DNA Methylation in T-Cell Acute Lymphoblastic Leukemia: In Search for Clinical and Biological Meaning
Abstract
1. Introduction
DNA Methylation: Basic Aspects and Biological Roles
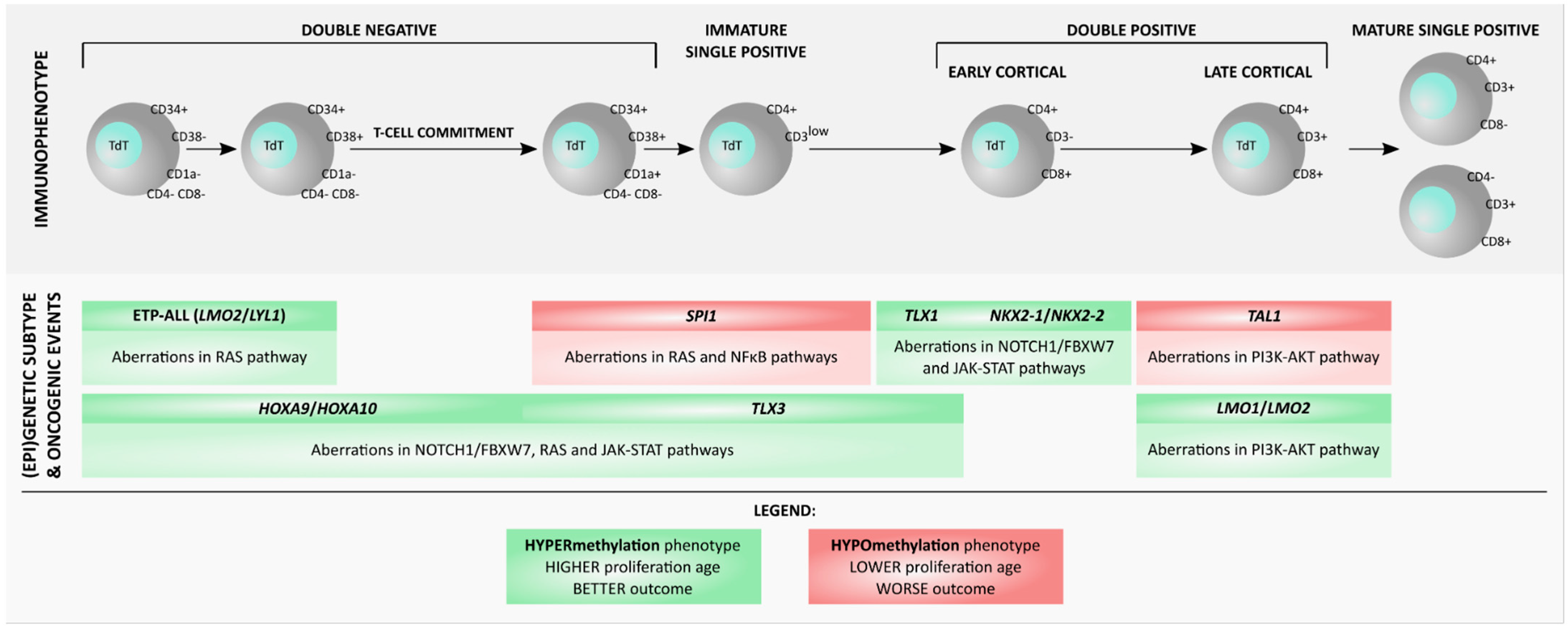
2. DNA Methylation in Normal Hematopoiesis and T-Cell Development
2.1. DNA Methylation Writers Involved in T-Cell Development and Malignant Transformation
2.2. DNA Methylation Erasers Involved in T-Cell Development and Malignant Transformation
3. Investigating Methylation Phenotypes in T-ALL Patients: In Search for Clinical Meaning
4. Investigating Mechanisms behind Methylation Phenotypes: In Search for Biological Meaning
5. Summary and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Feinberg, A.P.; Koldobskiy, M.A.; Göndör, A. Epigenetic modulators, modifiers and mediators in cancer aetiology and progression. Nat. Rev. Genet. 2016, 17, 284–299. [Google Scholar] [CrossRef]
- Greenberg, M.V.C.; Bourc’his, D. The diverse roles of DNA methylation in mammalian development and disease. Nat. Rev. Mol. Cell Biol. 2019, 20, 590–607. [Google Scholar] [CrossRef]
- Belver, L.; Ferrando, A. The genetics and mechanisms of T cell acute lymphoblastic leukaemia. Nat. Rev. Cancer 2016, 16, 494–507. [Google Scholar] [CrossRef]
- Gujar, H.; Weisenberger, D.J.; Liang, G. The Roles of Human DNA Methyltransferases and Their Isoforms in Shaping the Epigenome. Genes 2019, 10, 172. [Google Scholar] [CrossRef]
- Riggs, A.D. X inactivation, differentiation, and DNA methylation. Cytogenet. Cell Genet. 1975, 14, 9–25. [Google Scholar] [CrossRef]
- Patil, V.; Ward, R.L.; Hesson, L.B. The evidence for functional non-CpG methylation in mammalian cells. Epigenetics 2014, 9, 823–828. [Google Scholar] [CrossRef]
- Patil, V.; Cuenin, C.; Chung, F.; Aguilera, J.R.R.; Fernandez-Jimenez, N.; Romero-Garmendia, I.; Bilbao, J.R.; Cahais, V.; Rothwell, J.; Herceg, Z. Human mitochondrial DNA is extensively methylated in a non-CpG context. Nucleic Acids Res. 2019, 47, 10072–10085. [Google Scholar] [CrossRef]
- Rošić, S.; Amouroux, R.; Requena, C.E.; Gomes, A.; Emperle, M.; Beltran, T.; Rane, J.K.; Linnett, S.; Selkirk, M.E.; Schiffer, P.H.; et al. Evolutionary analysis indicates that DNA alkylation damage is a byproduct of cytosine DNA methyltransferase activity. Nat. Genet. 2018, 50, 452–459. [Google Scholar] [CrossRef]
- He, Y.F.; Li, B.Z.; Li, Z.; Liu, P.; Wang, Y.; Tang, Q.; Ding, J.; Jia, Y.; Chen, Z.; Li, L.; et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 2011, 333, 1303–1307. [Google Scholar] [CrossRef]
- Ito, S.; D’Alessio, A.C.; Taranova, O.V.; Hong, K.; Sowers, L.C.; Zhang, Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 2010, 466, 1129–1133. [Google Scholar] [CrossRef]
- Wu, X.; Zhang, Y. TET-mediated active DNA demethylation: Mechanism, function and beyond. Nat. Rev. Genet. 2017, 18, 517–534. [Google Scholar] [CrossRef] [PubMed]
- Lorsbach, R.B.; Moore, J.; Mathew, S.; Raimondi, S.C.; Mukatira, S.T.; Downing, J.R. TET1, a member of a novel protein family, is fused to MLL in acute myeloid leukemia containing the t(10;11)(q22;q23). Leukemia 2003, 17, 637–641. [Google Scholar] [CrossRef] [PubMed]
- Tahiliani, M.; Koh, K.P.; Shen, Y.; Pastor, W.A.; Bandukwala, H.; Brudno, Y.; Agarwal, S.; Iyer, L.M.; Liu, D.R.; Aravind, L.; et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 2009, 324, 930–935. [Google Scholar] [CrossRef]
- Ito, S.; Shen, L.; Dai, Q.; Wu, S.C.; Collins, L.B.; Swenberg, J.A.; He, C.; Zhang, Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 2011, 333, 1300–1303. [Google Scholar] [CrossRef]
- Maiti, A.; Drohat, A.C. Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: Potential implications for active demethylation of CpG sites. J. Biol. Chem. 2011, 286, 35334–35338. [Google Scholar] [CrossRef]
- Bird, A.P.; Taggart, M.H. Variable patterns of total DNA and rDNA methylation in animals. Nucleic Acids Res. 1980, 8, 1485–1497. [Google Scholar] [CrossRef]
- Cooper, D.N.; Krawczak, M. Cytosine methylation and the fate of CpG dinucleotides in vertebrate genomes. Hum. Genet. 1989, 83, 181–188. [Google Scholar] [CrossRef]
- Gardiner-Garden, M.; Frommer, M. CpG islands in vertebrate genomes. J. Mol. Biol. 1987, 196, 261–282. [Google Scholar] [CrossRef]
- Straussman, R.; Nejman, D.; Roberts, D.; Steinfeld, I.; Blum, B.; Benvenisty, N.; Simon, I.; Yakhini, Z.; Cedar, H. Developmental programming of CpG island methylation profiles in the human genome. Nat. Struct. Mol. Biol. 2009, 16, 564–571. [Google Scholar] [CrossRef]
- Maunakea, A.K.; Nagarajan, R.P.; Bilenky, M.; Ballinger, T.J.; D’Souza, C.; Fouse, S.D.; Johnson, B.E.; Hong, C.; Nielsen, C.; Zhao, Y.; et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature 2010, 466, 253–257. [Google Scholar] [CrossRef]
- Maunakea, A.K.; Chepelev, I.; Cui, K.; Zhao, K. Intragenic DNA methylation modulates alternative splicing by recruiting MeCP2 to promote exon recognition. Cell Res. 2013, 23, 1256–1269. [Google Scholar] [CrossRef] [PubMed]
- Angeloni, A.; Bogdanovic, O. Enhancer DNA methylation: Implications for gene regulation. Essays Biochem. 2019, 63, 707–715. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.; Tan, P.P.; Portales-Casamar, E.; Pavlidis, P. Meta-analysis of human methylomes reveals stably methylated sequences surrounding CpG islands associated with high gene expression. Epigenet. Chromatin 2014, 7, 28. [Google Scholar] [CrossRef] [PubMed]
- Irizarry, R.A.; Ladd-Acosta, C.; Wen, B.; Wu, Z.; Montano, C.; Onyango, P.; Cui, H.; Gabo, K.; Rongione, M.; Webster, M.; et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat. Genet. 2009, 41, 178–186. [Google Scholar] [CrossRef]
- Ira, G. CpG islands--the only unmethylated fragments of DNA in the vertebrate genome. Postepy Biochem. 1997, 43, 189–198. [Google Scholar]
- Li, E.; Zhang, Y. DNA methylation in mammals. Cold Spring Harb. Perspect. Biol. 2014, 6, a019133. [Google Scholar] [CrossRef]
- Lister, R.; Pelizzola, M.; Dowen, R.H.; Hawkins, R.D.; Hon, G.; Tonti-Filippini, J.; Nery, J.R.; Lee, L.; Ye, Z.; Ngo, Q.M.; et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 2009, 462, 315–322. [Google Scholar] [CrossRef]
- Reik, W.; Collick, A.; Norris, M.L.; Barton, S.C.; Surani, M.A. Genomic imprinting determines methylation of parental alleles in transgenic mice. Nature 1987, 328, 248–251. [Google Scholar] [CrossRef]
- Blecua, P.; Martinez-Verbo, L.; Esteller, M. The DNA methylation landscape of hematological malignancies: An update. Mol. Oncol. 2020, 14, 1616–1639. [Google Scholar] [CrossRef]
- Baylin, S.B.; Jones, P.A. A decade of exploring the cancer epigenome—Biological and translational implications. Nat. Rev. Cancer 2011, 11, 726–734. [Google Scholar] [CrossRef]
- Yu, V.W.C.; Yusuf, R.Z.; Oki, T.; Wu, J.; Saez, B.; Wang, X.; Cook, C.; Baryawno, N.; Ziller, M.J.; Lee, E.; et al. Epigenetic Memory Underlies Cell-Autonomous Heterogeneous Behavior of Hematopoietic Stem Cells. Cell 2016, 167, 1310–1322.e17. [Google Scholar] [CrossRef] [PubMed]
- Ji, H.; Ehrlich, L.I.; Seita, J.; Murakami, P.; Doi, A.; Lindau, P.; Lee, H.; Aryee, M.J.; Irizarry, R.A.; Kim, K.; et al. Comprehensive methylome map of lineage commitment from haematopoietic progenitors. Nature 2010, 467, 338–342. [Google Scholar] [CrossRef] [PubMed]
- Chiang, Y.J.; Hodes, R.J. T-cell development is regulated by the coordinated function of proximal and distal Lck promoters active at different developmental stages. Eur. J. Immunol. 2016, 46, 2401–2408. [Google Scholar] [CrossRef] [PubMed]
- Lipka, D.B.; Wang, Q.; Cabezas-Wallscheid, N.; Klimmeck, D.; Weichenhan, D.; Herrmann, C.; Lier, A.; Brocks, D.; von Paleske, L.; Renders, S.; et al. Identification of DNA methylation changes at cis-regulatory elements during early steps of HSC differentiation using tagmentation-based whole genome bisulfite sequencing. Cell Cycle 2014, 13, 3476–3487. [Google Scholar] [CrossRef] [PubMed]
- Farlik, M.; Halbritter, F.; Müller, F.; Choudry, F.A.; Ebert, P.; Klughammer, J.; Farrow, S.; Santoro, A.; Ciaurro, V.; Mathur, A.; et al. DNA Methylation Dynamics of Human Hematopoietic Stem Cell Differentiation. Cell Stem Cell 2016, 19, 808–822. [Google Scholar] [CrossRef] [PubMed]
- Cabezas-Wallscheid, N.; Klimmeck, D.; Hansson, J.; Lipka, D.B.; Reyes, A.; Wang, Q.; Weichenhan, D.; Lier, A.; von Paleske, L.; Renders, S.; et al. Identification of regulatory networks in HSCs and their immediate progeny via integrated proteome, transcriptome, and DNA methylome analysis. Cell Stem Cell 2014, 15, 507–522. [Google Scholar] [CrossRef] [PubMed]
- Kramer, A.C.; Kothari, A.; Wilson, W.C.; Celik, H.; Nikitas, J.; Mallaney, C.; Ostrander, E.L.; Eultgen, E.; Martens, A.; Valentine, M.C.; et al. Dnmt3a regulates T-cell development and suppresses T-ALL transformation. Leukemia 2017, 31, 2479–2490. [Google Scholar] [CrossRef]
- Challen, G.A.; Sun, D.; Jeong, M.; Luo, M.; Jelinek, J.; Berg, J.S.; Bock, C.; Vasanthakumar, A.; Gu, H.; Xi, Y.; et al. Dnmt3a is essential for hematopoietic stem cell differentiation. Nat. Genet. 2011, 44, 23–31. [Google Scholar] [CrossRef]
- Pham, D.; Yu, Q.; Walline, C.C.; Muthukrishnan, R.; Blum, J.S.; Kaplan, M.H. Opposing roles of STAT4 and Dnmt3a in Th1 gene regulation. J. Immunol. 2013, 191, 902–911. [Google Scholar] [CrossRef]
- Grossmann, V.; Haferlach, C.; Weissmann, S.; Roller, A.; Schindela, S.; Poetzinger, F.; Stadler, K.; Bellos, F.; Kern, W.; Haferlach, T.; et al. The molecular profile of adult T-cell acute lymphoblastic leukemia: Mutations in RUNX1 and DNMT3A are associated with poor prognosis in T-ALL. Genes Chromosomes Cancer 2013, 52, 410–422. [Google Scholar] [CrossRef]
- Neumann, M.; Heesch, S.; Schlee, C.; Schwartz, S.; Gökbuget, N.; Hoelzer, D.; Konstandin, N.P.; Ksienzyk, B.; Vosberg, S.; Graf, A.; et al. Whole-exome sequencing in adult ETP-ALL reveals a high rate of DNMT3A mutations. Blood 2013, 121, 4749–4752. [Google Scholar] [CrossRef] [PubMed]
- Shiba, N.; Taki, T.; Park, M.J.; Shimada, A.; Sotomatsu, M.; Adachi, S.; Tawa, A.; Horibe, K.; Tsuchida, M.; Hanada, R.; et al. DNMT3A mutations are rare in childhood acute myeloid leukaemia, myelodysplastic syndromes and juvenile myelomonocytic leukaemia. Br. J. Haematol. 2012, 156, 413–414. [Google Scholar] [CrossRef] [PubMed]
- Paganin, M.; Pigazzi, M.; Bresolin, S.; Masetti, R.; Fagioli, F.; Chiaretti, S.; Cazzaniga, G.; Locatelli, F.; Pession, A.; te Kronnie, G.; et al. DNA methyltransferase 3a hot-spot locus is not mutated in pediatric patients affected by acute myeloid or T-cell acute lymphoblastic leukemia: An Italian study. Haematologica 2011, 96, 1886–1887. [Google Scholar] [CrossRef] [PubMed]
- Szarzyńska-Zawadzka, B.; Kosmalska, M.; Sędek, Ł.; Sonsala, A.; Twardoch, M.; Kowalczyk, J.R.; Szczepański, T.; Witt, M.; Dawidowska, M. Cost-effective screening of DNMT3A coding sequence identifies somatic mutation in pediatric T-cell acute lymphoblastic leukemia. Eur. J. Haematol. 2017, 99, 514–519. [Google Scholar] [CrossRef]
- Hayette, S.; Thomas, X.; Jallades, L.; Chabane, K.; Charlot, C.; Tigaud, I.; Gazzo, S.; Morisset, S.; Cornillet-Lefebvre, P.; Plesa, A.; et al. High DNA methyltransferase DNMT3B levels: A poor prognostic marker in acute myeloid leukemia. PLoS ONE 2012, 7, e51527. [Google Scholar] [CrossRef]
- Amara, K.; Ziadi, S.; Hachana, M.; Soltani, N.; Korbi, S.; Trimeche, M. DNA methyltransferase DNMT3b protein overexpression as a prognostic factor in patients with diffuse large B-cell lymphomas. Cancer Sci. 2010, 101, 1722–1730. [Google Scholar] [CrossRef]
- Trowbridge, J.J.; Snow, J.W.; Kim, J.; Orkin, S.H. DNA methyltransferase 1 is essential for and uniquely regulates hematopoietic stem and progenitor cells. Cell Stem Cell 2009, 5, 442–449. [Google Scholar] [CrossRef]
- Bröske, A.M.; Vockentanz, L.; Kharazi, S.; Huska, M.R.; Mancini, E.; Scheller, M.; Kuhl, C.; Enns, A.; Prinz, M.; Jaenisch, R.; et al. DNA methylation protects hematopoietic stem cell multipotency from myeloerythroid restriction. Nat. Genet. 2009, 41, 1207–1215. [Google Scholar] [CrossRef]
- Lee, P.P.; Fitzpatrick, D.R.; Beard, C.; Jessup, H.K.; Lehar, S.; Makar, K.W.; Pérez-Melgosa, M.; Sweetser, M.T.; Schlissel, M.S.; Nguyen, S.; et al. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity 2001, 15, 763–774. [Google Scholar] [CrossRef]
- Poole, C.J.; Zheng, W.; Lodh, A.; Yevtodiyenko, A.; Liefwalker, D.; Li, H.; Felsher, D.W.; van Riggelen, J. DNMT3B overexpression contributes to aberrant DNA methylation and MYC-driven tumor maintenance in T-ALL and Burkitt’s lymphoma. Oncotarget 2017, 8, 76898–76920. [Google Scholar] [CrossRef]
- Christman, J.K. 5-Azacytidine and 5-aza-2′-deoxycytidine as inhibitors of DNA methylation: Mechanistic studies and their implications for cancer therapy. Oncogene 2002, 21, 5483–5495. [Google Scholar] [CrossRef] [PubMed]
- Roberti, A.; Valdes, A.F.; Torrecillas, R.; Fraga, M.F.; Fernandez, A.F. Epigenetics in cancer therapy and nanomedicine. Clin. Epigenet. 2019, 11, 81. [Google Scholar] [CrossRef] [PubMed]
- Palii, S.S.; Van Emburgh, B.O.; Sankpal, U.T.; Brown, K.D.; Robertson, K.D. DNA methylation inhibitor 5-Aza-2′-deoxycytidine induces reversible genome-wide DNA damage that is distinctly influenced by DNA methyltransferases 1 and 3B. Mol. Cell Biol. 2008, 28, 752–771. [Google Scholar] [CrossRef] [PubMed]
- Chiappinelli, K.B.; Strissel, P.L.; Desrichard, A.; Li, H.; Henke, C.; Akman, B.; Hein, A.; Rote, N.S.; Cope, L.M.; Snyder, A.; et al. Inhibiting DNA Methylation Causes an Interferon Response in Cancer via dsRNA Including Endogenous Retroviruses. Cell 2015, 162, 974–986. [Google Scholar] [CrossRef] [PubMed]
- Rahmat, L.T.; Nguyen, A.; Abdulhaq, H.; Prakash, S.; Logan, A.C.; Mannis, G.N. Venetoclax in Combination with Decitabine for Relapsed T-Cell Acute Lymphoblastic Leukemia after Allogeneic Hematopoietic Cell Transplant. Case Rep. Hematol. 2018, 2018, 6092646. [Google Scholar] [CrossRef]
- Cimmino, L.; Dawlaty, M.M.; Ndiaye-Lobry, D.; Yap, Y.S.; Bakogianni, S.; Yu, Y.; Bhattacharyya, S.; Shaknovich, R.; Geng, H.; Lobry, C.; et al. TET1 is a tumor suppressor of hematopoietic malignancy. Nat. Immunol. 2015, 16, 653–662. [Google Scholar] [CrossRef]
- Bamezai, S.; Demir, D.; Pulikkottil, A.J.; Ciccarone, F.; Fischbein, E.; Sinha, A.; Borga, C.; Te Kronnie, G.; Meyer, L.H.; Mohr, F.; et al. TET1 promotes growth of T-cell acute lymphoblastic leukemia and can be antagonized via PARP inhibition. Leukemia 2020. [Google Scholar] [CrossRef]
- Poole, C.J.; Lodh, A.; Choi, J.H.; van Riggelen, J. MYC deregulates TET1 and TET2 expression to control global DNA (hydroxy)methylation and gene expression to maintain a neoplastic phenotype in T-ALL. Epigenet. Chromatin 2019, 12, 41. [Google Scholar] [CrossRef]
- Delhommeau, F.; Dupont, S.; Della Valle, V.; James, C.; Trannoy, S.; Massé, A.; Kosmider, O.; Le Couedic, J.P.; Robert, F.; Alberdi, A.; et al. Mutation in TET2 in myeloid cancers. N. Engl. J. Med. 2009, 360, 2289–2301. [Google Scholar] [CrossRef]
- Ko, M.; Huang, Y.; Jankowska, A.M.; Pape, U.J.; Tahiliani, M.; Bandukwala, H.S.; An, J.; Lamperti, E.D.; Koh, K.P.; Ganetzky, R.; et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature 2010, 468, 839–843. [Google Scholar] [CrossRef]
- Pronier, E.; Delhommeau, F. Role of TET2 mutations in myeloproliferative neoplasms. Curr. Hematol. Malig. Rep. 2012, 7, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Kalender Atak, Z.; De Keersmaecker, K.; Gianfelici, V.; Geerdens, E.; Vandepoel, R.; Pauwels, D.; Porcu, M.; Lahortiga, I.; Brys, V.; Dirks, W.G.; et al. High accuracy mutation detection in leukemia on a selected panel of cancer genes. PLoS ONE 2012, 7, e38463. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.G.; Cho, S.Y.; Kim, M.J.; Oh, S.H.; Cho, E.H.; Lee, S.; Baek, E.J.; Choi, J.H.; Bohlander, S.K.; Lode, L.; et al. Genomic breakpoints and clinical features of MLL-TET1 rearrangement in acute leukemias. Haematologica 2013, 98, e55–e57. [Google Scholar] [CrossRef] [PubMed]
- Ciccarone, F.; Valentini, E.; Bacalini, M.G.; Zampieri, M.; Calabrese, R.; Guastafierro, T.; Mariano, G.; Reale, A.; Franceschi, C.; Caiafa, P. Poly(ADP-ribosyl)ation is involved in the epigenetic control of TET1 gene transcription. Oncotarget 2014, 5, 10356–10367. [Google Scholar] [CrossRef] [PubMed]
- Scopim-Ribeiro, R.; Machado-Neto, J.A.; de Melo Campos, P.; Niemann, F.S.; Lorand-Metze, I.; Costa, F.F.; Olalla Saad, S.T.; Traina, F. Low Ten-eleven-translocation 2 (TET2) transcript level is independent of TET2 mutation in patients with myeloid neoplasms. Diagn. Pathol. 2016, 11, 28. [Google Scholar] [CrossRef] [PubMed]
- Marçais, A.; Waast, L.; Bruneau, J.; Hanssens, K.; Asnafi, V.; Gaulard, P.; Suarez, F.; Dubreuil, P.; Gessain, A.; Hermine, O.; et al. Adult T cell leukemia aggressivenness correlates with loss of both 5-hydroxymethylcytosine and TET2 expression. Oncotarget 2017, 8, 52256–52268. [Google Scholar] [CrossRef]
- Paz, M.F.; Fraga, M.F.; Avila, S.; Guo, M.; Pollan, M.; Herman, J.G.; Esteller, M. A systematic profile of DNA methylation in human cancer cell lines. Cancer Res. 2003, 63, 1114–1121. [Google Scholar]
- Pfeifer, G.P. Defining Driver DNA Methylation Changes in Human Cancer. Int. J. Mol. Sci. 2018, 19, 1166. [Google Scholar] [CrossRef]
- Shiramizu, B.; Mick, P. Epigenetic changes in the DAP-kinase CpG island in pediatric lymphoma. Med. Pediatr. Oncol. 2003, 41, 527–531. [Google Scholar] [CrossRef]
- Galm, O.; Wilop, S.; Lüders, C.; Jost, E.; Gehbauer, G.; Herman, J.G.; Osieka, R. Clinical implications of aberrant DNA methylation patterns in acute myelogenous leukemia. Ann. Hematol. 2005, 84 (Suppl. 1), 39–46. [Google Scholar] [CrossRef]
- Strathdee, G.; Sim, A.; Soutar, R.; Holyoake, T.L.; Brown, R. HOXA5 is targeted by cell-type-specific CpG island methylation in normal cells and during the development of acute myeloid leukaemia. Carcinogenesis 2007, 28, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Roman-Gomez, J.; Jimenez-Velasco, A.; Agirre, X.; Prosper, F.; Heiniger, A.; Torres, A. Lack of CpG island methylator phenotype defines a clinical subtype of T-cell acute lymphoblastic leukemia associated with good prognosis. J. Clin. Oncol. 2005, 23, 7043–7049. [Google Scholar] [CrossRef]
- Roman-Gomez, J. Lack of CpG Island Methylator Phenotype Defines a Clinical Subtype of T-Cell Acute Lymphoblastic Leukemia Associated With Good Prognosis. J. Clin. Oncol. 2013, 31, 979. [Google Scholar] [CrossRef]
- Roman-Gomez, J.; Jimenez-Velasco, A.; Agirre, X.; Castillejo, J.A.; Navarro, G.; Garate, L.; Jose-Eneriz, E.S.; Cordeu, L.; Barrios, M.; Prosper, F.; et al. Promoter hypermethylation and global hypomethylation are independent epigenetic events in lymphoid leukemogenesis with opposing effects on clinical outcome. Leukemia 2006, 20, 1445–1448. [Google Scholar] [CrossRef] [PubMed]
- Borssén, M.; Palmqvist, L.; Karrman, K.; Abrahamsson, J.; Behrendtz, M.; Heldrup, J.; Forestier, E.; Roos, G.; Degerman, S. Promoter DNA methylation pattern identifies prognostic subgroups in childhood T-cell acute lymphoblastic leukemia. PLoS ONE 2013, 8, e65373. [Google Scholar] [CrossRef]
- Borssén, M.; Haider, Z.; Landfors, M.; Norén-Nyström, U.; Schmiegelow, K.; Åsberg, A.E.; Kanerva, J.; Madsen, H.O.; Marquart, H.; Heyman, M.; et al. DNA Methylation Adds Prognostic Value to Minimal Residual Disease Status in Pediatric T-Cell Acute Lymphoblastic Leukemia. Pediatr. Blood Cancer 2016, 63, 1185–1192. [Google Scholar] [CrossRef]
- Kimura, S.; Seki, M.; Kawai, T.; Goto, H.; Yoshida, K.; Isobe, T.; Sekiguchi, M.; Watanabe, K.; Kubota, Y.; Nannya, Y.; et al. DNA methylation-based classification reveals difference between pediatric T-cell acute lymphoblastic leukemia and normal thymocytes. Leukemia 2020, 34, 1163–1168. [Google Scholar] [CrossRef]
- Touzart, A.; Boissel, N.; Belhocine, M.; Smith, C.; Graux, C.; Latiri, M.; Lhermitte, L.; Mathieu, E.L.; Huguet, F.; Lamant, L.; et al. Low level CpG island promoter methylation predicts a poor outcome in adult T-cell acute lymphoblastic leukemia. Haematologica 2020, 105, 1575–1581. [Google Scholar] [CrossRef]
- Roels, J.; Thénoz, M.; Szarzyńska, B.; Landfors, M.; De Coninck, S.; Demoen, L.; Provez, L.; Kuchmiy, A.; Strubbe, S.; Reunes, L.; et al. Aging of preleukemic thymocytes drives CpG island hypermethylation in T-cell acute lymphoblastic leukemia. Blood Cancer Discov. 2020, 1, 274–289. [Google Scholar] [CrossRef]
- Kraszewska, M.D.; Dawidowska, M.; Larmonie, N.S.; Kosmalska, M.; Sędek, Ł.; Szczepaniak, M.; Grzeszczak, W.; Langerak, A.W.; Szczepański, T.; Witt, M.; et al. DNA methylation pattern is altered in childhood T-cell acute lymphoblastic leukemia patients as compared with normal thymic subsets: Insights into CpG island methylator phenotype in T-ALL. Leukemia 2012, 26, 367–371. [Google Scholar] [CrossRef]
- De Smedt, R.; Morscio, J.; Goossens, S.; Van Vlierberghe, P. Targeting steroid resistance in T-cell acute lymphoblastic leukemia. Blood Rev. 2019, 38, 100591. [Google Scholar] [CrossRef] [PubMed]
- Kimura, S.; Mullighan, C.G. Molecular markers in ALL: Clinical implications. Best Pract. Res. Clin. Haematol. 2020, 33, 101193. [Google Scholar] [CrossRef] [PubMed]
- Haider, Z.; Larsson, P.; Landfors, M.; Köhn, L.; Schmiegelow, K.; Flaegstad, T.; Kanerva, J.; Heyman, M.; Hultdin, M.; Degerman, S. An integrated transcriptome analysis in T-cell acute lymphoblastic leukemia links DNA methylation subgroups to dysregulated TAL1 and ANTP homeobox gene expression. Cancer Med. 2019, 8, 311–324. [Google Scholar] [CrossRef] [PubMed]
- Clappier, E.; Collette, S.; Grardel, N.; Girard, S.; Suarez, L.; Brunie, G.; Kaltenbach, S.; Yakouben, K.; Mazingue, F.; Robert, A.; et al. NOTCH1 and FBXW7 mutations have a favorable impact on early response to treatment, but not on outcome, in children with T-cell acute lymphoblastic leukemia (T-ALL) treated on EORTC trials 58881 and 58951. Leukemia 2010, 24, 2023–2031. [Google Scholar] [CrossRef] [PubMed]
- Natarajan, V.; Bandapalli, O.R.; Rajkumar, T.; Sagar, T.G.; Karunakaran, N. NOTCH1 and FBXW7 mutations favor better outcome in pediatric South Indian T-cell acute lymphoblastic leukemia. J. Pediatr. Hematol. Oncol. 2015, 37, e23–e30. [Google Scholar] [CrossRef]
- Hu, Y.; Su, H.; Liu, C.; Wang, Z.; Huang, L.; Wang, Q.; Liu, S.; Chen, S.; Zhou, J.; Li, P.; et al. DEPTOR is a direct NOTCH1 target that promotes cell proliferation and survival in T-cell leukemia. Oncogene 2017, 36, 1038–1047. [Google Scholar] [CrossRef]
- Seki, M.; Kimura, S.; Isobe, T.; Yoshida, K.; Ueno, H.; Nakajima-Takagi, Y.; Wang, C.; Lin, L.; Kon, A.; Suzuki, H.; et al. Recurrent SPI1 (PU.1) fusions in high-risk pediatric T cell acute lymphoblastic leukemia. Nat. Genet. 2017, 49, 1274–1281. [Google Scholar] [CrossRef]
- Szarzyńska-Zawadzka, B.; Kunz, J.B.; Sędek, Ł.; Kosmalska, M.; Zdon, K.; Biecek, P.; Bandapalli, O.R.; Kraszewska-Hamilton, M.; Jaksik, R.; Drobna, M.; et al. PTEN abnormalities predict poor outcome in children with T-cell acute lymphoblastic leukemia treated according to ALL IC-BFM protocols. Am. J. Hematol. 2019, 94, E93–E96. [Google Scholar] [CrossRef]
| Ref. | T-ALL Patients (n) | Treatment Protocol | Classifier | Methylation Subgroup | Methylation Phenotype | Outcome | Survival and Relapse Rates # | Classifier Based on Genomic Regions | Method |
|---|---|---|---|---|---|---|---|---|---|
| Roman-Gomez et al. 2005 [72,73] | n = 50 (19 pediatric and 31 adults) | PETHEMA ALL-89 PETHEMA ALL-93 | CIMP | CIMP− | hypo | better | DFS12yr = 100% OS13yr = 91% | 27 selected gene promoters with potential or documented impact on ALL development | MS-PCR |
| CIMP+ | hyper | worse | DFS12yr = 20% OS13yr = 17% | ||||||
| Roman-Gomez et al. 2006 [74] | n = 307 ALL (T- and B-) | PETHEMA ALL-89 PETHEMA ALL-93 | CIMP | CIMP− | hypo | better | DFS12yr = 68% OS12yr = 63% | 39 selected gene promoters with potential or documented impact on ALL development | MS-PCR |
| CIMP+ | hyper | worse | DFS12yr = 32% OS12yr = 32% | ||||||
| Borssen et al. 2013 [75] | n = 43 (pediatric) | NOPHO ALL 1992 NOPHO ALL 2000 | CIMP | CIMP− | hypo | better | EFS5yr = 86% OS5yr = 86% | 1038 gene promoters mainly corresponding to PRC2 target genes | Infinium® HumanMeth27K BeadArray (Illumina Inc., San Diego, CA, USA) |
| CIMP+ | hyper | worse | EFS5yr = 36% OS5yr = 45% | ||||||
| Borssen et al. 2016 [76] | n = 113 (pediatric) | NOPHO ALL 2008 | CIMP | CIMP + _MRD < 0.01% | hyper | better | CIR3yr = 0% OS3yr = 100% | 1038 gene promoters mainly corresponding to PRC2 target genes | Infinium® HumanMethylation450 BeadChip (Illumina Inc., San Diego, CA, USA) |
| CIMP − _MRD < 0.01% | hypo | better | CIR3yr = 0% OS3yr = 100% | ||||||
| CIMP + _MRD ≥ 0.01% | hyper | bette | CIR3yr = 12% OS3yr = 83% | ||||||
| CIMP − _MRD ≥ 0.01% | hypo | worse | CIR3yr = 50% OS3yr = 45% | ||||||
| Kimura et al. 2020 [77] ## | n = 79 (pediatric) | n.a. | methylation clusters | Cluster 1 | intermediate | worse | DFS5yr ~ 68% OS5yr = 70% | 939 most differentially methylated sites within clusters | Infinium® MethylationEPIC BeadChip (Illumina Inc., San Diego, CA, USA) |
| Cluster 2 | better | DFS5yr ~ 95% OS5yr = 88% | |||||||
| Cluster 3 | hyper | the best | DFS5yr = 100% OS5yr = 100% | ||||||
| Cluster 4 | hypo | dismal | DFS5yr = 0% OS5yr = 0% | ||||||
| Touzart et al. 2020 [78] | n = 24 (adult) * n = 168 (adult) ** | GRAALL-2003 & GRAALL-2005 | nine-promoter classifier ** | Inter/High ** | hyper | better | CIR5yr = 27% OS5yr = 68% | ** 300 differentially methylated gene promoters | * Me-DIP and custom human promoter arrays (Agilent, Santa Clara, CA, USA) |
| Low ** | hypo | worse | CIR5yr = 45% OS5yr = 50% | ** 9 differentially methylated gene promoters selected from Me-DIP results | ** MS-MLPA with custom probes | ||||
| Roels et al. 2020 [79] | n = 109 (pediatric) | ALL IC-BFM 2002/2009 | COSMe & CIMP | CIMP− | hypo | worse | CIR3yr = 31% | Region A of EPIC array: mainly promoters of PRC2 target genes | Infinium® MethylationEPIC BeadChip (Illumina Inc., San Diego, CA, USA) |
| CIMP+ | hyper | better | CIR3yr = 15% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maćkowska, N.; Drobna-Śledzińska, M.; Witt, M.; Dawidowska, M. DNA Methylation in T-Cell Acute Lymphoblastic Leukemia: In Search for Clinical and Biological Meaning. Int. J. Mol. Sci. 2021, 22, 1388. https://doi.org/10.3390/ijms22031388
Maćkowska N, Drobna-Śledzińska M, Witt M, Dawidowska M. DNA Methylation in T-Cell Acute Lymphoblastic Leukemia: In Search for Clinical and Biological Meaning. International Journal of Molecular Sciences. 2021; 22(3):1388. https://doi.org/10.3390/ijms22031388
Chicago/Turabian StyleMaćkowska, Natalia, Monika Drobna-Śledzińska, Michał Witt, and Małgorzata Dawidowska. 2021. "DNA Methylation in T-Cell Acute Lymphoblastic Leukemia: In Search for Clinical and Biological Meaning" International Journal of Molecular Sciences 22, no. 3: 1388. https://doi.org/10.3390/ijms22031388
APA StyleMaćkowska, N., Drobna-Śledzińska, M., Witt, M., & Dawidowska, M. (2021). DNA Methylation in T-Cell Acute Lymphoblastic Leukemia: In Search for Clinical and Biological Meaning. International Journal of Molecular Sciences, 22(3), 1388. https://doi.org/10.3390/ijms22031388







