Antioxidant vs. Prooxidant Properties of the Flavonoid, Kaempferol, in the Presence of Cu(II) Ions: A ROS-Scavenging Activity, Fenton Reaction and DNA Damage Study
Abstract
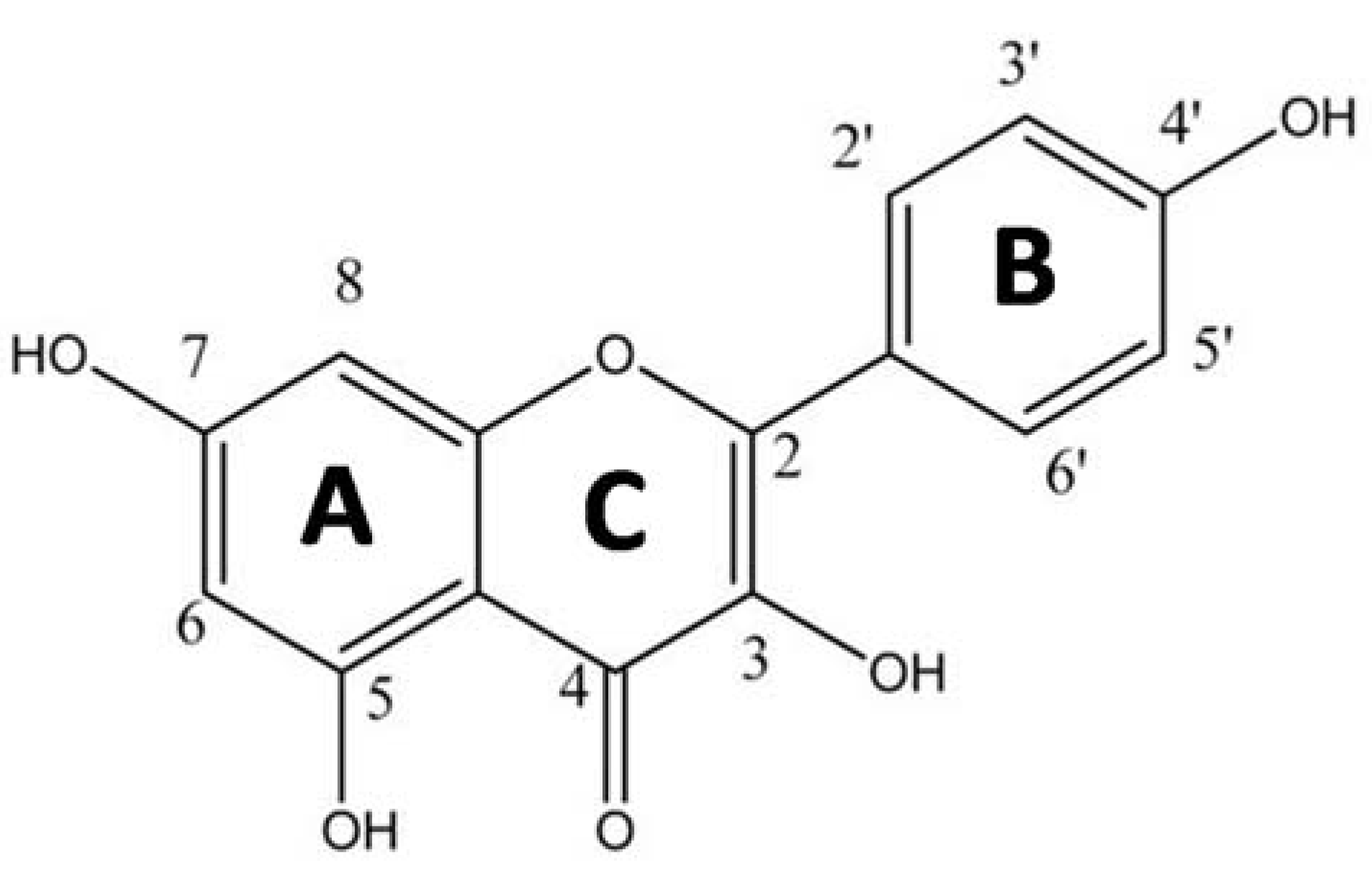
1. Introduction
2. Results and Discussion
2.1. EPR Spectroscopy
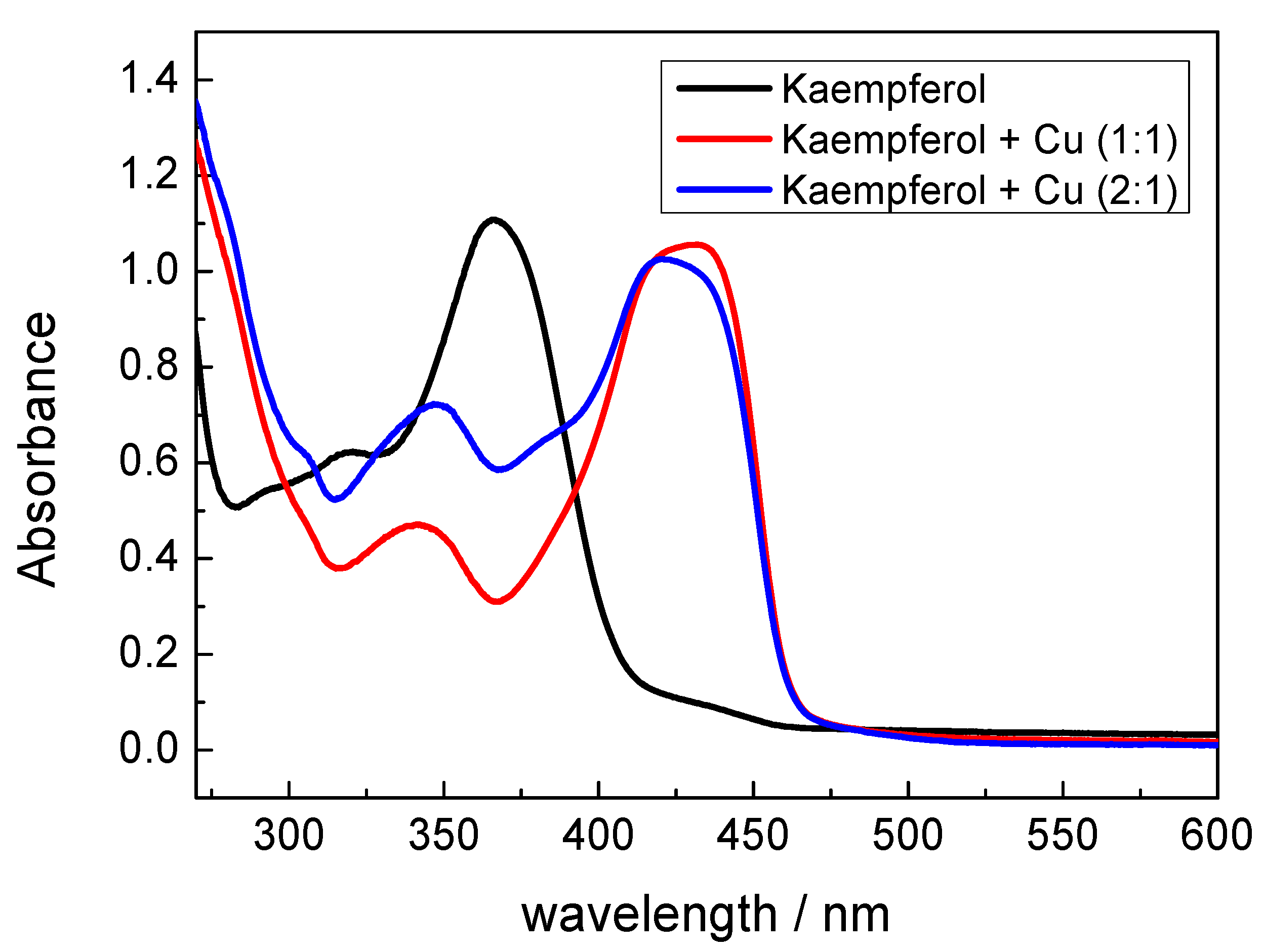
2.2. UV-Vis Spectroscopy
2.3. Radical Scavenging Activity of Kaempferol and Cu(II)-Kaempferol (1:1 and 1:2) Complexes
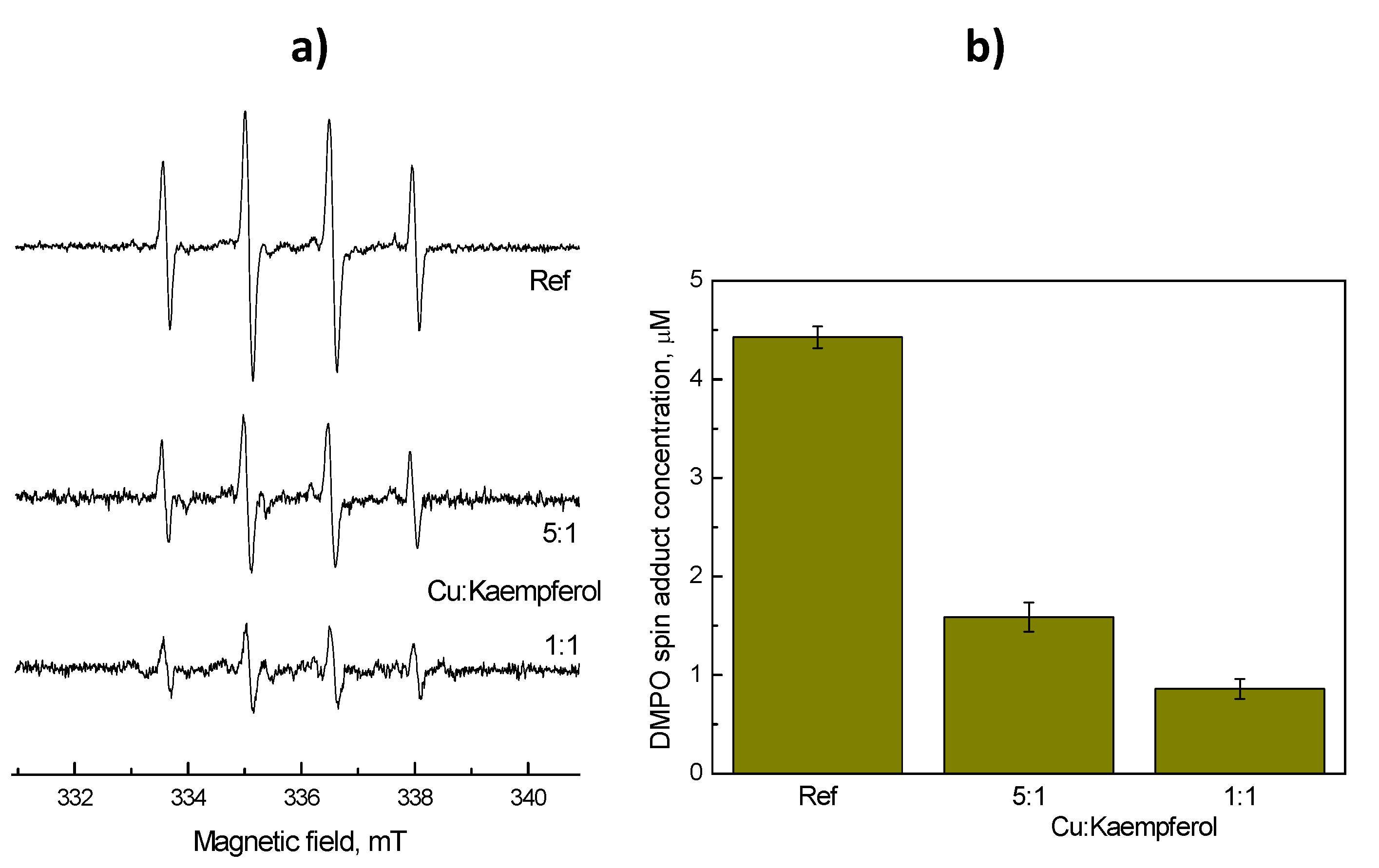
2.4. Protective Role of Kaempferol in Cu Fenton Raection: EPR Spin Trapping Experiment
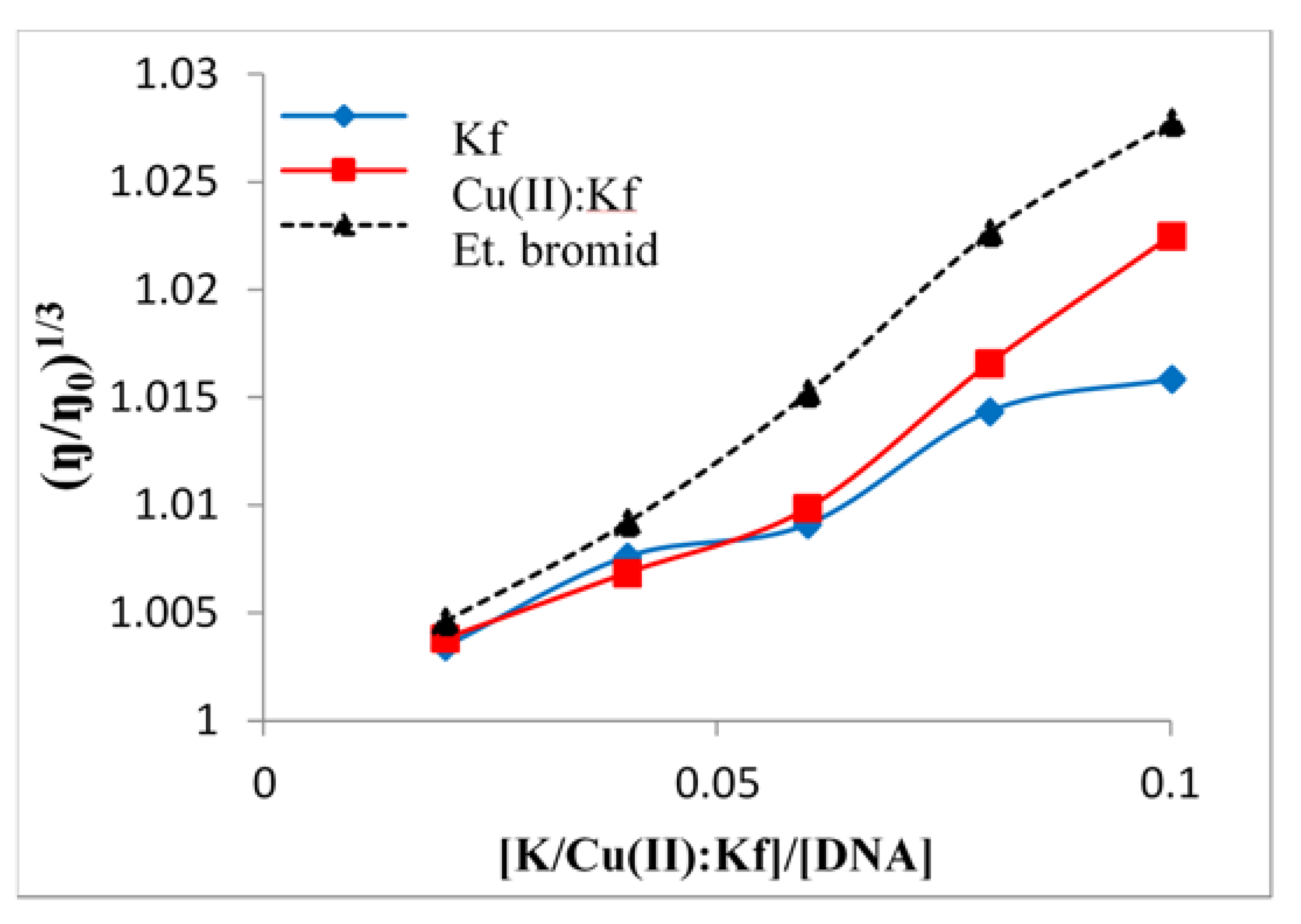
2.5. Viscometric Study of the Interaction between Kaempferol/Cu-Kaempferol Complex with DNA
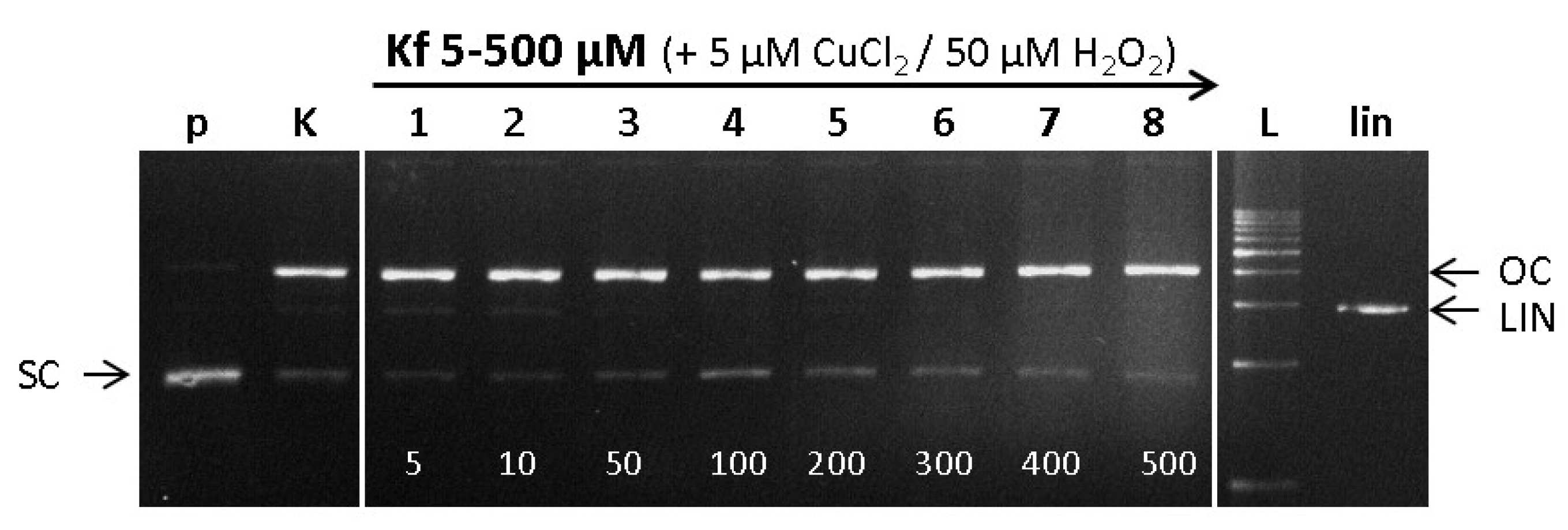
2.6. DNA Damage/Protection Study Induced by Cu-Kaempferol Complex in Fenton Reaction
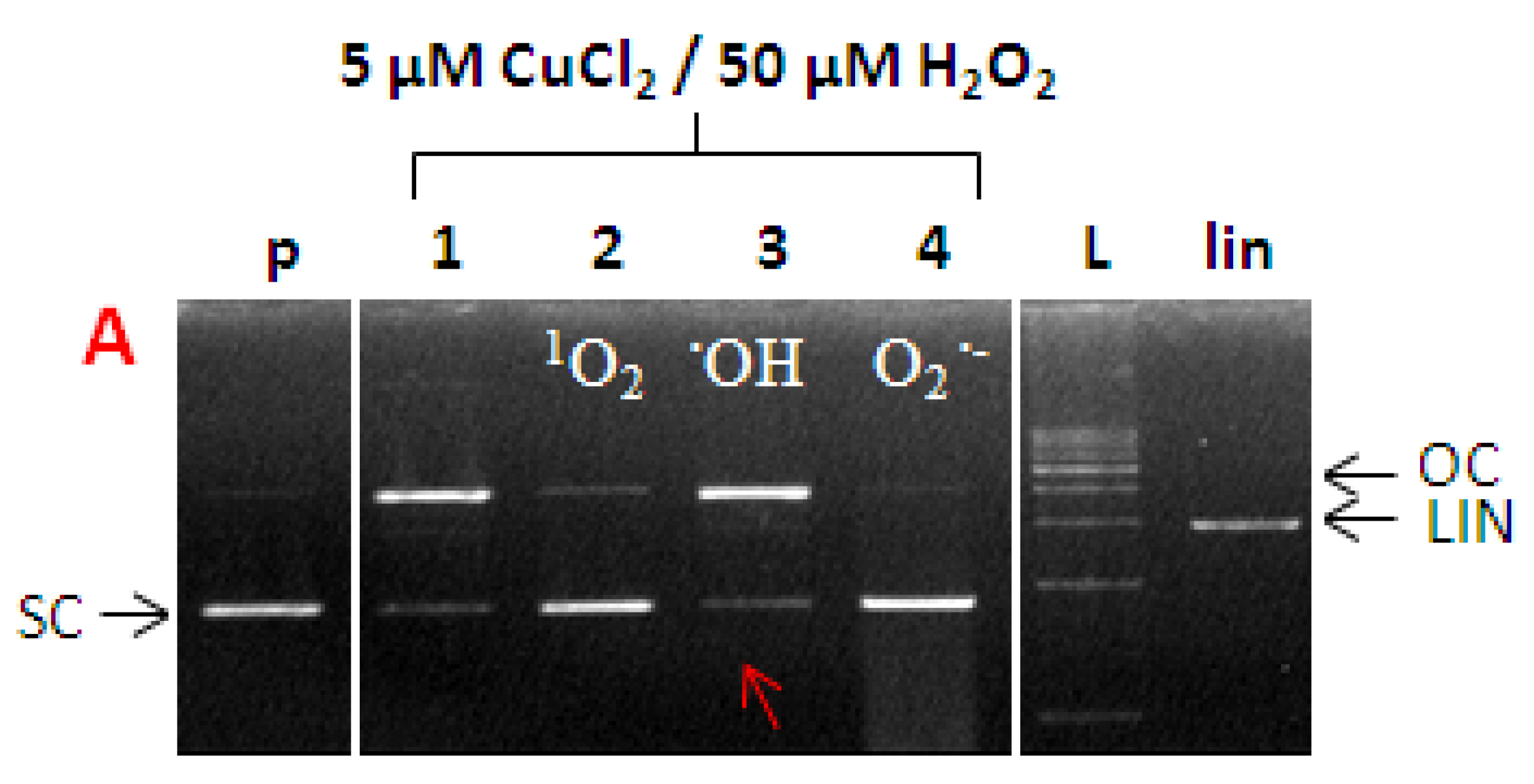
2.7. ROS Involved in DNA Damage by Cu-Kamferol Fenton System
3. Materials and Methods
3.1. Materials
3.2. EPR Spectroscopy
3.3. UV-Vis Spectroscopy
3.4. Radical Svavenging Activity of Kamepferol and Its Cu(II) Complexes
3.5. EPR Spin Trapping Experiment
3.6. Viscosity Measurements
3.7. DNA Damage Study
3.7.1. Isolation of Plasmid DNA
3.7.2. Analysis of DNA Damage by Gel-Electrophoresis
3.7.3. Identification of ROS
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rice-Evans, C.A.; Miller, N.J.; Paganga, G. Structure-antioxidant activity relationships of flavonoids and phenolic acids. Free Radic. Biol. Med. 1996, 20, 933–956. [Google Scholar] [CrossRef]
- Williams, R.J.L.; Spencer, J.P.; Rice-Evans, C. Flavonoids: Antioxidants or signalling molecules? Free Radic. Biol. Med. 2004, 36, 838–849. [Google Scholar] [CrossRef] [PubMed]
- Prochazkova, D.; Bousova, I.; Wilhelmova, N. Antioxidant and prooxidant properties of flavonoids. Fitoterapia 2011, 82, 513–523. [Google Scholar] [CrossRef]
- Jomova, K.; Lawson, M.; Drostinova, L.; Lauro, P.; Poprac, P.; Brezova, V.; Michalik, M.; Lukes, V.; Valko, M. Protective role of quercetin against copper(II)-induced oxidative stress: A spectroscopic, theoretical and DNA damage study. Food Chem. Toxicol. 2017, 110, 340–350. [Google Scholar] [CrossRef]
- Bukhari, S.B.; Memon, S.; Mahroof-Tahir, M.; Bhanger, M.I. Synthesis, characterization and antioxidant activity copper-quercetin complex. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2009, 71, 1901–1906. [Google Scholar] [CrossRef]
- Ravichandran, R.; Rajendran, M.; Devapiriam, D. Antioxidant study of quercetin and their metal complex and determination of stability constant by spectrophotometry method. Food Chem. 2014, 146, 472–478. [Google Scholar] [CrossRef]
- Pattubala, A.N.R.; Munirathinam, N.; Akhil, R.C. Hydrolytic cleavage of DNA by ternary amino acid schiff base copper (II) complexes having planar heterocyclic ligands. Eur. J. Inorg. Chem. 2004, 7, 1440–1446. [Google Scholar] [CrossRef]
- Cherrak, S.A.; Mokhtari-Soulimane, N.; Berroukeche, F.; Bensenane, B.; Cherbonnel, A.; Merzouk, H.; Elhabiri, M. In Vitro Antioxidant versus Metal Ion Chelating Properties of Flavonoids: A Structure-Activity Investigation. PLoS ONE 2016, 11, 0165575. [Google Scholar] [CrossRef] [PubMed]
- Jomova, L.; Hudecova, L.; Lauro, P.; Simunkova, M.; Alwasel, S.H.; Alhazza, I.M.; Valko, M. A Switch between Antioxidant and Prooxidant Properties of the Phenolic Compounds Myricetin, Morin, 3′,4′-Dihydroxyflavone, Taxifolin and 4-Hydroxy-Coumarin in the Presence of Copper(II) Ions: A Spectroscopic, Absorption Titration and DNA Damage Study. Molecules 2019, 23, 4335. [Google Scholar] [CrossRef] [PubMed]
- Hadi, S.M.; Asad, S.F.; Singh, S.; Ahmad, A. Putative Mechanism for Anticancer and Apoptosis-Inducing Properties of Plant-Derived Polyphenolic Compounds. IUBMB Life 2000, 50, 167–171. [Google Scholar] [CrossRef] [PubMed]
- Arif, H.; Rehmani, N.; Farhan, M.; Ahmad, A.; Hadi, S.M. Mobilization of Copper ions by Flavonoids in Human Peripheral Lymphocytes Leads to Oxidative DNA Breakage: A Structure Activity Study. Int. J. Mol. Sci. 2015, 16, 26754–26769. [Google Scholar] [CrossRef]
- Borowska, S.; Tomczyk, M.; Strawa, J.W.; Brzóska, M.M. Estimation of the Chelating Ability of an Extract from Aronia melanocarpa L. Berries and Its Main Polyphenolic Ingredients Towards Ions of Zinc and Copper. Molecules 2020, 25, 1507. [Google Scholar] [CrossRef]
- Tan, J.; Wang, B.; Zhu, L. DNA binding and oxidative DNA damage induced by a quercetin copper(II) complex: Potential mechanism of its antitumor properties. J. Biol. Inorg. Chem. 2009, 14, 727–739. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Kim, Y.S.; Choi, J.G.; Li, W.; Lee, E.J.; Park, J.W.; Song, J.; Chung, H.S. Kaempferol and Its Glycoside, Kaempferol 7-O-rhamnoside, Inhibit PD-1/PD-L1 Interaction In Vitro. Int. J. Mol. Sci. 2020, 21, 3239. [Google Scholar] [CrossRef] [PubMed]
- Deng, S.P.; Yang, Y.L.; Cheng, X.X.; Li, W.R.; Cai, J.Y. Synthesis, Spectroscopic Study and Radical Scavenging Activity of Kaempferol Derivatives: Enhanced Water Solubility and Antioxidant Activity. Int. J. Mol. Sci. 2019, 20, 975. [Google Scholar] [CrossRef] [PubMed]
- Budisan, L.; Gulei, D.; Jurj, A.; Braicu, C.; Zanoaga, O.; Cojocneanu, R.; Pop, L.; Raduly, L.; Barbat, A.; Moldovan, A.; et al. Inhibitory Effect of CAPE and Kaempferol in Colon Cancer Cell LinesPossible Implications in New Therapeutic Strategies. Int. J. Mol. Sci. 2019, 20, 1199. [Google Scholar] [CrossRef] [PubMed]
- Chobot, V.; Kubicova, L.; Bachmann, G.; Hadacek, F. Versatile Redox Chemistry Complicates Antioxidant Capacity Assessment: Flavonoids as Milieu-Dependent Anti- and Pro-Oxidants. Int. J. Mol. Sci. 2013, 14, 11830–11841. [Google Scholar] [CrossRef] [PubMed]
- Chobot, V.; Hadacek, F.; Bachmann, G.; Weckwerth, W.; Kubicova, L. Pro- and Antioxidant Activity of Three Selected Flavan Type Flavonoids: Catechin, Eriodictyol and Taxifolin. Int. J. Mol. Sci. 2016, 17, 1986. [Google Scholar] [CrossRef] [PubMed]
- Alam, W.; Khan, H.; Shah, M.A.; Cauli, O.; Saso, L. Kaempferol as a Dietary Anti-Inflammatory Agent: Current Therapeutic Standing. Molecules 2020, 25, 4073. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.Y.; Qian, L.L.; Xu, Y.; Song, M.T.; Liu, C.; Han, R.M.; Zhang, J.P.; Skibsted, L.H. Kinetic Studies on Radical Scavenging Activity of Kaempferol Decreased by Sn(II) Binding. Molecules 2020, 25, 1975. [Google Scholar] [CrossRef]
- Imran, M.; Salehi, B.; Sharifi-Rad, J.; Aslam Gondal, T.; Saeed, F.; Imran, A.; Shahbaz, M.; Tsouh Fokou, P.V.; Umair Arshad, M.; Khan, H.; et al. Kaempferol: A Key Emphasis to Its Anticancer Potential. Molecules 2019, 24, 2277. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.; Meng, X.; Zheng, H.; Zeng, Q.; Chen, T.; Wang, W.; Zhang, X.; Su, J. Kaempferol Attenuates ROS-Induced Hemolysis and the Molecular Mechanism of Its Induction of Apoptosis on Bladder Cancer. Molecules 2018, 23, 2592. [Google Scholar] [CrossRef] [PubMed]
- Kuckova, L.; Jomova, K.; Svorcova, A.; Valko, M.; Segla, P.; Moncol, P.; Kozisek, J. Synthesis, crystal structure, spectroscopic properties and potential biological activities of salicylate-neocuproine ternary copper(II) complexes. Molecules 2015, 20, 2115–2137. [Google Scholar] [CrossRef] [PubMed]
- Woźnicka, E.; Kopacz, M.; Umbreit, M.; Kłos, J. New complexes of La(III), Ce(III), Pr(III), Nd(III), Sm(III), Eu(III) and Gd(III) ions with morin. J. Inorg. Biochem. 2007, 101, 774–782. [Google Scholar] [CrossRef]
- Kasprzask, M.M.; Erxleben, A.; Ochocki, J. Properties and applications of flavonoid metal complexes. RSC Adv. 2015, 5, 45853–45877. [Google Scholar] [CrossRef]
- Malesev, D.; Kuntic, V. Investigation of metal-flavonoid chelates and the determination of flavonoids via metal-flavonoid complexing reactions. J. Serb. Chem. Soc. 2007, 72, 921–939. [Google Scholar] [CrossRef]
- Jovanovic, S.V.; Steenken, S.; Tosic, M.; Marjanovic, B.; Simic, M.G. Flavonoids as Antioxidants. J. Am. Chem. Soc. 1994, 116, 4846–4851. [Google Scholar] [CrossRef]
- Wang, T.Y.; Li, Q.; Bi, K.S. Bioactive flavonoids in medicinal plants: Structure, activity and biological fate. Asian J. Pharmacol. Sci. 2018, 13, 12–23. [Google Scholar] [CrossRef]
- Panhwar, Q.K.; Memon, S.; Bhanger, M.I. Synthesis, characterization, spectroscopic and antioxidation studies of Cu(II)–morin complex. J. Mol. Struct. 2010, 967, 47–53. [Google Scholar] [CrossRef]
- Li, X.; Jiang, Q.; Chen, B.; Luo, X.; Chen, D. Structure Activity Relationship and Prediction of the Electron-Transfer Potential of the Xanthones Series. Chem. Open 2018, 7, 730–736. [Google Scholar] [CrossRef] [PubMed]
- Cramer, R.E.; Bopp, T.T. Graphical display of the enthalpies of adduct formation for Lewis acids and bases. J. Chem. Educ. 1977, 54, 612–613. [Google Scholar] [CrossRef]
- Pekal, A.; Biesaga, M.; Pyrzynska, K. Interaction of quercetin with copper ions: Complexation, oxidation and reactivity towards radicals. Biometals 2011, 24, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Szabados-Furjesi, P.; Pajtas, D.; Barta, D.; Czepanyi, E.; Kiss-Szikszai, A.; Tosaki, A.; Bak, I. Synthesis, in vitro evaluation, and oxidative transformation of new flavonoid derivatives: The possible role of the phenyl-N,N dimethylamino group. Molecules 2018, 23, 3161. [Google Scholar] [CrossRef]
- Soltes, L.; Brezová, V.; Stankovská, M.; Kogan, G.; Gemeiner, P. Degradation of high-molecular-weight hyaluronan by hydrogen peroxide in the presence of cupric ions. Carbohydr Res. 2006, 341, 639–644. [Google Scholar] [CrossRef] [PubMed]
- Hricovíni, M.; Mazúr, M.; Sîrbu, A.; Palamarciuc, O.; Arion, V.B.; Brezová, V. Copper(II) thiosemicarbazone complexes and their proligands upon UVA irradiation: An EPR and spectrophotometric steady-state study. Molecules 2018, 23, 721. [Google Scholar] [CrossRef]
- Pham, A.N.; Xing, G.; Miller, C.J.; Waite, T.D. Fenton-like copper redox chemistry revisited: Hydrogen peroxide and superoxide mediation of copper-catalyzed oxidant production. J. Catal. 2013, 301, 54–64. [Google Scholar] [CrossRef]
- Mitroka, S.; Zimmeck, S.; Troya, D.; Tanko, J. How Solvent Modulates Hydroxyl Radical Reactivity in Hydrogen Atom Abstractions. J. Am. Chem. Soc. 2010, 132, 2907–2913. [Google Scholar] [CrossRef]
- Buxton, G.; Greenstock, C.; Helman, W.; Ross, A. Critical-review of rate constants for reactions of hydrated electrons, hydrogen-atoms and hydroxyl radicals (•OH/•O–) in aqueous-solution. J. Phys. Chem. Ref. Data 1988, 17, 513–886. [Google Scholar] [CrossRef]
- Eghbaliferiz, S.; Iranshahi, M. Prooxidant Activity of Polyphenols, Flavonoids, Anthocyanins and Carotenoids: Updated Review of Mechanisms and Catalyzing Metals. Phytother. Res. 2016, 30, 1379–1391. [Google Scholar] [CrossRef] [PubMed]
- Selvaraj, S.; Krishnaswamy, S.; Devashya, V.; Sethuraman, S.; Krishnan, U.M. Flavonoid-metal ion complexes: A novel class of therapeutic agents. Med. Res. Rev. 2014, 34, 677–702. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, S.; Kanai, S. Superoxide scavenging effects of some novel bis-ligands and their solvated metal complexes prepared by the reaction of ligands with aluminum, copper and lanthanum ions. Molecules 2013, 18, 6128–6141. [Google Scholar] [CrossRef] [PubMed]
- Barolli, M.G.; Werner, R.A.; Slep, L.D.; Pomilio, A.B. Formation of complexes of flavonoids and metals. Determination of the Stoichiometry and Stability Constants. Molecules 2000, 5, 516–517. [Google Scholar] [CrossRef]
- Liu, Y.; Guo, M. Studies on transition metal-quercetin complexes using electrospray ionization tandem mass spectrometry. Molecules 2015, 20, 8583–8594. [Google Scholar] [CrossRef] [PubMed]
- Hirohama, T.; Kuranuki, Y.; Ebina, E.; Sugizaki, T.; Arii, H.; Chikira, M.; Selvi, P.T.; Palaniandavar, M. Copper(II) complexes of 1,10-phenanthroline-derived ligands: Studies on DNA binding properties and nuclease activity. J. Inorg. Biochem. 2005, 99, 1205–1219. [Google Scholar] [CrossRef]
- Chalkidou, E.; Perdih, F.; Turel, I.; Kessissoglou, D.P.; Psomas, G. Copper(II) complexes with antimicrobial drug flumequine: Structure and biological evaluation. J. Inorg. Biochem. 2012, 113, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Dimiza, F.; Perdih, F.; Tangoulis, V.; Turel, I.; Kessissoglou, D.P.; Psomas, G. Interaction of copper(II) with the non-steroidal anti-inflammatory drugs naproxenand diclofenac: Synthesis, structure, DNA- and albumin-binding. J. Inorg. Biochem. 2011, 105, 476–489. [Google Scholar] [CrossRef] [PubMed]
- Fountoulaki, S.; Perdih, F.; Turel, I.; Kessissoglou, D.P.; Psomas, G. Non-steroidal anti-inflammatory drug diflunisal interacting with Cu(II). Structure and biological features. J. Inorg. Biochem. 2011, 105, 1645–1655. [Google Scholar] [CrossRef] [PubMed]
- Turel, I. The interactions of metal ions with quinolone antibacterial agents. Coord. Chem. Rev. 2002, 232, 24–47. [Google Scholar] [CrossRef]
- Kanakis, C.D.; Tarantilis, P.A.; Pappas, C.; Bariyanga, J.; Tajmir-Riahi, H.A.; Polissiou, M.G. An overview of structural features of DNA and RNA complexes withsaffron compounds: Models and antioxidant activity. J. Photochem. Photobiol. B 2009, 95, 204–212. [Google Scholar] [CrossRef]
- Battin, E.E.; Brumaghim, J.L. Antioxidant activity of sulfur and selenium: A review of reactive oxygen species scavenging, glutathione peroxidase, and metal-binding antioxidant mechanisms. Cell. Biochem. Biophys. 2009, 55, 1–23. [Google Scholar] [CrossRef]
- Lerner, A.; Kornweitz, H.; Zilberman, I.; Yardeni, G.; Saphier, M.; Bar, R.; Meyerstein, D. Radicals in ‘biologically relevant’ concentrations behave differently: Uncovering new radical reactions following the reaction of hydroxyl radicals with DMSO. Free Radic. Biol. Med. 2020, in press. [Google Scholar] [CrossRef]
- Brezova, V.; Valko, M.; Breza, M.; Morris, H.; Telser, J.; Dvoranova, D.; Kaiserova, K.; Varecka, L.; Mazur, M.; Leibfritz, D. Role of Radicals and Singlet Oxygen in Photoactivated DNA Cleavage by the Anticancer Drug Camptothecin: An Electron Paramagnetic Resonance Study. J. Phys. Chem. B 2003, 107, 2415–2425. [Google Scholar] [CrossRef]
- Poprac, P.; Jomova, K.; Simunkova, M.; Kollar, V.; Rhodes, C.J.; Valko, M. Targeting Free Radicals in Oxidative Stress-Related Human Diseases. Trends Pharmacol. Sci. 2017, 38, 592–607. [Google Scholar] [CrossRef] [PubMed]
- Mazúr, M.; Valko, M.; Morris, H. Analysis of the radial and longitudinal effect in a double TE(104) and a single TE(102) rectangular cavity. J. Magn. Reson. 2000, 142, 37–56. [Google Scholar] [CrossRef]
- Ciulu, M.; Cádiz-Gurrea, M.L.; Segura-Carretero, A. Extraction and Analysis of Phenolic Compounds in Rice: A Review. Molecules 2018, 23, 2890. [Google Scholar] [CrossRef] [PubMed]
- Duling, D.R. Simulation of multiple isotropic spin-trap EPR spectra. J. Magn. Reson. B 1994, 104, 105–110. [Google Scholar] [CrossRef] [PubMed]
- Stoll, S.; Schweiger, A. EasySpin, a comprehensive software package for spectral simulation and analysis in EPR. J. Magn. Reson. 2006, 178, 42–55. [Google Scholar] [CrossRef] [PubMed]
- Perron, N.R.; García, C.R.; Pinzón, J.R.; Chaur, M.N.; Brumaghim, J.L. Antioxidant and prooxidant effects of polyphenol compounds on copper-mediated DNA damage. J. Inorg. Biochem. 2011, 105, 745–753. [Google Scholar] [CrossRef]
- Kubatka, P.; Kapinova, A.; Kello, M.; Kruzliak, P.; Kajo, P.; Vybohova, D. Fruit peel polyphenols demonstrate substantial anti-tumour effects in the model of breast cancer. Eur. J. Nutr. 2016, 55, 955–965. [Google Scholar] [CrossRef]
- Bhullar, K.S.; Rupasinghe, H.P.V. Polyphenols: Multipotent therapeutic agents in neurodegenerative diseases. Oxid. Med. Cell. Longev. 2013, 2013, 891748. [Google Scholar] [CrossRef] [PubMed]
- Brunetti, C.; Di Ferdinando, M.; Fini, A.; Pollastri, S.; Tattini, M. Flavonoids as Antioxidants and Developmental Regulators: Relative Significance in Plants and Humans. Int. J. Mol. Sci. 2013, 14, 3540–3555. [Google Scholar] [CrossRef] [PubMed]
- Kostecka-Gugała, A.; Kruczek, M.; Ledwożyw-Smoleń, I.; Kaszycki, P. Antioxidants and Health-Beneficial Nutrients in Fruits of Eighteen Cucurbita Cultivars: Analysis of Diversity and Dietary Implications. Molecules 2020, 25, 1792. [Google Scholar] [CrossRef] [PubMed]
- Romanucci, V.; Di Fabio, G.; Zarrelli, A. A new class of synthetic flavonolignan-like dimers: Still few molecules, but with attractive properties. Molecules 2019, 24, 108. [Google Scholar] [CrossRef]
- Rodríguez-García, C.; Sánchez-Quesada, C.; Toledo, E.; Delgado-Rodríguez, M.; Gaforio, J.J. Naturally lignan-rich foods: A dietary tool for health promotion? Molecules 2019, 24, 917. [Google Scholar] [CrossRef] [PubMed]
- Hussain, G.; Zhang, L.; Rasul, A.; Anwar, H.; Sohail, M.U.; Razzaq, A.; Aziz, N.; Shabbir, A.; Ali, M.; Sun, T. Role of plant-derived flavonoids and their mechanism in attenuation of Alzheimer’s and Parkinson’s diseases: An update of recent data. Molecules 2018, 23, 814. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Gonzalez, A.I.; Díaz-Sánchez, A.G.; De La Rosa, L.A.; Vargas-Requena, C.L.; Bustos-Jaimes, I.; Alvarez-Parrilla, E. Polyphenolic compounds and digestive enzymes: In vitro non-covalent interactions. Molecules 2017, 22, 669. [Google Scholar] [CrossRef]
- Kachlicki, P.; Piasecka, A.; Stobiecki, M.; Marczak, Ł. Structural characterization of flavonoid glycoconjugates and their derivatives with mass spectrometric techniques. Molecules 2016, 21, 1494. [Google Scholar] [CrossRef] [PubMed]
- Cirmi, S.; Ferlazzo, N.; Lombardo, G.E.; Ventura-Spagnolo, E.; Gangemi, S.; Calapai, G.; Navarra, M. Neurodegenerative diseases: Might citrus flavonoids play a protective role? Molecules 2016, 21, 1312. [Google Scholar] [CrossRef] [PubMed]
- Mierziak, J.; Kostyn, K.; Kulma, A. Flavonoids as important molecules of plant interactions with the environment. Molecules 2014, 19, 16240–16265. [Google Scholar] [CrossRef] [PubMed]
- Kejík, Z.; Kaplánek, R.; Masařík, M.; Babula, P.; Matkowski, A.; Filipenský, P.; Vesela, K.; Gburek, J.; Sýkora, D.; Martasek, P.; et al. Iron complexes of flavonoids—Antioxidant capacity and beyond. Int. J. Mol. Sci. 2021, 22, 646. [Google Scholar] [CrossRef] [PubMed]



| System | g⊥ | g‖ a | A‖ (Gauss) a |
|---|---|---|---|
| Cu–kaempferol | 2.073 | 2.291 | 125 |
| 2.242 | 118 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Simunkova, M.; Barbierikova, Z.; Jomova, K.; Hudecova, L.; Lauro, P.; Alwasel, S.H.; Alhazza, I.; Rhodes, C.J.; Valko, M. Antioxidant vs. Prooxidant Properties of the Flavonoid, Kaempferol, in the Presence of Cu(II) Ions: A ROS-Scavenging Activity, Fenton Reaction and DNA Damage Study. Int. J. Mol. Sci. 2021, 22, 1619. https://doi.org/10.3390/ijms22041619
Simunkova M, Barbierikova Z, Jomova K, Hudecova L, Lauro P, Alwasel SH, Alhazza I, Rhodes CJ, Valko M. Antioxidant vs. Prooxidant Properties of the Flavonoid, Kaempferol, in the Presence of Cu(II) Ions: A ROS-Scavenging Activity, Fenton Reaction and DNA Damage Study. International Journal of Molecular Sciences. 2021; 22(4):1619. https://doi.org/10.3390/ijms22041619
Chicago/Turabian StyleSimunkova, Miriama, Zuzana Barbierikova, Klaudia Jomova, Lenka Hudecova, Peter Lauro, Saleh H. Alwasel, Ibrahim Alhazza, Christopher J. Rhodes, and Marian Valko. 2021. "Antioxidant vs. Prooxidant Properties of the Flavonoid, Kaempferol, in the Presence of Cu(II) Ions: A ROS-Scavenging Activity, Fenton Reaction and DNA Damage Study" International Journal of Molecular Sciences 22, no. 4: 1619. https://doi.org/10.3390/ijms22041619
APA StyleSimunkova, M., Barbierikova, Z., Jomova, K., Hudecova, L., Lauro, P., Alwasel, S. H., Alhazza, I., Rhodes, C. J., & Valko, M. (2021). Antioxidant vs. Prooxidant Properties of the Flavonoid, Kaempferol, in the Presence of Cu(II) Ions: A ROS-Scavenging Activity, Fenton Reaction and DNA Damage Study. International Journal of Molecular Sciences, 22(4), 1619. https://doi.org/10.3390/ijms22041619







