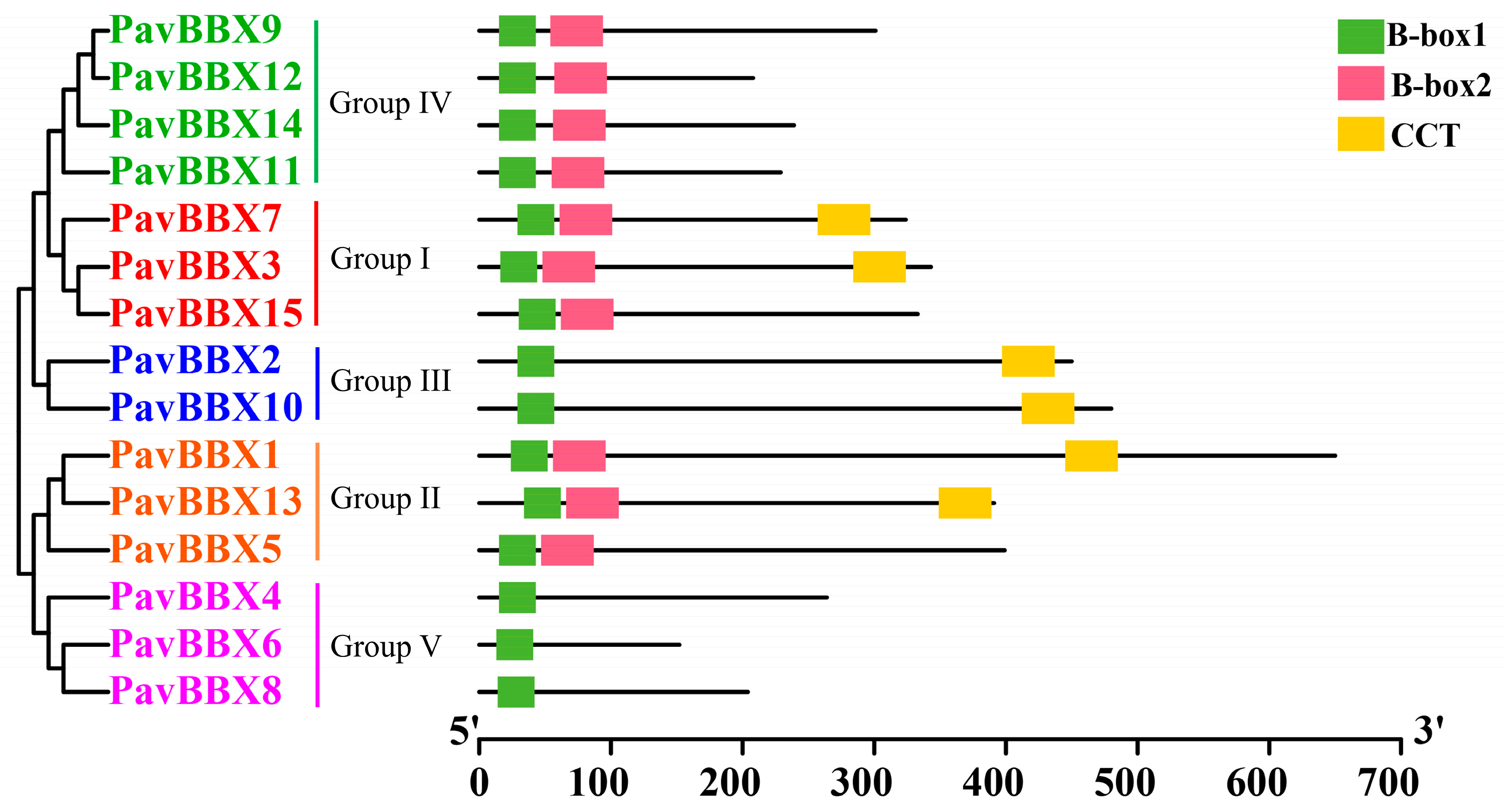
2.2. Protein Sequence Alignment and Phylogenetic Analysis of the PavBBX Gene Family
2.3. Cis-Elements in the Promoters of Sweet Cherry BBX Genes
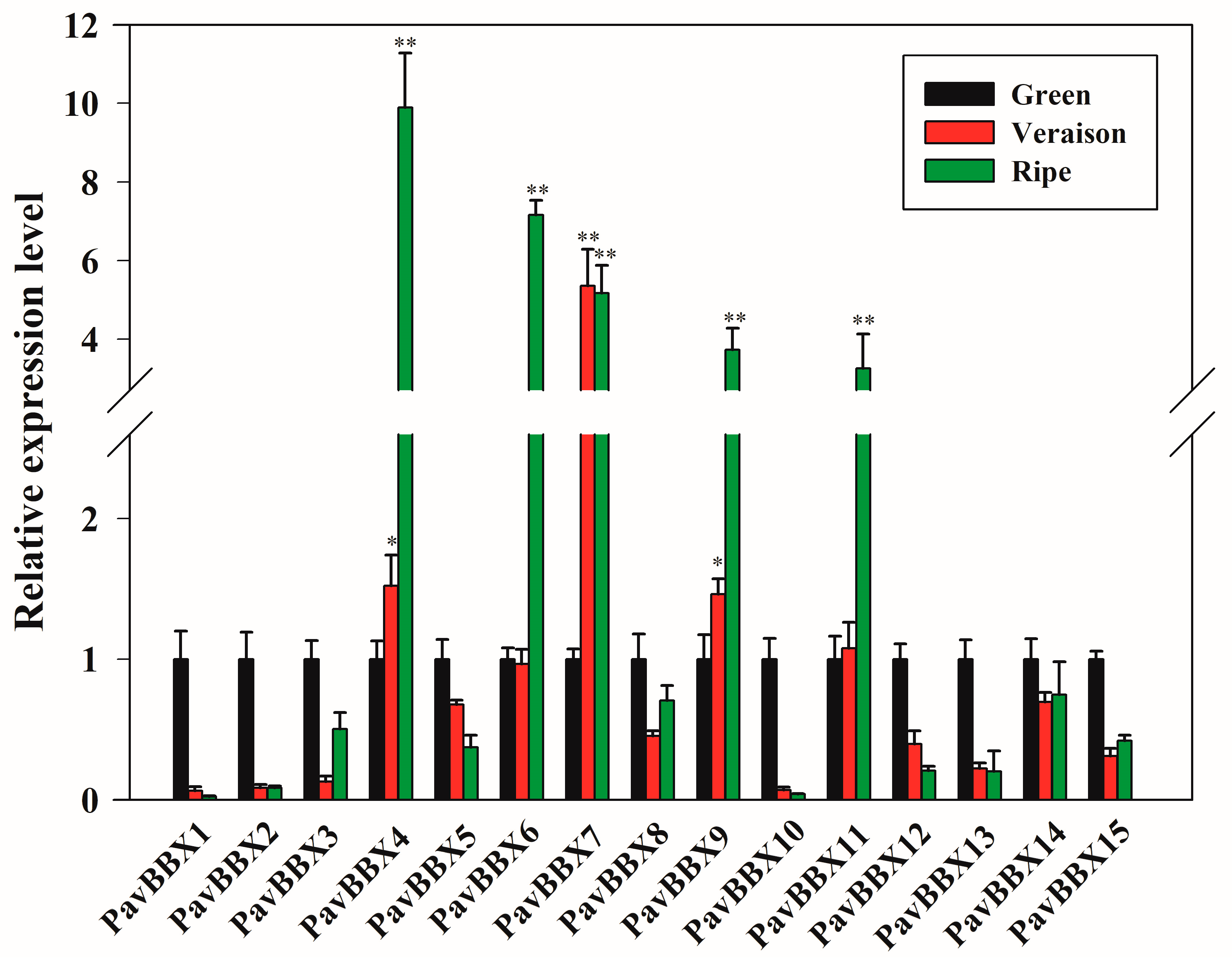
2.4. Expression Patterns of PavBBX Genes during Sweet Cherry Fruit Development and Ripening
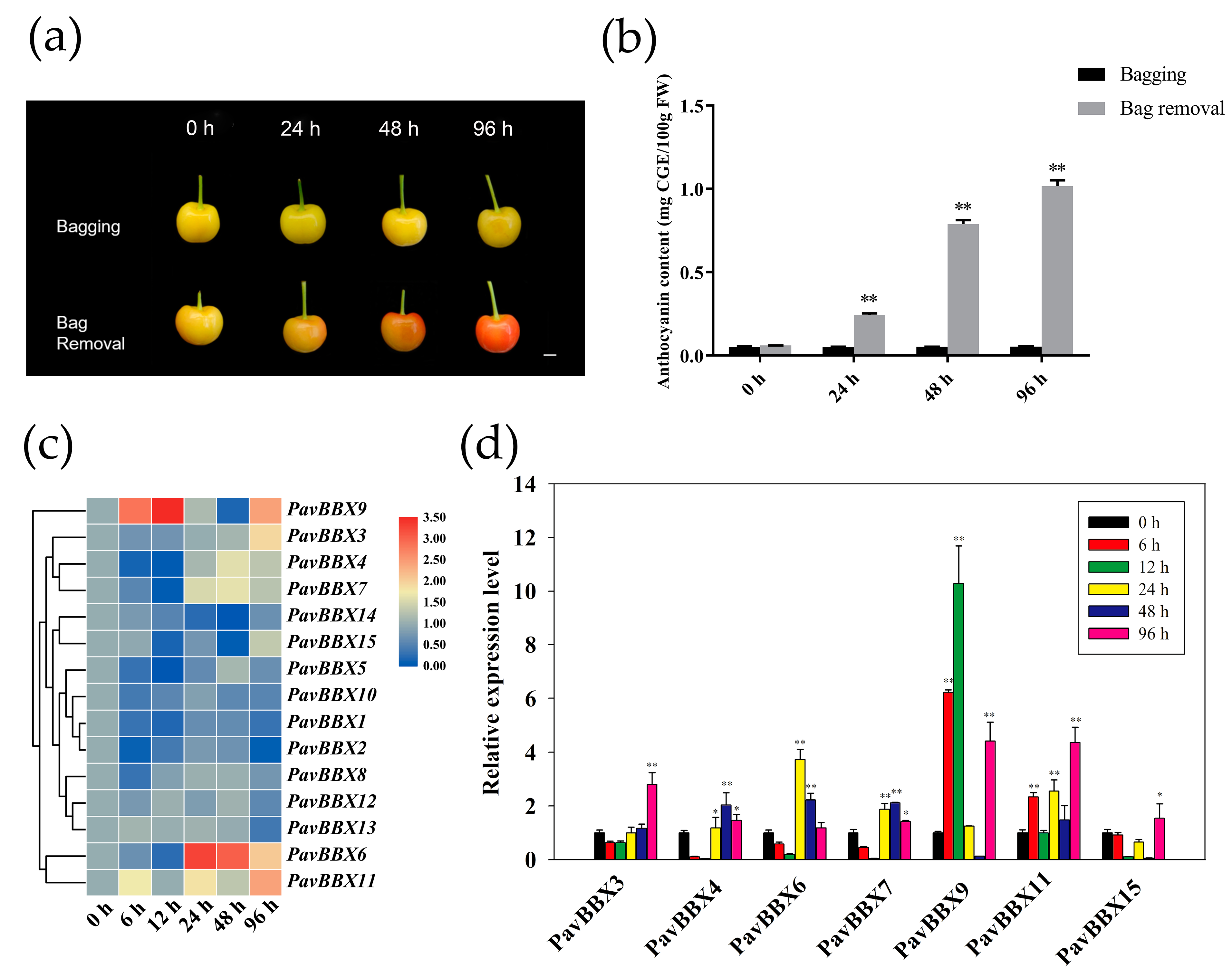
2.5. Regulation of PavBBX Gene Expression during Light Induction
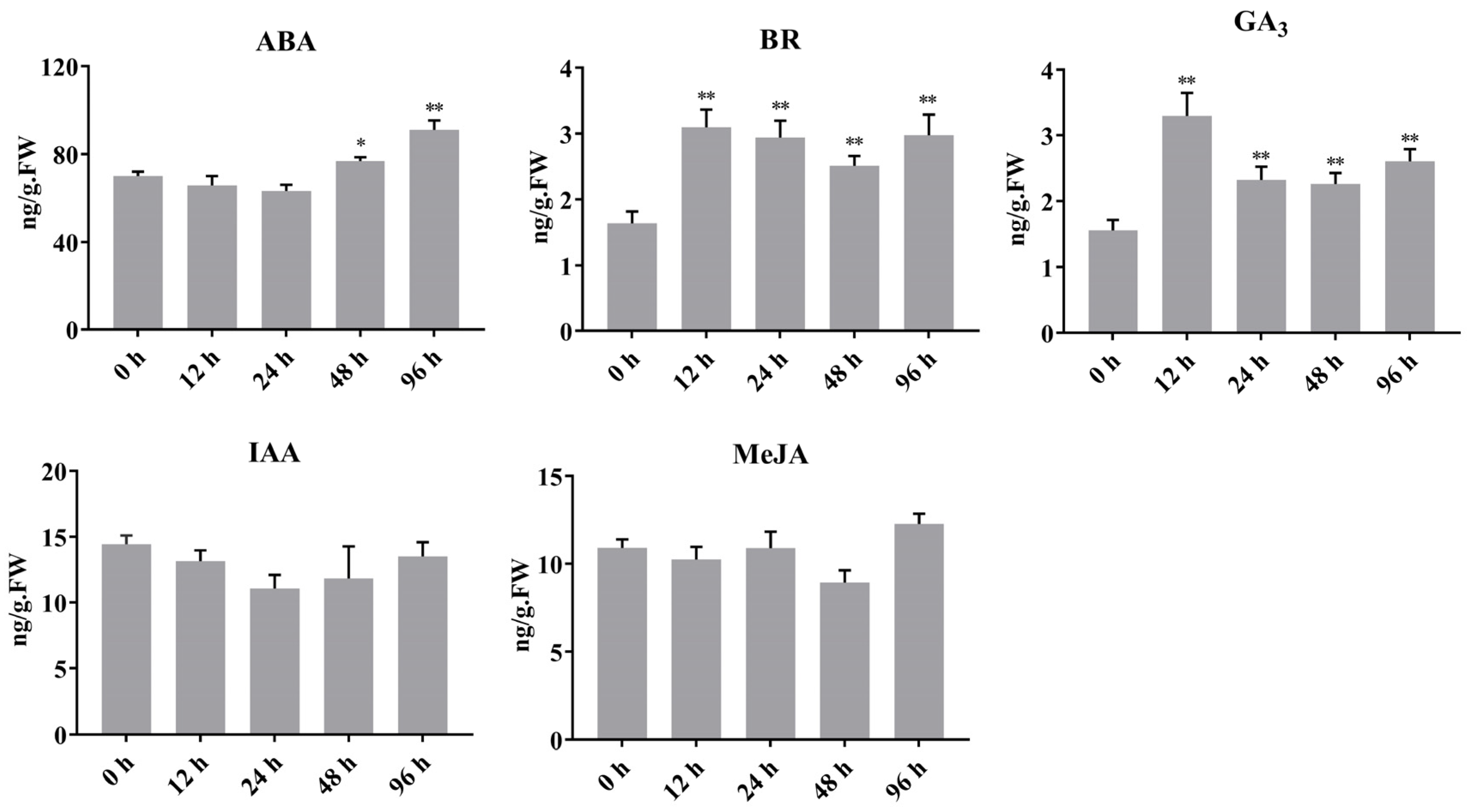
2.6. Light-Induced Regulation of Hormone Content in Sweet Cherry Fruit
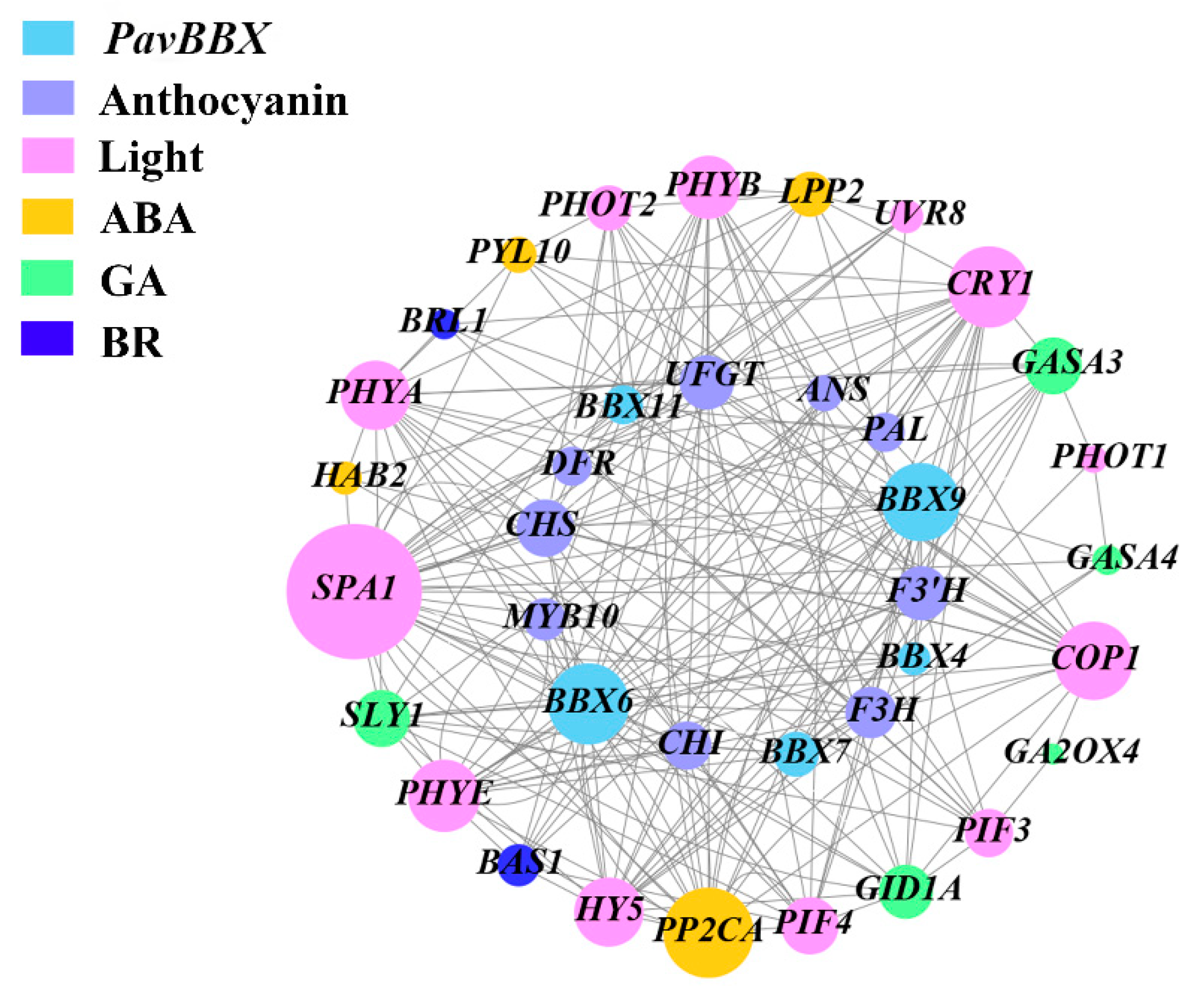
2.7. Co-Expression Network Analysis of PavBBX Genes with Anthocyanin Biosynthesis Genes, Light Signaling Genes, and Hormone Signaling Genes
2.8. Regulation of PavBBX Family Gene Expression during Hormone Treatment
2.9. Subcellular Localization of Sweet Cherry BBX Proteins
3. Discussion
3.1. Evolutionary Analysis of Sweet Cherry BBX Genes
3.2. The Expression Patterns of PavBBX Genes in Sweet Cherry Fruit Development and Ripening
3.3. The Expression Patterns of PavBBX Genes in Plant Responses to Light and Multiple Hormones
4. Materials and Methods
4.1. Plant Growth Conditions and Hormone Treatments
4.2. Quantification of Total Anthocyanin Content
4.3. Identification of BBX Genes in the Prunus avium Genome
4.4. Phylogenetic Analysis and Gene Structure Analysis
4.5. Cis-Element Prediction in the BBX Gene Promoters
4.6. qRT-PCR Analysis
4.7. Measuring ABA, IAA, MeJA, GA3, and BR Contents
4.8. Co-Expression Network Construction and Visualization
4.9. Subcellular Localization Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BBX | B-box |
| ABA | Abscisic acid |
| MeJA | Methyl jasmonate |
| BR | Brassinolide |
| GA | Gibberellic acid |
| IAA | Indole-3-acetic acid |
| CCT | CO, CO-like, TOC1 |
| DAF | Days after flowering |
| AA | amino acid residues |
| MW | molecular weight |
| pI | theoretical isoelectric point |
| GRAVY | grand average of hydropathicity |
| MS | Murashige and Skoog Medium |
| 6BA | N-(Phenylmethyl)-9H-purin-6-amine |
| 2,4-D | 2,4-Dichlorophenoxyacetic acid |
| ELISA | Enzyme-linked immunosorbent assay |
| FPKM | Fragments per kilobase of exon per million mapped fragments |
| ORF | Open reading frame |
| GFP | Green fluorescent protein |
| MIQE | Minimum information stablished for qRT-PCR experiments |
References
- Riechmann, J.L.; Heard, J.; Martin, G.; Reuber, L.; Jiang, C.Z.; Keddie, J.; Adam, L.; Pineda, O.; Ratcliffe, O.J.; Samaha, R.R.; et al. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 2000, 290, 2105–2110. [Google Scholar] [CrossRef]
- Takatsuji, H. Zinc-finger transcription factors in plants. Cell Mol. Life Sci. 1998, 54, 582–596. [Google Scholar] [CrossRef]
- Xu, D.Q.; Jiang, Y.; Li, J.G.; Lin, F.; Holm, M.; Deng, X.W. BBX21, an Arabidopsis B-box protein, directly activates HY5 and is targeted by COP1 for 26S proteasome-mediated degradation. Proc. Natl. Acad. Sci. USA 2016, 113, 7655–7660. [Google Scholar] [CrossRef]
- Wei, H.R.; Wang, P.P.; Chen, J.Q.; Li, C.J.; Wang, Y.Z.; Yuan, Y.B.; Fang, J.G.; Leng, X.P. Genome-wide identification and analysis of B-BOX gene family in grapevine reveal its potential functions in berry development. BMC Plant Biol. 2020, 20, 72. [Google Scholar] [CrossRef] [PubMed]
- Khanna, R.; Kronmiller, B.; Maszle, D.R.; Coupland, G.; Holm, M.; Mizuno, T.; Wu, S.H. The Arabidopsis B-box zinc finger family. Plant Cell 2009, 21, 3416–3420. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, S.; Dunford, R.P.; Coupland, G.; Laurie, D.A. The evolution of constans-like gene families in barley, rice, and Arabidopsis. Plant Physiol. 2003, 131, 1855–1867. [Google Scholar] [CrossRef]
- Gangappa, S.N.; Botto, J.F. The BBX family of plant transcription factors. Trends Plant Sci. 2014, 19, 460–470. [Google Scholar] [CrossRef]
- Putterill, J.; Robson, F.; Lee, K.; Simon, R.; Coupland, G. The constans gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc-finger transcription factors. Cell 1995, 80, 847–857. [Google Scholar] [CrossRef]
- Robson, F.; Costa, M.M.R.; Hepworth, S.R.; Vizir, I.; Pineiro, M.; Reeves, P.H.; Putterill, J.; Coupland, G. Functional importance of conserved domains in the flowering-time gene constans demonstrated by analysis of mutant alleles and transgenic plants. Plant J. 2001, 28, 619–631. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.J.; Ma, C.; Xu, Y.J.; Wei, Q.; Imtiaz, M.; Lan, H.B.; Gao, S.; Cheng, L.N.; Wang, M.Y.; Fei, Z.J.; et al. A zinc finger protein regulates flowering time and abiotic stress tolerance in chrysanthemum by modulating gibberellin biosynthesis. Plant Cell 2014, 26, 2038–2054. [Google Scholar] [CrossRef]
- Datta, S.; Hettiarachchi, G.H.C.M.; Deng, X.W.; Holm, M. Arabidopsis constans-Like3 is a positive regulator of red light signaling and root growth. Plant Cell 2006, 18, 70–84. [Google Scholar] [CrossRef]
- Cheng, X.F.; Wang, Z.Y. Overexpression of COL9, a CONSTANS-LIKE gene, delays flowering by reducing expression of CO and FT in A. thaliana. Plant J. 2005, 43, 758–768. [Google Scholar] [CrossRef]
- Zhao, X.; Heng, Y.; Wang, X.; Deng, X.W.; Xu, D. A positive feedback loop of BBX11-BBX21-HY5 promotes photomorphogenic development in Arabidopsis. Plant Commun. 2020, 1, 100045. [Google Scholar] [CrossRef]
- Chang, C.S.; Li, Y.H.; Chen, L.T.; Chen, W.C.; Hsieh, W.P.; Shin, J.; Jane, W.N.; Chou, S.J.; Choi, G.; Hu, J.M.; et al. LZF1, a HY5-regulated transcriptional factor, functions in Arabidopsis de-etiolation. Plant J. 2008, 54, 205–219. [Google Scholar] [CrossRef]
- Zhang, X.; Huai, J.; Shang, F.; Xu, G.; Tang, W.; Jing, Y.; Lin, R. A PIF1/PIF3-HY5-BBX23 transcription factor cascade affects photomorphogenesis. Plant Physiol. 2017, 174, 2487–2500. [Google Scholar] [CrossRef]
- Job, N.; Yadukrishnan, P.; Bursch, K.; Datta, S.; Johansson, H. Two B-box proteins regulate photomorphogenesis by oppositely modulating HY5 through their diverse C-terminal domains. Plant Physiol. 2018, 176, 2963–2976. [Google Scholar] [CrossRef] [PubMed]
- Gangappa, S.N.; Holm, M.; Botto, J.F. Molecular interactions of BBX24 and BBX25 with HYH, HY5 homolog, to modulate Arabidopsis seedling development. Plant Sign. Behav. 2013, 8, e25208. [Google Scholar] [CrossRef] [PubMed]
- Gangappa, S.N.; Crocco, C.D.; Johansson, H.; Datta, S.; Hettiarachchi, C.; Holm, M.; Botto, J.F. The Arabidopsis B-box protein BBX25 interacts with HY5, negatively regulating BBX22 expression to suppress seedling photomorphogenesis. Plant Cell 2013, 25, 1243–1257. [Google Scholar] [CrossRef]
- Lin, F.; Jiang, Y.; Li, J.; Yan, T.T.; Fan, L.M.; Liang, J.S.; Chen, Z.J.; Xu, D.Q.; Deng, X.W. B-box domain Protein28 negatively regulates photomorphogenesis by repressing the activity of transcription factor HY5 and undergoes COP1-mediated degradation. Plant Cell 2018, 30, 2006–2019. [Google Scholar] [CrossRef] [PubMed]
- Holtan, H.E.; Bandong, S.; Marion, C.M.; Adam, L.; Tiwari, S.; Shen, Y.; Maloof, J.N.; Maszle, D.R.; Ohto, M.A.; Preuss, S.; et al. BBX32, an Arabidopsis B-Box protein, functions in light signaling by suppressing HY5-regulated gene expression and interacting with STH2/BBX21. Plant Physiol. 2011, 156, 2109–2123. [Google Scholar] [CrossRef] [PubMed]
- Osterlund, M.T.; Hardtke, C.S.; Wei, N.; Deng, X.W. Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 2000, 405, 462–466. [Google Scholar] [CrossRef] [PubMed]
- Heng, Y.; Lin, F.; Jiang, Y.; Ding, M.; Yan, T.; Lan, H.; Zhou, H.; Zhao, X.; Xu, D.; Deng, X.W. B-Box Containing Proteins BBX30 and BBX31, acting downstream of HY5, negatively regulate photomorphogenesis in Arabidopsis. Plant Physiol. 2019, 180, 497–508. [Google Scholar] [CrossRef]
- Yadav, A.; Bakshi, S.; Yadukrishnan, P.; Lingwan, M.; Dolde, U.; Wenkel, S.; Masakapalli, S.K.; Datta, S. The B-box-containing microprotein miP1a/BBX31 regulates photomorphogenesis and UV-B protection. Plant Physiol. 2019, 179, 1876–1892. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Bian, Y.; Liu, J.; Sun, Y.; Xu, D. B-box proteins: Pivotal players in light-mediated development in plants. J. Integr. Plant Biol. 2020, 62, 1293–1309. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Yan, T.; Liu, J.; Bian, Y.; Heng, Y.; Lin, F.; Jiang, Y.; Wang Deng, X.; Xu, D. BBX28/BBX29, HY5 and BBX30/31 form a feedback loop to fine-tune photomorphogenic development. Plant J. 2020. [Google Scholar] [CrossRef] [PubMed]
- Nagaoka, S.; Takano, T. Salt tolerance-related protein STO binds to a Myb transcription factor homologue and confers salt tolerance in Arabidopsis. J. Exp. Bot. 2003, 54, 2231–2237. [Google Scholar] [CrossRef] [PubMed]
- Fang, H.; Dong, Y.; Yue, X.; Hu, J.; Jiang, S.; Xu, H.; Wang, Y.; Su, M.; Zhang, J.; Zhang, Z.; et al. The B-box zinc finger protein MdBBX20 integrates anthocyanin accumulation in response to ultraviolet radiation and low temperature. Plant Cell Environ. 2019, 42, 2090–2104. [Google Scholar] [CrossRef]
- An, J.P.; Wang, X.F.; Espley, R.V.; Lin-Wang, K.; Bi, S.Q.; You, C.X.; Hao, Y.J. An apple B-Box protein MdBBX37 modulates anthocyanin biosynthesis and hypocotyl elongation synergistically with MdMYBs and MdHY5. Plant Cell Physiol. 2020, 61, 130–143. [Google Scholar] [CrossRef]
- Bai, S.; Tao, R.; Tang, Y.; Yin, L.; Ma, Y.; Ni, J.; Yan, X.; Yang, Q.; Wu, Z.; Zeng, Y.; et al. BBX16, a B-box protein, positively regulates light-induced anthocyanin accumulation by activating MYB10 in red pear. Plant Biotechnol. J. 2019, 17, 1985–1997. [Google Scholar] [CrossRef]
- Xu, Y.; Zhao, X.; Aiwaili, P.; Mu, X.; Zhao, M.; Zhao, J.; Cheng, L.; Ma, C.; Gao, J.; Hong, B. A zinc finger protein BBX19 interacts with ABF3 to affect drought tolerance negatively in chrysanthemum. Plant J. 2020, 103, 1783–1795. [Google Scholar] [CrossRef]
- Min, J.H.; Chung, J.S.; Lee, K.H.; Kim, C.S. The constans-like 4 transcription factor, AtCOL4, positively regulates abiotic stress tolerance through an abscisic acid-dependent manner in Arabidopsis. J. Integr. Plant Biol. 2015, 57, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.M.; Zeng, J.X.; Deng, K.Q.; Tu, X.J.; Zhao, X.Y.; Tang, D.Y.; Liu, X.M. DBB1a, involved in gibberellin homeostasis, functions as a negative regulator of blue light-mediated hypocotyl elongation in Arabidopsis. Planta 2011, 233, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.Y.; Sun, Y.; Cao, D.M.; Bai, M.Y.; Luo, X.M.; Yang, H.J.; Wei, C.Q.; Zhu, S.W.; Sun, Y.; Chong, K.; et al. BZS1, a B-box protein, promotes photomorphogenesis downstream of both brassinosteroid and light signaling pathways. Mol. Plant 2012, 5, 591–600. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, Q.; Zhai, H.; Gao, S.P.; Yang, L.; Wang, Z.; Xu, Y.T.; Huo, J.X.; Ren, Z.T.; Zhao, N.; et al. IbBBX24 promotes the jasmonic acid pathway and enhances fusarium wilt resistance in sweet potato. Plant Cell 2020, 32, 1102–1123. [Google Scholar] [CrossRef]
- Esti, M.; Cinquanta, L.; Sinesio, F.; Moneta, E.; Di Matteo, M. Physicochemical and sensory fruit characteristics of two sweet cherry cultivars after cool storage. Food Chem. 2002, 76, 399–405. [Google Scholar] [CrossRef]
- Guo, X.; Wang, Y.T.; Zhai, Z.F.; Huang, T.J.; Zhao, D.; Peng, X.; Feng, C.; Xiao, Y.H.; Li, T.H. Transcriptomic analysis of light-dependent anthocyanin accumulation in bicolored cherry fruits. Plant Physiol. Bioch. 2018, 130, 663–677. [Google Scholar] [CrossRef]
- Chu, Z.N.; Wang, X.; Li, Y.; Yu, H.Y.; Li, J.H.; Lu, Y.G.; Li, H.X.; Ouyang, B. Genomic organization, phylogenetic and expression analysis of the B-BOX gene family in tomato. Front. Plant Sci. 2016, 7, 1552. [Google Scholar] [CrossRef]
- Cao, Y.P.; Han, Y.H.; Meng, D.D.; Li, D.H.; Jiao, C.Y.; Jin, Q.; Lin, Y.; Cai, Y.P. B-box genes: Genome-wide identification, evolution and their contribution to pollen growth in pear (Pyrus bretschneideri Rehd.). BMC Plant Biol. 2017, 17, 156. [Google Scholar] [CrossRef]
- Xu, D.Q.; Li, J.G.; Gangappa, S.N.; Hettiarachchi, C.; Lin, F.; Andersson, M.X.; Jiang, Y.; Deng, X.W.; Holm, M. Convergence of light and ABA signaling on the ABI5 promoter. Plos Genet. 2014, 10, e1004197. [Google Scholar] [CrossRef]
- Jaakola, L. New insights into the regulation of anthocyanin biosynthesis in fruits. Trends Plant Sci. 2013, 18, 477–483. [Google Scholar] [CrossRef]
- Huang, J.Y.; Zhao, X.B.; Weng, X.Y.; Wang, L.; Xie, W.B. The rice B-Box zinc finger gene family: Genomic identification, characterization, expression profiling and diurnal analysis. PLoS ONE 2012, 7, e48242. [Google Scholar] [CrossRef]
- Liu, X.; Li, R.; Dai, Y.Q.; Chen, X.S.; Wang, X.Y. Genome-wide identification and expression analysis of the B-box gene family in the apple (Malus domestica Borkh.) genome. Mol. Genet. Genomics 2018, 293, 303–315. [Google Scholar] [CrossRef]
- Shen, X.J.; Zhao, K.; Liu, L.L.; Zhang, K.C.; Yuan, H.Z.; Liao, X.; Wang, Q.; Guo, X.W.; Li, F.; Li, T.H. A role for PacMYBA in ABA-regulated anthocyanin biosynthesis in red-colored sweet cherry cv. Hong Deng (Prunus avium L.). Plant Cell Physiol. 2014, 55, 862–880. [Google Scholar] [CrossRef]
- Shin, D.H.; Choi, M.; Kim, K.; Bang, G.; Cho, M.; Choi, S.B.; Choi, G.; Park, Y.I. HY5 regulates anthocyanin biosynthesis by inducing the transcriptional activation of the MYB75/PAP1 transcription factor in Arabidopsis. Febs. Lett. 2013, 587, 1543–1547. [Google Scholar] [CrossRef]
- Fang, H.C.; Dong, Y.H.; Yue, X.X.; Chen, X.L.; He, N.B.; Hu, J.F.; Jiang, S.H.; Xu, H.F.; Wang, Y.C.; Su, M.; et al. MdCOL4 interaction mediates crosstalk between UV-B and high temperature to control fruit coloration in Apple. Plant Cell Physiol. 2019, 60, 1055–1066. [Google Scholar] [CrossRef]
- Kim, D.H.; Park, S.; Lee, J.Y.; Ha, S.H.; Lee, J.G.; Lim, S.H. A rice B-box protein, OsBBX14, finely regulates anthocyanin biosynthesis in rice. Int. J. Mol. Sci. 2018, 19, 2190. [Google Scholar] [CrossRef]
- Yamawaki, S.; Yamashino, T.; Nakamichi, N.; Nakanishi, H.; Mizuno, T. Light-responsive double B-box containing transcription factors are conserved in physcomitrella patens. Biosci. Biotech. Bioch. 2011, 75, 2037–2041. [Google Scholar] [CrossRef]
- Yeh, C.M.; Kobayashi, K.; Fujii, S.; Fukaki, H.; Mitsuda, N.; Ohme-Takagi, M. Blue light regulates phosphate deficiency-dependent primary root growth inhibition in Arabidopsis. Front. Plant. Sci. 2020, 10. [Google Scholar] [CrossRef]
- Bai, S.L.; Tao, R.Y.; Yin, L.; Ni, J.B.; Yang, Q.S.; Yan, X.H.; Yang, F.P.; Guo, X.P.; Li, H.X.; Teng, Y.W. Two B-box proteins, PpBBX18 and PpBBX21, antagonistically regulate anthocyanin biosynthesis via competitive association with Pyrus pyrifolia Elongated hypocotyl 5 in the peel of pear fruit. Plant J. 2019, 100, 1208–1223. [Google Scholar] [CrossRef]
- Lira, B.S.; Oliveira, M.J.; Shiose, L.; Wu, R.T.A.; Rosado, D.; Lupi, A.C.D.; Freschi, L.; Rossi, M. Light and ripening-regulated BBX protein-encoding genes in S. lycopersicum. Sci. Rep. 2020, 10, 19235. [Google Scholar] [CrossRef]
- Forlani, S.; Masiero, S.; Mizzotti, C. Fruit ripening: The role of hormones, cell wall modifications, and their relationship with pathogens. J. Exp. Bot. 2019, 70, 2993–3006. [Google Scholar] [CrossRef]
- Li, F.; Sun, J.J.; Wang, D.H.; Bai, S.N.; Clarke, A.K.; Holm, M. The B-Box family gene STO (BBX24) in A. thaliana. regulates flowering time in different pathways. PLoS ONE 2014, 9, e87544. [Google Scholar] [CrossRef]
- Vaishak, K.P.; Yadukrishnan, P.; Bakshi, S.; Kushwaha, A.K.; Ramachandran, H.; Job, N.; Babu, D.; Datta, S. The B-box bridge between light and hormones in plants. J. Photoch. Photobio. B. 2019, 191, 164–174. [Google Scholar] [CrossRef]
- Yadukrishnan, P.; Datta, S. Light and abscisic acid interplay in early seedling development. New Phytol. 2021, 229, 763–769. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Alkio, M.; Jonas, U.; Sprink, T.; van Nocker, S.; Knoche, M. Identification of putative candidate genes involved in cuticle formation in Prunus avium (sweet cherry) fruit. Ann. Bot. Lond. 2012, 110, 101–112. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-△△Ct method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Sun, L.A.; Zhang, M.; Ren, J.; Qi, J.X.; Zhang, G.J.; Leng, P. Reciprocity between abscisic acid and ethylene at the onset of berry ripening and after harvest. BMC Plant. Biol. 2010, 10, 257. [Google Scholar] [CrossRef]
- Sparkes, I.A.; Runions, J.; Kearns, A.; Hawes, C. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 2006, 1, 2019–2025. [Google Scholar] [CrossRef]




| Gene Name | Accession Number | Protein/AA | Chrom | Chr Start | Chr End | MW (Da) | pI | Aliphatic Index | GRAVY |
|---|---|---|---|---|---|---|---|---|---|
| PavBBX1 | Pav_sc0004527.1g080.1 | 650 | Chr1 | 17737787 | 17743220 | 71,169.18 | 5.68 | 74.68 | −0.458 |
| PavBBX2 | Pav_sc0001051.1g030.1 | 450 | Chr1 | 24564167 | 24566102 | 49,926.26 | 5.15 | 56.84 | −0.790 |
| PavBBX3 | Pav_sc0000131.1g850.1 | 343 | Chr1 | 29989611 | 29991028 | 38,307.77 | 5.47 | 65.95 | −0.539 |
| PavBBX4 | Pav_sc0000293.1g410.1 | 264 | Chr3 | 661262 | 662056 | 28,357.08 | 9.32 | 72.16 | −0.246 |
| PavBBX5 | Pav_sc0000405.1g160.1 | 399 | Chr3 | 16146055 | 16148421 | 42,967.79 | 5.07 | 61.13 | −0.485 |
| PavBBX6 | Pav_sc0001859.1g200.1 | 152 | Chr3 | 16934620 | 16935833 | 17,011.06 | 9.09 | 60.46 | −0.849 |
| PavBBX7 | Pav_sc0000051.1g110.1 | 324 | Chr3 | 18112949 | 18115389 | 35,601.87 | 5.78 | 67.25 | −0.482 |
| PavBBX8 | Pav_sc0000598.1g150.1 | 204 | Chr4 | 1931059 | 1932064 | 23,047.58 | 4.51 | 38.68 | −1.254 |
| PavBBX9 | Pav_sc0000352.1g070.1 | 301 | Chr4 | 11752866 | 11755126 | 31,786.49 | 5.53 | 62.86 | −0.317 |
| PavBBX10 | Pav_sc0000383.1g410.1 | 480 | Chr5 | 15713586 | 15715861 | 53,565.68 | 5.12 | 62.23 | −0.770 |
| PavBBX11 | Pav_sc0000044.1g810.1 | 229 | Chr7 | 13871819 | 13874309 | 25,370.08 | 8.51 | 74.10 | −0.387 |
| PavBBX12 | Pav_sc0001518.1g660.1 | 208 | Chr8 | 8383526 | 8384850 | 23,011.66 | 5.97 | 58.17 | −0.462 |
| PavBBX13 | Pav_sc0000848.1g470.1 | 391 | Chr8 | 19897720 | 19900725 | 43,198.97 | 6.16 | 58.90 | −0.661 |
| PavBBX14 | Pav_sc0001100.1g020.1 | 239 | ChrUn | 27735116 | 27739262 | 26,178.43 | 4.77 | 74.81 | −0.315 |
| PavBBX15 | Pav_sc0002706.1g040.1 | 333 | ChrUn | 55031701 | 55033349 | 35,617.19 | 4.95 | 82.97 | −0.046 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Zhai, Z.; Sun, Y.; Feng, C.; Peng, X.; Zhang, X.; Xiao, Y.; Zhou, X.; Wang, W.; Jiao, J.; et al. Genome-Wide Identification of the B-BOX Genes that Respond to Multiple Ripening Related Signals in Sweet Cherry Fruit. Int. J. Mol. Sci. 2021, 22, 1622. https://doi.org/10.3390/ijms22041622
Wang Y, Zhai Z, Sun Y, Feng C, Peng X, Zhang X, Xiao Y, Zhou X, Wang W, Jiao J, et al. Genome-Wide Identification of the B-BOX Genes that Respond to Multiple Ripening Related Signals in Sweet Cherry Fruit. International Journal of Molecular Sciences. 2021; 22(4):1622. https://doi.org/10.3390/ijms22041622
Chicago/Turabian StyleWang, Yanyan, Zefeng Zhai, Yueting Sun, Chen Feng, Xiang Peng, Xiang Zhang, Yuqin Xiao, Xin Zhou, Weili Wang, Jiale Jiao, and et al. 2021. "Genome-Wide Identification of the B-BOX Genes that Respond to Multiple Ripening Related Signals in Sweet Cherry Fruit" International Journal of Molecular Sciences 22, no. 4: 1622. https://doi.org/10.3390/ijms22041622
APA StyleWang, Y., Zhai, Z., Sun, Y., Feng, C., Peng, X., Zhang, X., Xiao, Y., Zhou, X., Wang, W., Jiao, J., & Li, T. (2021). Genome-Wide Identification of the B-BOX Genes that Respond to Multiple Ripening Related Signals in Sweet Cherry Fruit. International Journal of Molecular Sciences, 22(4), 1622. https://doi.org/10.3390/ijms22041622






