Ca2+-Dependent Protein Kinase 6 Enhances KAT2 Shaker Channel Activity in Arabidopsis thaliana
Abstract
1. Introduction
2. Results
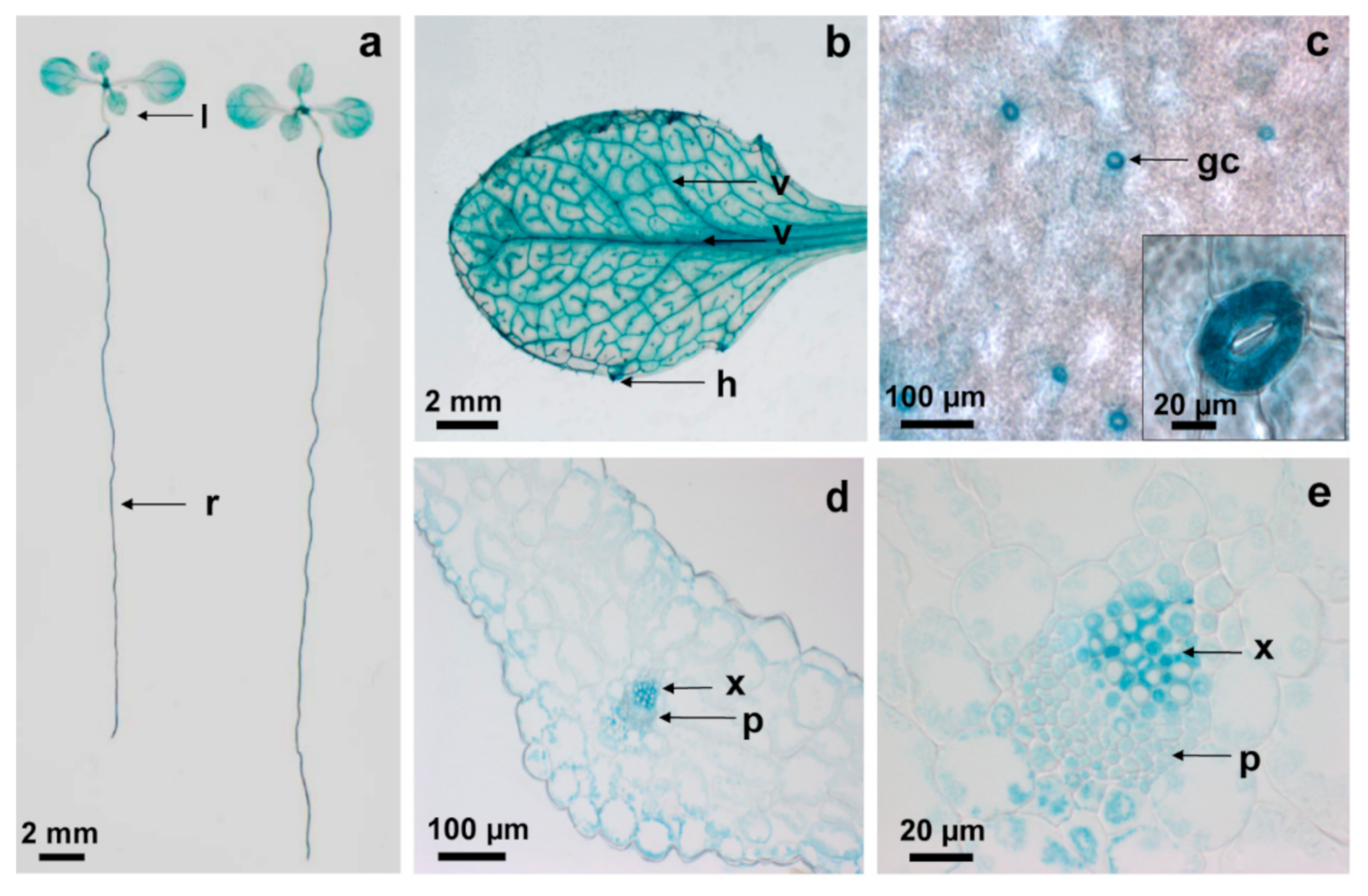
2.1. AtCPK6 Is Highly Expressed in Vascular Tissues
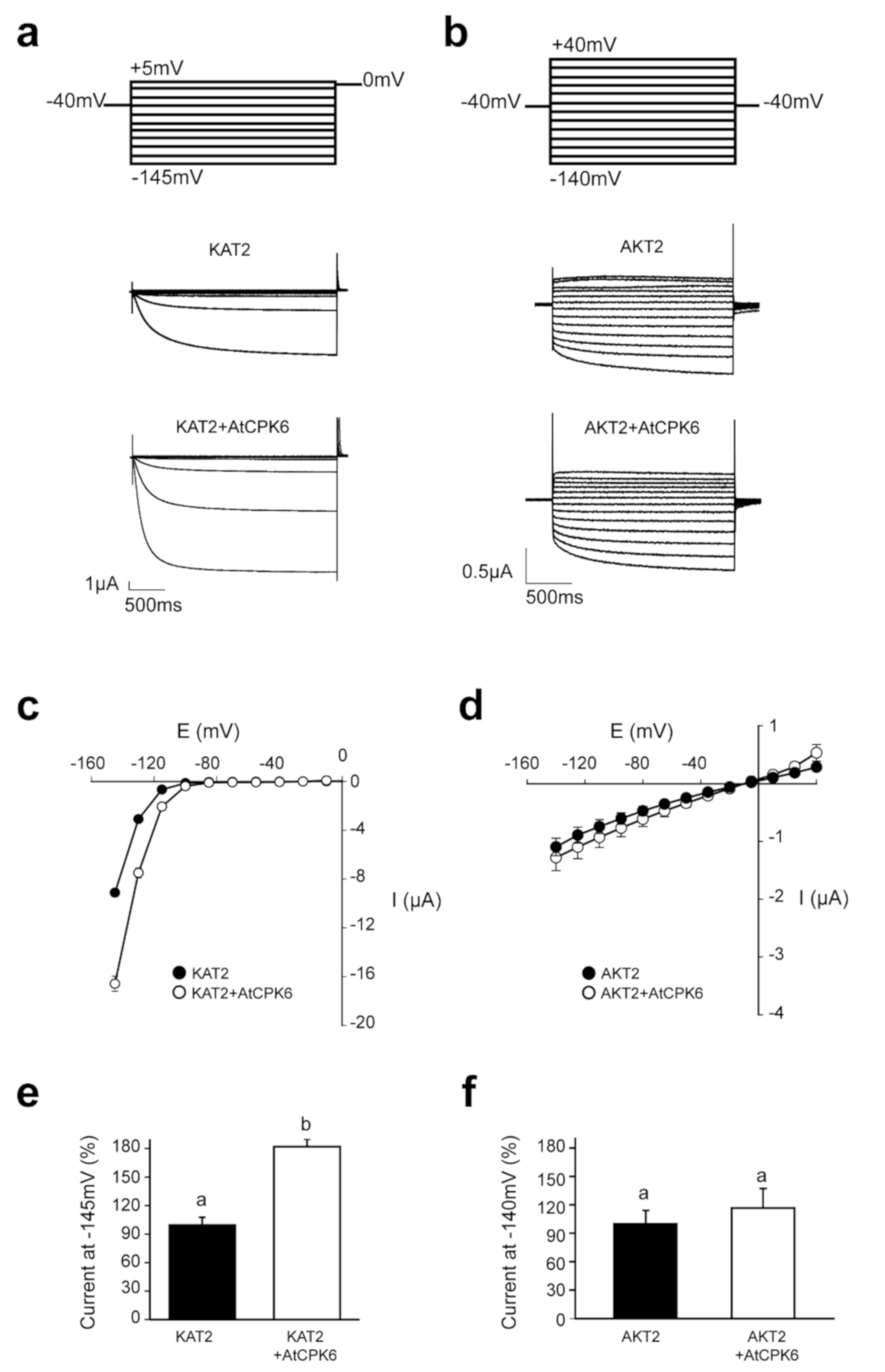
2.2. AtCPK6 Enhances Specifically KAT2 Channel Activity in Oocytes
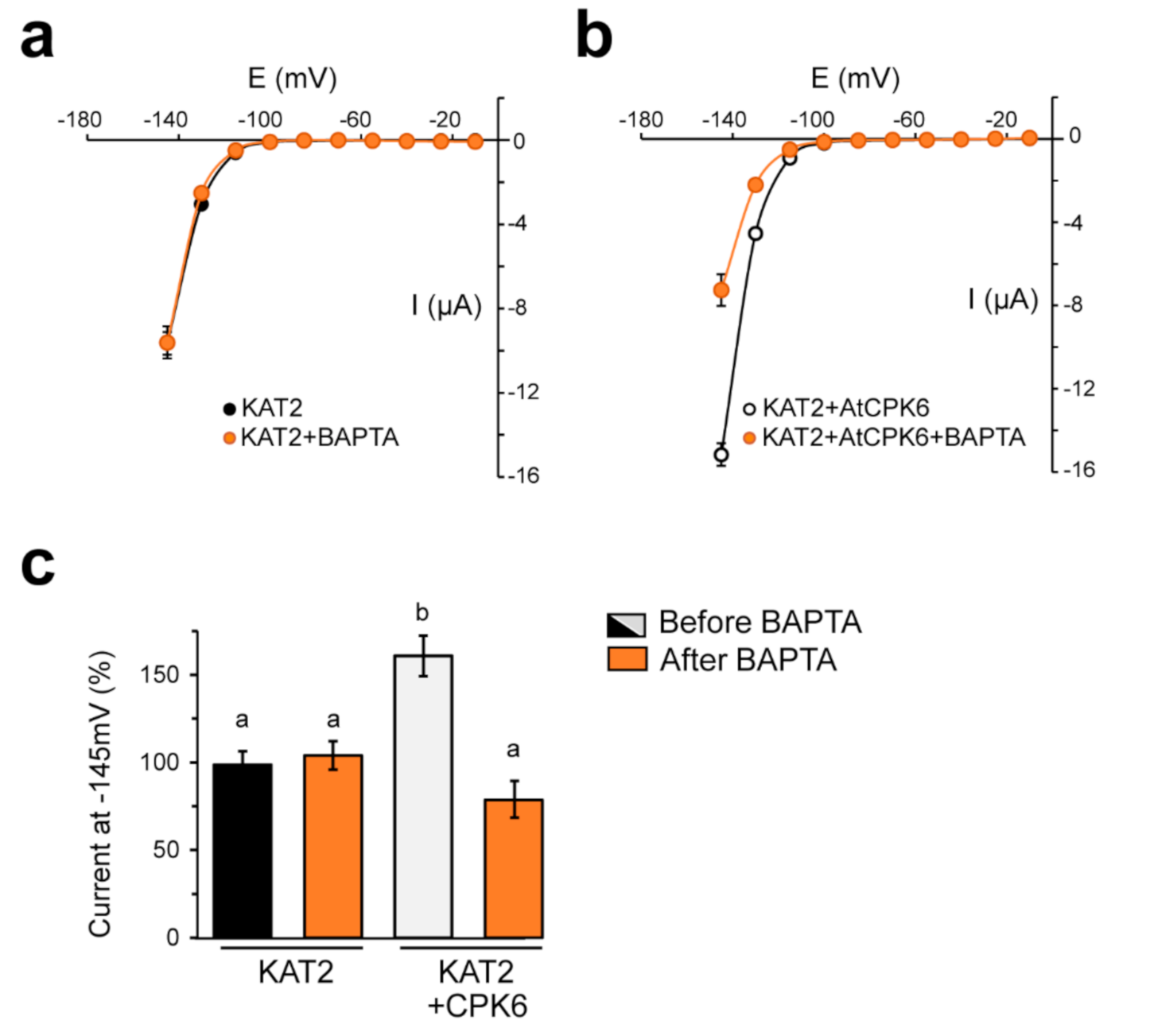
2.3. KAT2 Is Stimulated by AtCPK6 in Ca2+-Dependent Manner
2.4. AtCPK6 Physically Interacts with the KAT2 Subunit Channel in Planta
2.5. AtCPK6 Is Able to Phosphorylate KAT2 Peptides in the Presence of Free Ca2+
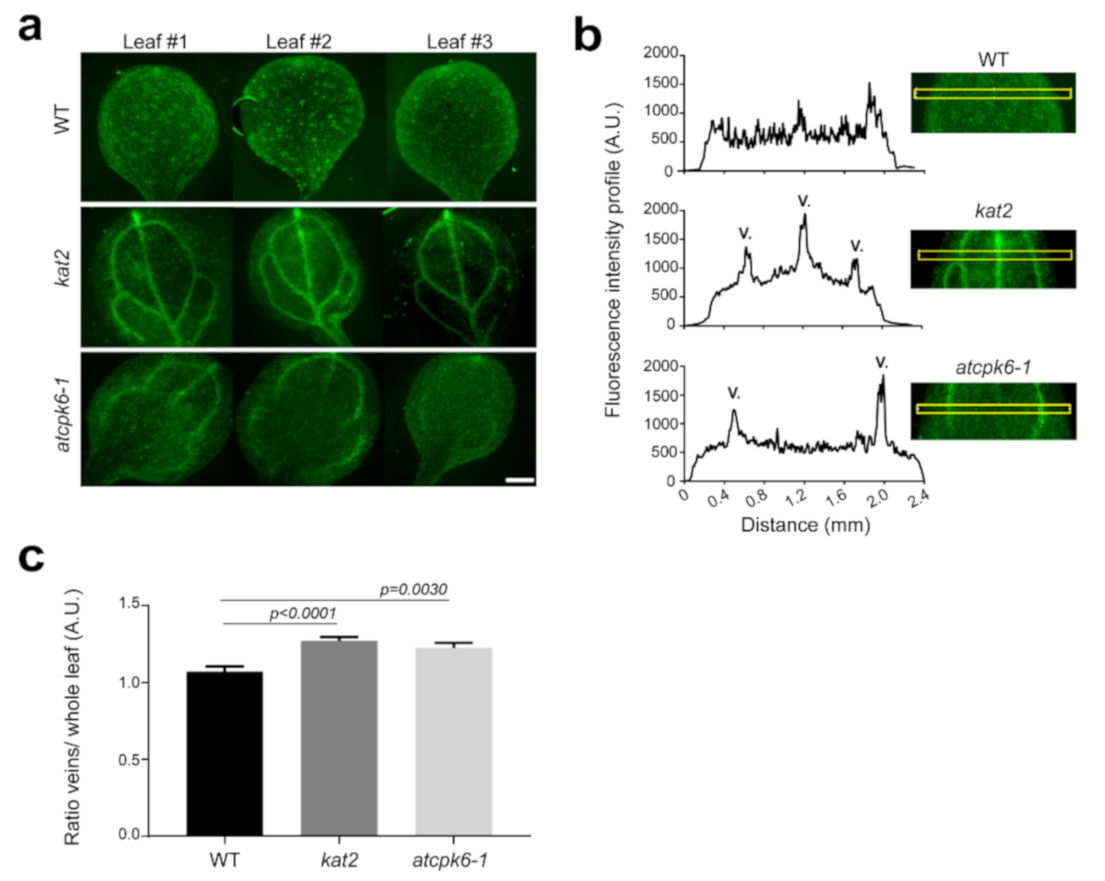
2.6. Potassium Distribution Is Perturbed in kat2 Knock-out Plants
3. Discussion
4. Materials and Methods
4.1. Wild-Type and Mutant Plants
4.2. Molecular Biology
4.3. Transgenic Plants
4.4. GUS Staining
4.5. Two Electrodes Voltage–Clamp (TEVC)
4.6. Fluorescence Lifetime Imaging (FLIM)
4.7. K+ Fluorescence Dye Staining
4.8. Recombinant Protein Production and Purification
4.9. In Vitro Phosphorylation
4.10. Peptide Array Phosphorylation
5. Conclusions and Perspective
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ache, P.; Becker, D.; Ivashikina, N.; Dietrich, P.; Roelfsema, M.R.; Hedrich, R. GORK, a delayed outward rectifier expressed in guard cells of Arabidopsis thaliana, is a K(+)-selective, K(+)-sensing ion channel. FEBS Lett. 2000, 486, 93–98. [Google Scholar] [CrossRef]
- Anderson, J.A.; Huprikar, S.S.; Kochian, L.V.; Lucas, W.J.; Gaber, R.F. Functional expression of a probable Arabidopsis thaliana potassium channel in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 1992, 89, 3736–3740. [Google Scholar] [CrossRef]
- Schachtman, D.P.; Schroeder, J.I.; Lucas, W.J.; Anderson, J.A.; Gaber, R.F. Expression of an inward-rectifying potassium channel by the Arabidopsis KAT1 cDNA. Science 1992, 258, 1654–1658. [Google Scholar] [CrossRef]
- Sentenac, H.; Bonneaud, N.; Minet, M.; Lacroute, F.; Salmon, J.M.; Gaymard, F.; Grignon, C. Cloning and expression in yeast of a plant potassium ion transport system. Science 1992, 256, 663–665. [Google Scholar] [CrossRef]
- Xicluna, J.; Lacombe, B.; Dreyer, I.; Alcon, C.; Jeanguenin, L.; Sentenac, H.; Thibaud, J.B.; Cherel, I. Increased functional diversity of plant K+ channels by preferential heteromerization of the shaker-like subunits AKT2 and KAT2. J. Biol. Chem. 2007, 282, 486–494. [Google Scholar] [CrossRef] [PubMed]
- Lacombe, B.; Pilot, G.; Michard, E.; Gaymard, F.; Sentenac, H.; Thibaud, J.B. A shaker-like K(+) channel with weak rectification is expressed in both source and sink phloem tissues of Arabidopsis. Plant Cell 2000, 12, 837–851. [Google Scholar] [CrossRef] [PubMed]
- Gajdanowicz, P.; Michard, E.; Sandmann, M.; Rocha, M.; Correa, L.G.; Ramirez-Aguilar, S.J.; Gomez-Porras, J.L.; Gonzalez, W.; Thibaud, J.B.; Van Dongen, J.T.; et al. Potassium (K+) gradients serve as a mobile energy source in plant vascular tissues. Proc. Natl. Acad. Sci. USA 2011, 108, 864–869. [Google Scholar] [CrossRef] [PubMed]
- Luan, S. The CBL-CIPK network in plant calcium signaling. Trends Plant Sci 2009, 14, 37–42. [Google Scholar] [CrossRef]
- Held, K.; Pascaud, F.; Eckert, C.; Gajdanowicz, P.; Hashimoto, K.; Corratge-Faillie, C.; Offenborn, J.N.; Lacombe, B.; Dreyer, I.; Thibaud, J.B.; et al. Calcium-dependent modulation and plasma membrane targeting of the AKT2 potassium channel by the CBL4/CIPK6 calcium sensor/protein kinase complex. Cell Res. 2011, 21, 1116–1130. [Google Scholar] [CrossRef]
- Pilot, G.; Gaymard, F.; Mouline, K.; Cherel, I.; Sentenac, H. Regulated expression of Arabidopsis shaker K+ channel genes involved in K+ uptake and distribution in the plant. Plant Mol. Biol. 2003, 51, 773–787. [Google Scholar] [CrossRef]
- Saito, S.; Uozumi, N. Calcium-Regulated Phosphorylation Systems Controlling Uptake and Balance of Plant Nutrients. Front Plant Sci. 2020, 11, 44. [Google Scholar] [CrossRef]
- Pilot, G.; Lacombe, B.; Gaymard, F.; Cherel, I.; Boucherez, J.; Thibaud, J.B.; Sentenac, H. Guard cell inward K+ channel activity in arabidopsis involves expression of the twin channel subunits KAT1 and KAT2. J. Biol. Chem. 2001, 276, 3215–3221. [Google Scholar] [CrossRef]
- Philippar, K.; Ivashikina, N.; Ache, P.; Christian, M.; Luthen, H.; Palme, K.; Hedrich, R. Auxin activates KAT1 and KAT2, two K+-channel genes expressed in seedlings of Arabidopsis thaliana. Plant J. 2004, 37, 815–827. [Google Scholar] [CrossRef]
- Sathyanarayanan, P.V.; Poovaiah, B.W. Decoding Ca2+ signals in plants. CRC Crit. Rev. Plant Sci. 2004, 23, 1–11. [Google Scholar] [CrossRef]
- Hetherington, A.M.; Brownlee, C. The generation of Ca(2+) signals in plants. Annu. Rev. Plant Biol. 2004, 55, 401–427. [Google Scholar] [CrossRef]
- Hashimoto, K.; Kudla, J. Calcium decoding mechanisms in plants. Biochimie 2011, 93, 2054–2059. [Google Scholar] [CrossRef] [PubMed]
- Ranty, B.; Aldon, D.; Cotelle, V.; Galaud, J.P.; Thuleau, P.; Mazars, C. Calcium Sensors as Key Hubs in Plant Responses to Biotic and Abiotic Stresses. Front Plant Sci. 2016, 7, 327. [Google Scholar] [CrossRef] [PubMed]
- Anil, V.S.; Harmon, A.C.; Rao, K.S. Spatio-temporal accumulation and activity of calcium-dependent protein kinases during embryogenesis, seed development, and germination in sandalwood. Plant Physiol. 2000, 122, 1035–1043. [Google Scholar] [CrossRef]
- Valmonte, G.R.; Arthur, K.; Higgins, C.M.; MacDiarmid, R.M. Calcium-dependent protein kinases in plants: Evolution, expression and function. Plant Cell Physiol. 2014, 55, 551–569. [Google Scholar] [CrossRef] [PubMed]
- Atif, R.M.; Shahid, L.; Waqas, M.; Ali, B.; Rashid, M.A.R.; Azeem, F.; Nawaz, M.A.; Wani, S.H.; Chung, G. Insights on Calcium-Dependent Protein Kinases (CPKs) Signaling for Abiotic Stress Tolerance in Plants. Int. J. Mol. Sci. 2019, 20, 5298. [Google Scholar] [CrossRef]
- Boudsocq, M.; Sheen, J. CDPKs in immune and stress signaling. Trends Plant Sci. 2013, 18, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Fantino, E.; Segretin, M.E.; Santin, F.; Mirkin, F.G.; Ulloa, R.M. Analysis of the potato calcium-dependent protein kinase family and characterization of StCDPK7, a member induced upon infection with Phytophthora infestans. Plant Cell Rep. 2017, 36, 1137–1157. [Google Scholar] [CrossRef]
- Asano, T.; Hayashi, N.; Kobayashi, M.; Aoki, N.; Miyao, A.; Mitsuhara, I.; Ichikawa, H.; Komatsu, S.; Hirochika, H.; Kikuchi, S.; et al. A rice calcium-dependent protein kinase OsCPK12 oppositely modulates salt-stress tolerance and blast disease resistance. Plant J. 2012, 69, 26–36. [Google Scholar] [CrossRef] [PubMed]
- Bundo, M.; Coca, M. Enhancing blast disease resistance by overexpression of the calcium-dependent protein kinase OsCPK4 in rice. Plant Biotechnol. J. 2016, 14, 1357–1367. [Google Scholar] [CrossRef]
- Ye, W.; Muroyama, D.; Munemasa, S.; Nakamura, Y.; Mori, I.C.; Murata, Y. Calcium-dependent protein kinase CPK6 positively functions in induction by yeast elicitor of stomatal closure and inhibition by yeast elicitor of light-induced stomatal opening in Arabidopsis. Plant Physiol. 2013, 163, 591–599. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Xu, C.; Zhu, Y.; Zhang, L.; Chen, T.; Zhou, F.; Chen, H.; Lin, Y. The calcium-dependent kinase OsCPK24 functions in cold stress responses in rice. J. Integr. Plant Biol. 2018, 60, 173–188. [Google Scholar] [CrossRef]
- Saijo, Y.; Hata, S.; Kyozuka, J.; Shimamoto, K.; Izui, K. Over-expression of a single Ca2+-dependent protein kinase confers both cold and salt/drought tolerance on rice plants. Plant J. 2000, 23, 319–327. [Google Scholar] [CrossRef]
- Xu, J.; Tian, Y.S.; Peng, R.H.; Xiong, A.S.; Zhu, B.; Jin, X.F.; Gao, F.; Fu, X.Y.; Hou, X.L.; Yao, Q.H. AtCPK6, a functionally redundant and positive regulator involved in salt/drought stress tolerance in Arabidopsis. Planta 2010, 231, 1251–1260. [Google Scholar] [CrossRef] [PubMed]
- Franz, S.; Ehlert, B.; Liese, A.; Kurth, J.; Cazale, A.C.; Romeis, T. Calcium-dependent protein kinase CPK21 functions in abiotic stress response in Arabidopsis thaliana. Mol. Plant 2011, 4, 83–96. [Google Scholar] [CrossRef] [PubMed]
- Chehab, E.W.; Patharkar, O.R.; Hegeman, A.D.; Taybi, T.; Cushman, J.C. Autophosphorylation and subcellular localization dynamics of a salt- and water deficit-induced calcium-dependent protein kinase from ice plant. Plant Physiol. 2004, 135, 1430–1446. [Google Scholar] [CrossRef] [PubMed]
- Corratge-Faillie, C.; Ronzier, E.; Sanchez, F.; Prado, K.; Kim, J.H.; Lanciano, S.; Leonhardt, N.; Lacombe, B.; Xiong, T.C. The Arabidopsis guard cell outward potassium channel GORK is regulated by CPK33. FEBS Lett. 2017, 591, 1982–1992. [Google Scholar] [CrossRef]
- Geiger, D.; Scherzer, S.; Mumm, P.; Marten, I.; Ache, P.; Matschi, S.; Liese, A.; Wellmann, C.; Al-Rasheid, K.A.; Grill, E.; et al. Guard cell anion channel SLAC1 is regulated by CDPK protein kinases with distinct Ca2+ affinities. Proc. Natl. Acad. Sci. USA 2010, 107, 8023–8028. [Google Scholar] [CrossRef] [PubMed]
- Gutermuth, T.; Lassig, R.; Portes, M.T.; Maierhofer, T.; Romeis, T.; Borst, J.W.; Hedrich, R.; Feijo, J.A.; Konrad, K.R. Pollen tube growth regulation by free anions depends on the interaction between the anion channel SLAH3 and calcium-dependent protein kinases CPK2 and CPK20. Plant Cell 2013, 25, 4525–4543. [Google Scholar] [CrossRef] [PubMed]
- Jaborsky, M.; Maierhofer, T.; Olbrich, A.; Escalante-Perez, M.; Muller, H.M.; Simon, J.; Krol, E.; Cuin, T.A.; Fromm, J.; Ache, P.; et al. SLAH3-type anion channel expressed in poplar secretory epithelia operates in calcium kinase CPK-autonomous manner. New Phytol. 2016, 210, 922–933. [Google Scholar] [CrossRef]
- Ronzier, E.; Corratge-Faillie, C.; Sanchez, F.; Prado, K.; Briere, C.; Leonhardt, N.; Thibaud, J.B.; Xiong, T.C. CPK13, a noncanonical Ca2+-dependent protein kinase, specifically inhibits KAT2 and KAT1 shaker K+ channels and reduces stomatal opening. Plant Physiol. 2014, 166, 314–326. [Google Scholar] [CrossRef]
- Li, J.; Lee, Y.R.; Assmann, S.M. Guard cells possess a calcium-dependent protein kinase that phosphorylates the KAT1 potassium channel. Plant Physiol 1998, 116, 785–795. [Google Scholar] [CrossRef] [PubMed]
- Berkowitz, G.; Zhang, X.; Mercie, R.; Leng, Q.; Lawton, M. Co-expression of calcium-dependent protein kinase with the inward rectified guard cell K+ channel KAT1 alters current parameters in Xenopus laevis oocytes. Plant Cell Physiol. 2000, 41, 785–790. [Google Scholar] [CrossRef]
- Brandt, B.; Brodsky, D.E.; Xue, S.; Negi, J.; Iba, K.; Kangasjarvi, J.; Ghassemian, M.; Stephan, A.B.; Hu, H.; Schroeder, J.I. Reconstitution of abscisic acid activation of SLAC1 anion channel by CPK6 and OST1 kinases and branched ABI1 PP2C phosphatase action. Proc. Natl. Acad. Sci. USA 2012, 109, 10593–10598. [Google Scholar] [CrossRef]
- Boudsocq, M.; Droillard, M.J.; Regad, L.; Lauriere, C. Characterization of Arabidopsis calcium-dependent protein kinases: Activated or not by calcium? Biochem. J. 2012, 447, 291–299. [Google Scholar] [CrossRef]
- Kawamoto, N.; Sasabe, M.; Endo, M.; Machida, Y.; Araki, T. Calcium-dependent protein kinases responsible for the phosphorylation of a bZIP transcription factor FD crucial for the florigen complex formation. Sci. Rep. 2015, 5, 8341. [Google Scholar] [CrossRef]
- Pollok, B.A.; Heim, R. Using GFP in FRET-based applications. Trends Cell Biol. 1999, 9, 57–60. [Google Scholar] [CrossRef]
- Lebaudy, A.; Pascaud, F.; Very, A.A.; Alcon, C.; Dreyer, I.; Thibaud, J.B.; Lacombe, B. Preferential KAT1-KAT2 heteromerization determines inward K+ current properties in Arabidopsis guard cells. J. Biol. Chem. 2010, 285, 6265–6274. [Google Scholar] [CrossRef] [PubMed]
- Xiong, T.C.; Ronzier, E.; Sanchez, F.; Corratge-Faillie, C.; Mazars, C.; Thibaud, J.B. Imaging long distance propagating calcium signals in intact plant leaves with the BRET-based GFP-aequorin reporter. Front Plant Sci. 2014, 5, 43. [Google Scholar] [CrossRef]
- Whalley, H.J.; Sargeant, A.W.; Steele, J.F.; Lacoere, T.; Lamb, R.; Saunders, N.J.; Knight, H.; Knight, M.R. Transcriptomic analysis reveals calcium regulation of specific promoter motifs in Arabidopsis. Plant Cell 2011, 23, 4079–4095. [Google Scholar] [CrossRef]
- Earley, K.W.; Haag, J.R.; Pontes, O.; Opper, K.; Juehne, T.; Song, K.; Pikaard, C.S. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 2006, 45, 616–629. [Google Scholar] [CrossRef]
- Nakagawa, T.; Suzuki, T.; Murata, S.; Nakamura, S.; Hino, T.; Maeo, K.; Tabata, R.; Kawai, T.; Tanaka, K.; Niwa, Y.; et al. Improved Gateway binary vectors: High-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci. Biotechnol. Biochem. 2007, 71, 2095–2100. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Chen, Z.H.; Liu, X.; Colmer, T.D.; Zhou, M.; Shabala, S. Tissue-specific root ion profiling reveals essential roles of the CAX and ACA calcium transport systems in response to hypoxia in Arabidopsis. J. Exp. Bot. 2016, 67, 3747–3762. [Google Scholar] [CrossRef]
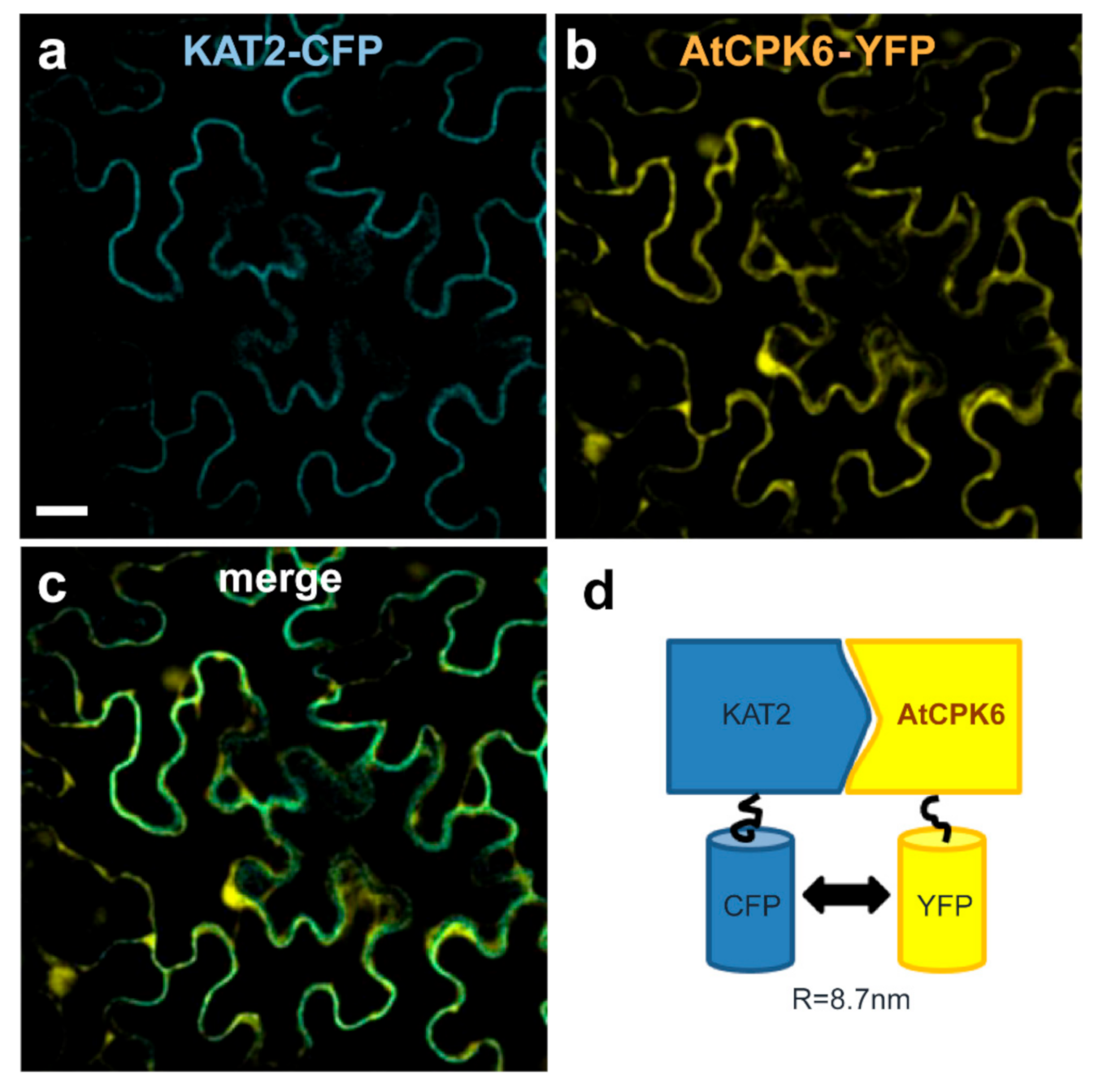

| Fluorescence Lifetime of CFP (ns) ± s.e. | Change in CFP Fluorescence Lifetime (%) | Number of Measures | |
|---|---|---|---|
| KAT2-CFP | 2.23 ± 0.02 | -- | 22 |
| KAT2-CFP + AtCPK6-YFP | 2.05 ± 0.03 | 8.1 | 25 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ronzier, E.; Corratgé-Faillie, C.; Sanchez, F.; Brière, C.; Xiong, T.C. Ca2+-Dependent Protein Kinase 6 Enhances KAT2 Shaker Channel Activity in Arabidopsis thaliana. Int. J. Mol. Sci. 2021, 22, 1596. https://doi.org/10.3390/ijms22041596
Ronzier E, Corratgé-Faillie C, Sanchez F, Brière C, Xiong TC. Ca2+-Dependent Protein Kinase 6 Enhances KAT2 Shaker Channel Activity in Arabidopsis thaliana. International Journal of Molecular Sciences. 2021; 22(4):1596. https://doi.org/10.3390/ijms22041596
Chicago/Turabian StyleRonzier, Elsa, Claire Corratgé-Faillie, Frédéric Sanchez, Christian Brière, and Tou Cheu Xiong. 2021. "Ca2+-Dependent Protein Kinase 6 Enhances KAT2 Shaker Channel Activity in Arabidopsis thaliana" International Journal of Molecular Sciences 22, no. 4: 1596. https://doi.org/10.3390/ijms22041596
APA StyleRonzier, E., Corratgé-Faillie, C., Sanchez, F., Brière, C., & Xiong, T. C. (2021). Ca2+-Dependent Protein Kinase 6 Enhances KAT2 Shaker Channel Activity in Arabidopsis thaliana. International Journal of Molecular Sciences, 22(4), 1596. https://doi.org/10.3390/ijms22041596







