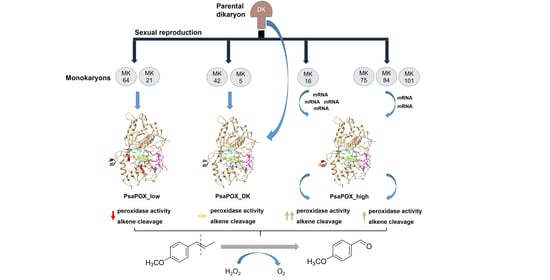
Monokaryotic Pleurotus sapidus Strains with Intraspecific Variability of an Alkene Cleaving DyP-Type Peroxidase Activity as a Result of Gene Mutation and Differential Gene Expression
Abstract
1. Introduction
2. Results and Discussion
2.1. Analysis of Monokaryons
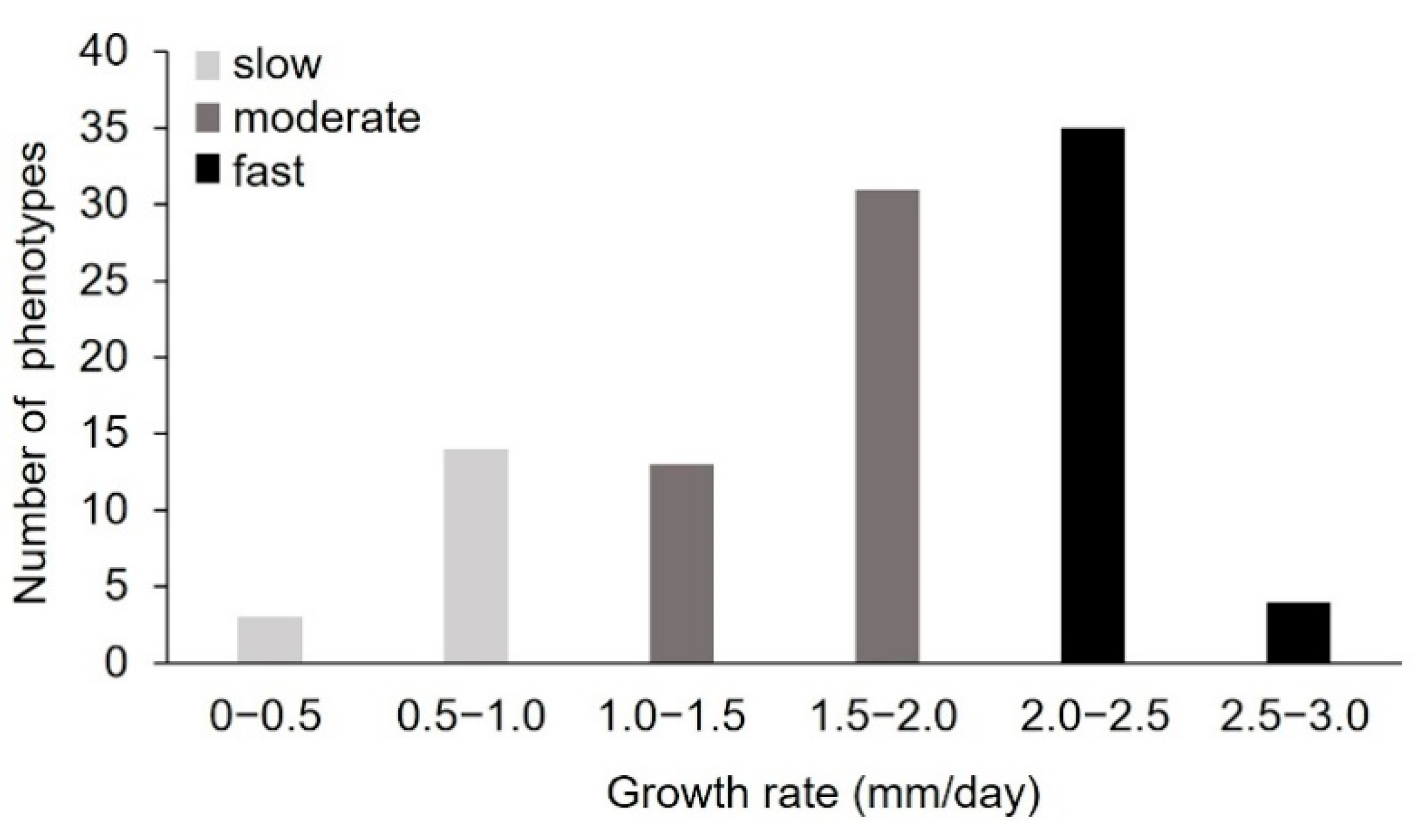
2.1.1. Pre-Selection of Monokaryons by Analysis of the Radial Growth Rate
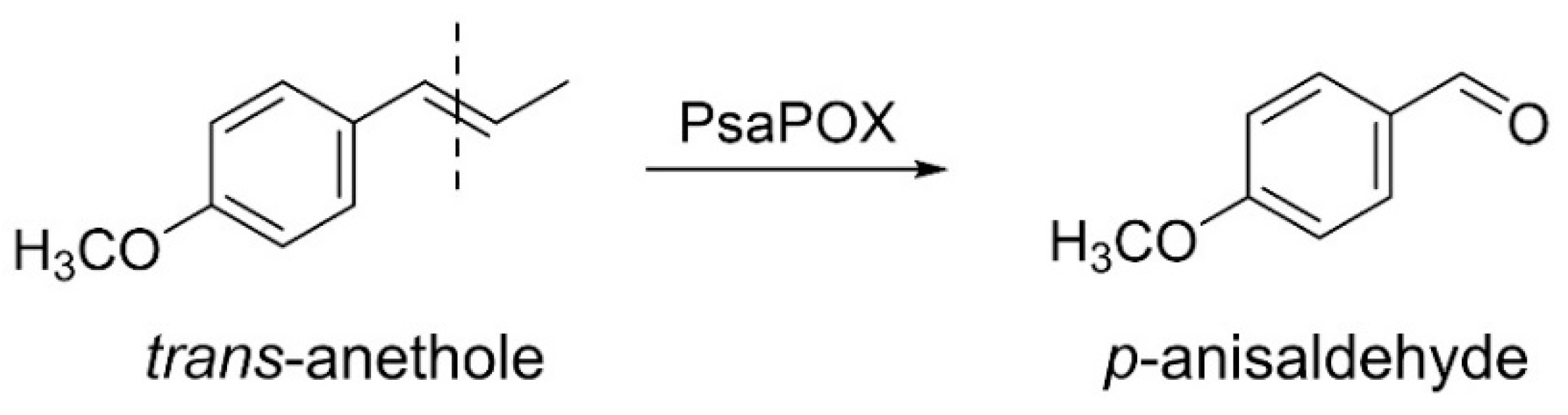
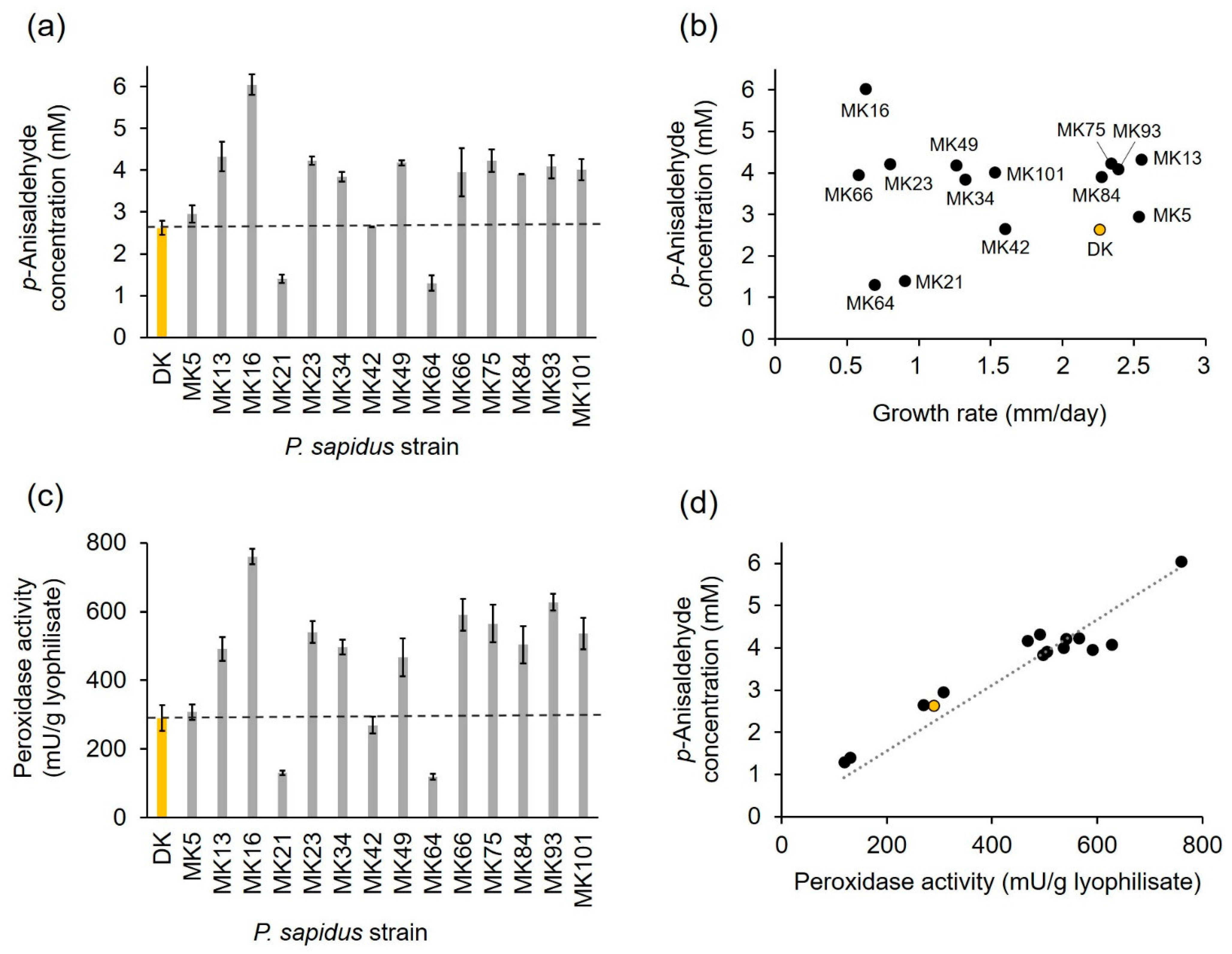
2.1.2. Profiling of Alkene Cleavage and Peroxidase Activity in Monokaryons
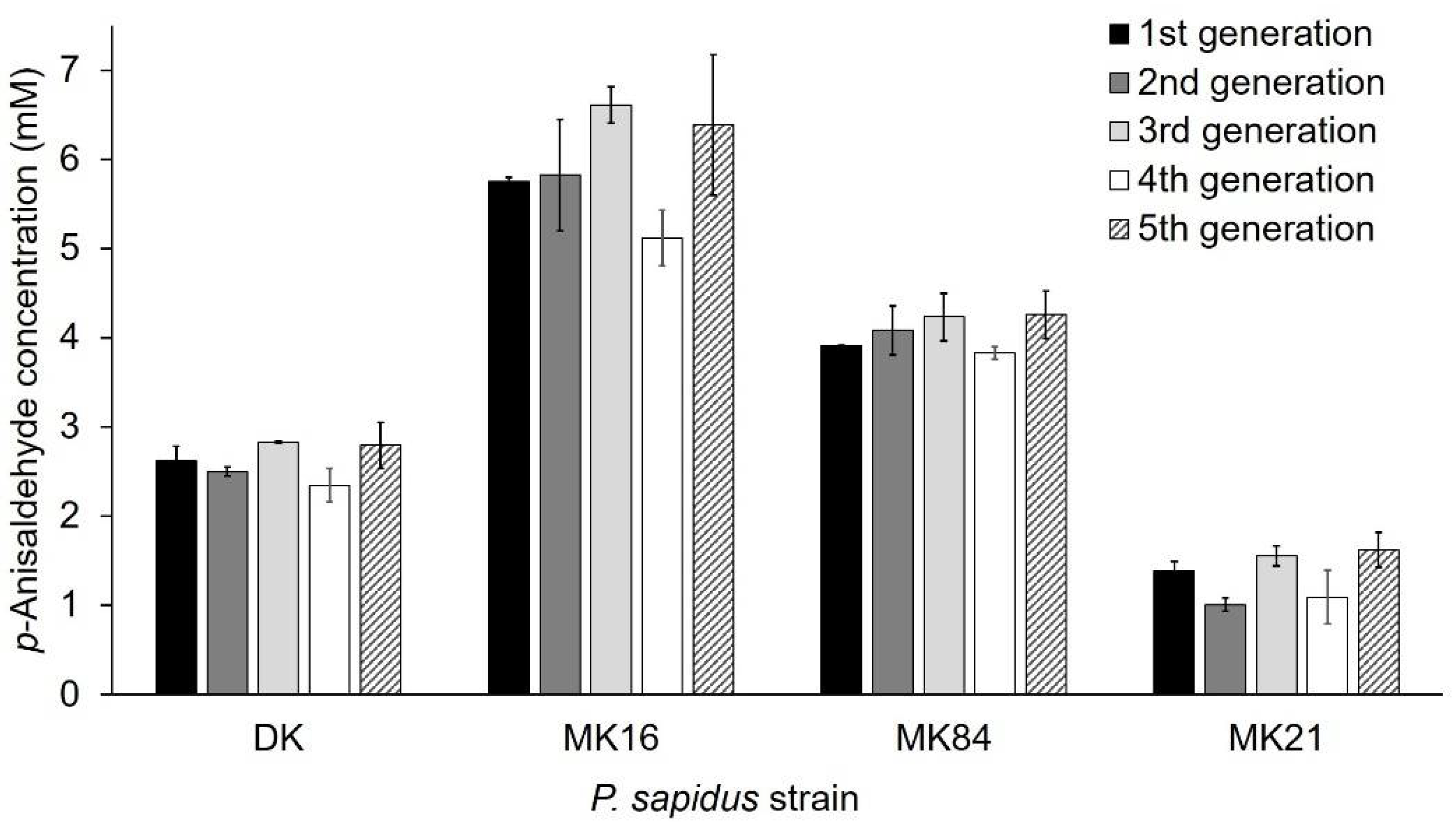
2.1.3. Phenotypic Stability of Alkene Cleavage Activity in Sequential Cultivations
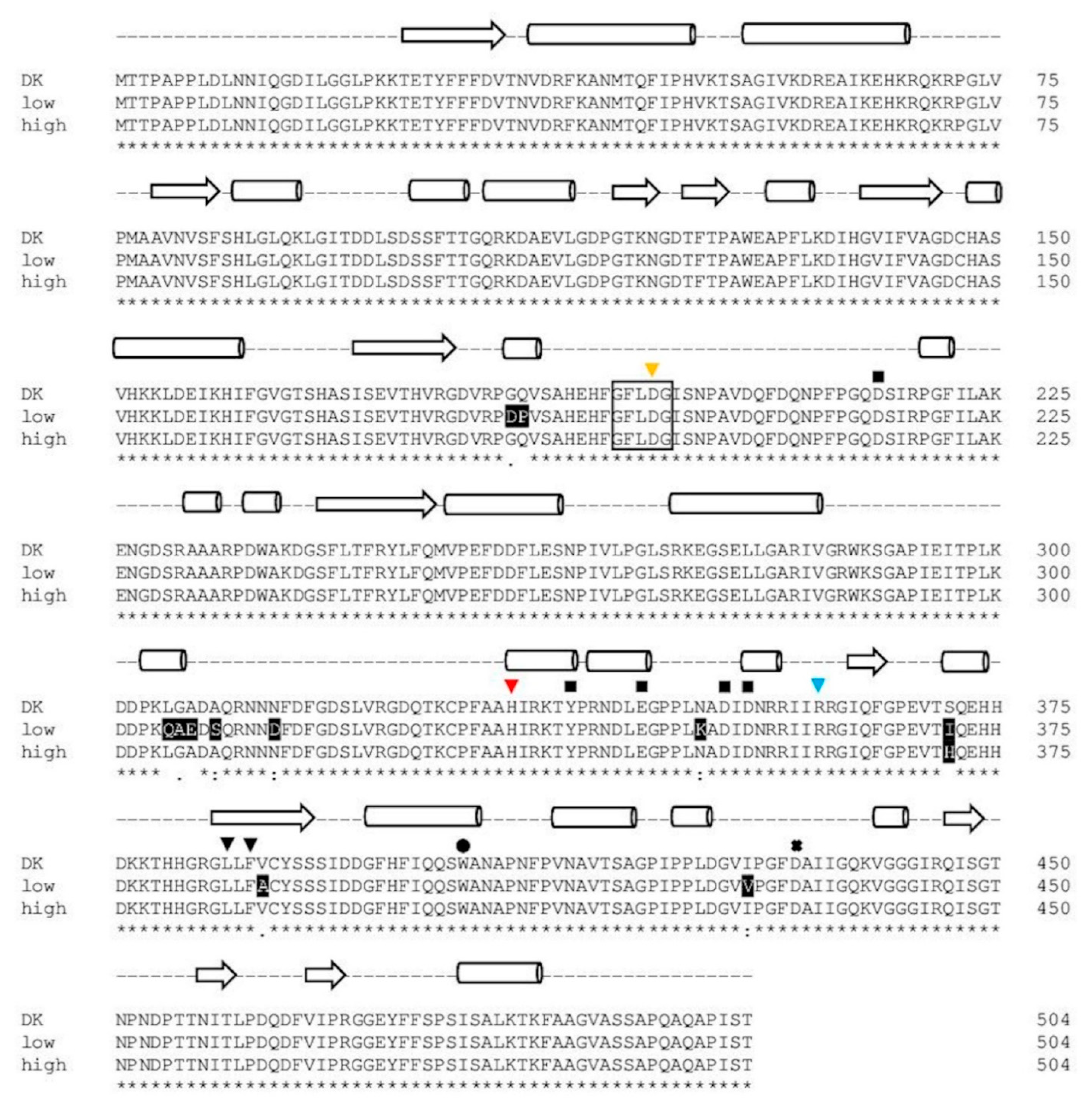
2.1.4. Comparison of PsaPOX from Selected P. sapidus Strains
2.2. Analysis of the Recombinant PsaPOX Variants
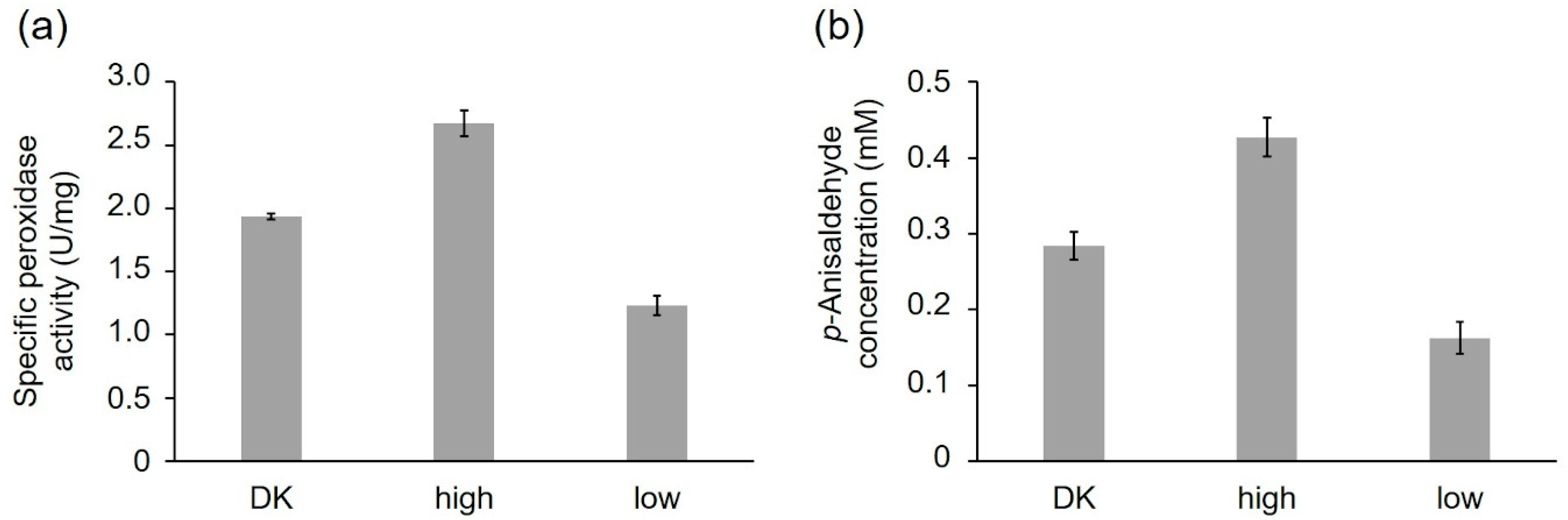
2.2.1. Activity of the Recombinant PsaPOX Variants
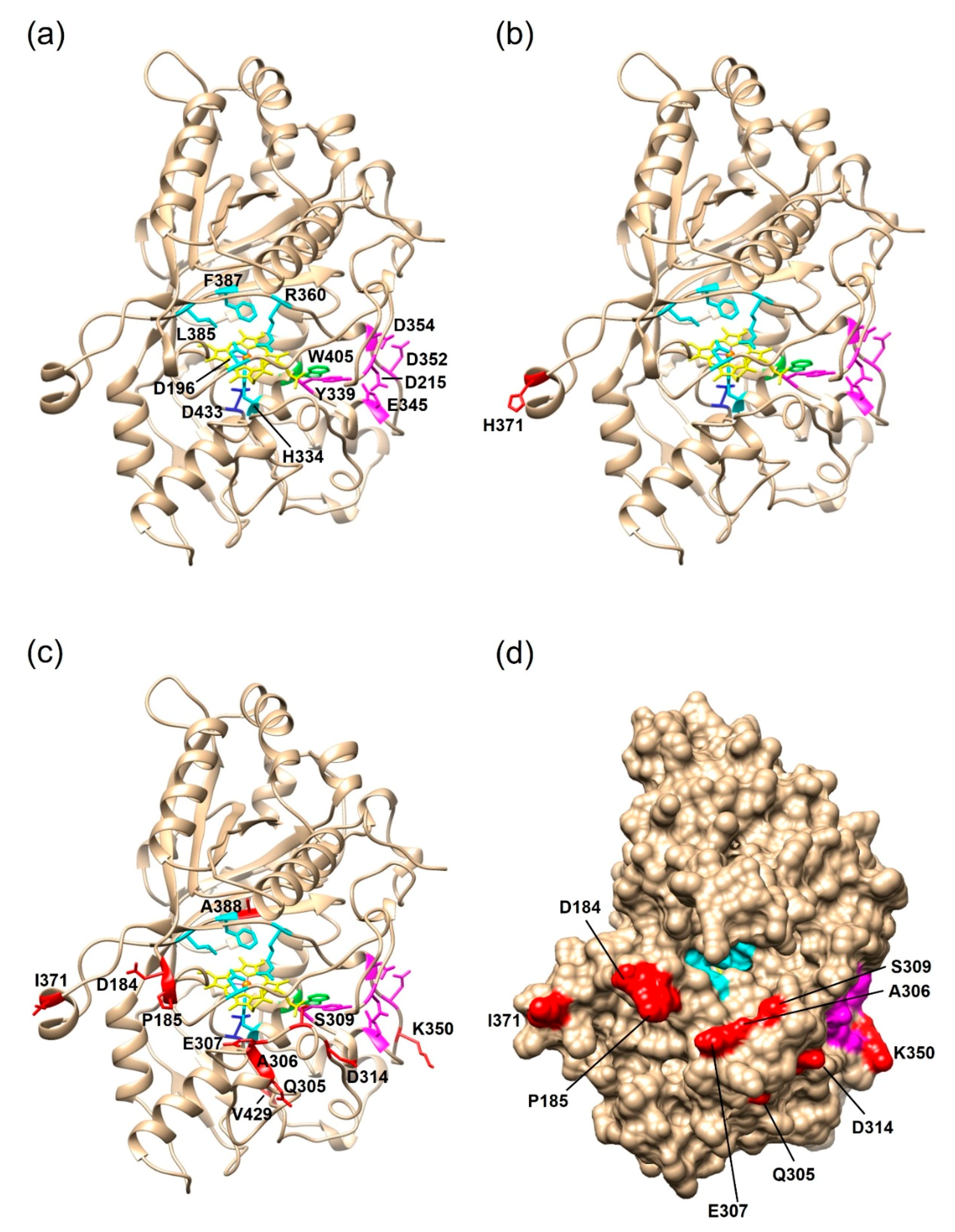
2.2.2. Structural Analysis of the PsaPOX Variants
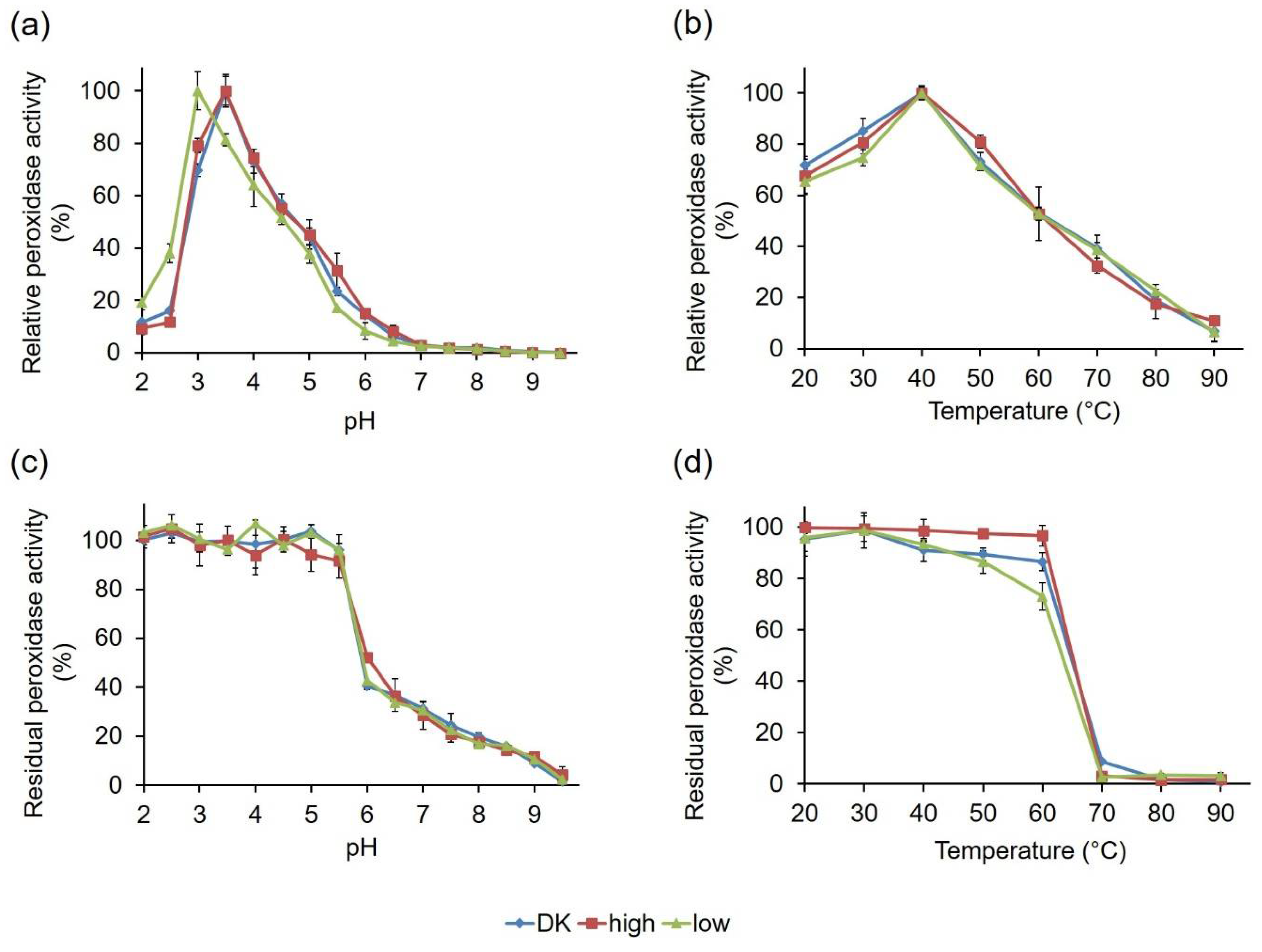
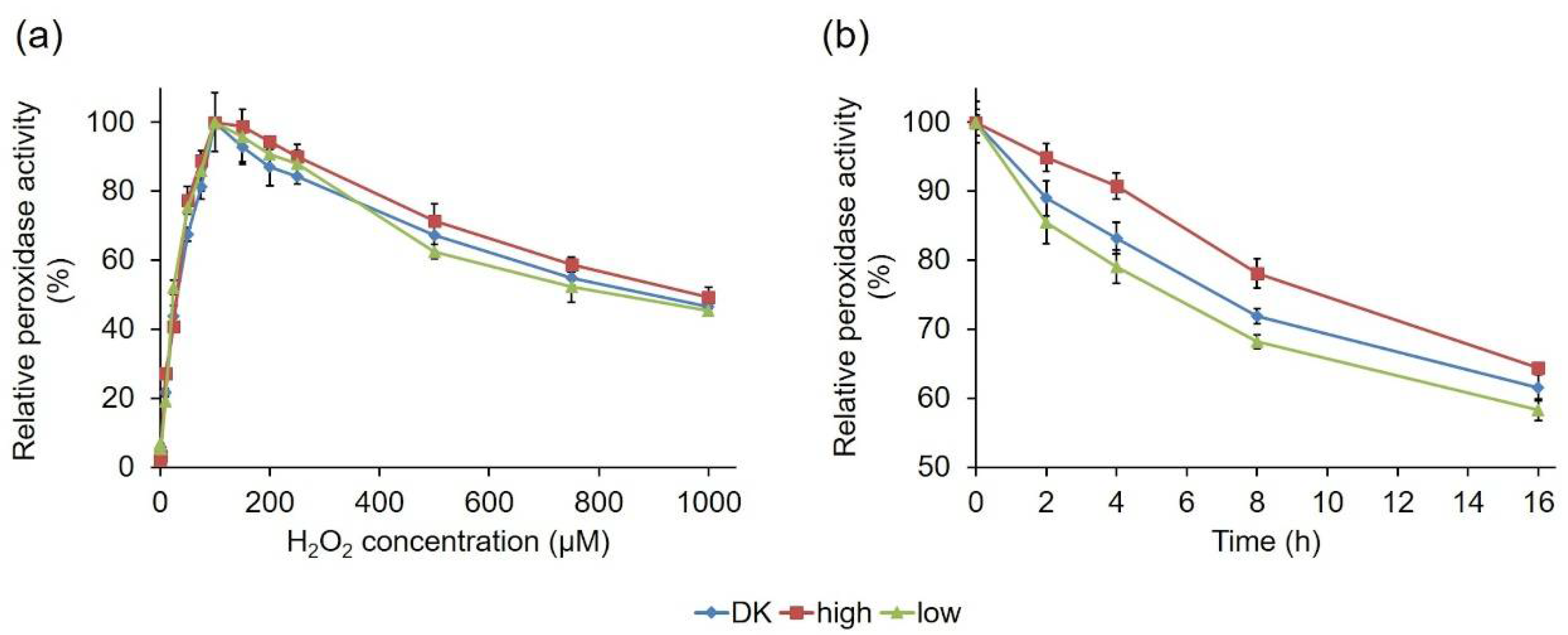
2.2.3. Comparative Biochemical Characterization of the PsaPOX Variants
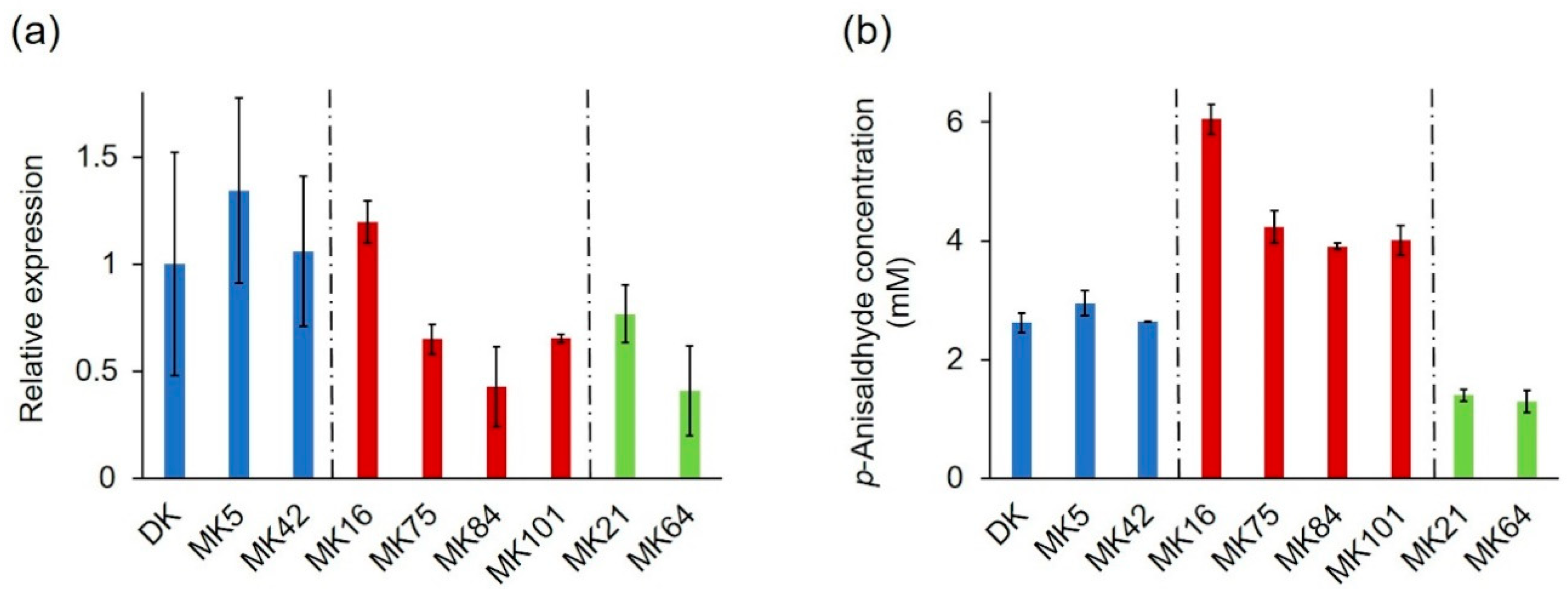
2.3. Expression Profile of the PsaPOX Gene from Different P. sapidus Strains
3. Materials and Methods
3.1. Chemicals and Materials
3.2. Fructification of P. sapidus, Isolation of Basidiospores, and Screening of Monokaryons
3.3. Submerged Cultivation of P. sapidus Strains
3.4. Phenotype Stability of Different P. sapidus Strains
3.5. cDNA Synthesis and Gene Amplification
3.6. Heterologous Expression of the PsaPOX Variants in Komagataella phaffii
3.7. His-Tag Purification of the Recombinant PsaPOX Variants
3.8. Biotransformation
3.9. Peroxidase Activity
3.10. Comparative Biochemical Characterization of the PsaPOX Variants
3.11. Quantitative Real-Time PCR
3.12. Sequence Accession Numbers
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ABTS | 2,2′-azino-bis(3-ethylbenzthiazoline-6-sulphonic acid) |
| DK | Dikaryon |
| DSMZ | Deutsche Sammlung von Mikroorganismen und Zellkulturen |
| DyP | Dye-decolorizing peroxidase |
| gpd3 | Glyceraldehyd-3-phosphate dehydrogenase |
| MK | Monokaryon |
| phos | Purine phosphorylase |
| PsaPOX | DyP of P. sapidus |
| PsaPOX_DK | Parental PsaPOX variant |
| PsaPOX_high | PsaPOX variant with higher activity than PsaPOX_DK |
| PsaPOX_low | PsaPOX variant with lower activity than PsaPOX_DK |
| RT-qPCR | Quantitative real-time PCR |
| SNL | Standard nutrient liquid |
References
- Fahlbusch, K.-G.; Hammerschmidt, F.-J.; Panten, J.; Pickenhagen, W.; Schatkowski, D.; Bauer, K.; Garbe, D.; Surburg, H. Flavors and Fragrances. In Ullmann’s Encyclopedia of Industrial Chemistry; Wiley-VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2003; pp. 73–140. [Google Scholar]
- Rajagopalan, A.; Lara, M.; Kroutil, W. Oxidative Alkene Cleavage by Chemical and Enzymatic Methods. Adv. Synth. Catal. 2013, 355, 3321–3335. [Google Scholar] [CrossRef]
- Spannring, P.; Bruijnincx, P.C.A.; Weckhuysen, B.M.; Klein Gebbink, R.J.M. Transition metal-catalyzed oxidative double bond cleavage of simple and bio-derived alkenes and unsaturated fatty acids. Catal. Sci. Technol. 2014, 4, 2182–2209. [Google Scholar] [CrossRef]
- Berger, R.G. Biotechnology of flavours—The next generation. Biotechnol. Lett. 2009, 31, 1651–1659. [Google Scholar] [CrossRef]
- Bel-Rhlid, R.; Berger, R.G.; Blank, I. Bio-mediated generation of food flavors—Towards sustainable flavor production inspired by nature. Trends Food Sci. Technol. 2018, 78, 134–143. [Google Scholar] [CrossRef]
- Kun, R.S.; Gomes, A.C.S.; Hildén, K.S.; Salazar Cerezo, S.; Mäkelä, M.R.; de Vries, R.P. Developments and opportunities in fungal strain engineering for the production of novel enzymes and enzyme cocktails for plant biomass degradation. Biotechnol. Adv. 2019, 37, 107361. [Google Scholar] [CrossRef] [PubMed]
- Nannemann, D.P.; Birmingham, W.R.; Scism, R.A.; Bachmann, B.O. Assessing directed evolution methods for the generation of biosynthetic enzymes with potential in drug biosynthesis. Future Med. Chem. 2011, 3, 809–819. [Google Scholar] [CrossRef] [PubMed]
- Del Vecchio, C.; Lettera, V.; Pezzella, C.; Piscitelli, A.; Leo, G.; Birolo, L.; Sannia, G. Classical breeding in Pleurotus ostreatus: A natural approach for laccase production improvement. Biocatal. Biotransform. 2012, 30, 78–85. [Google Scholar] [CrossRef]
- Ramírez, L.; Larraya, L.M.P.A. Molecular tools for breeding basidimycetes. Int. J. Microbiol. 2000, 3, 147–152. [Google Scholar]
- Kües, U. Life History and Developmental Processes in the Basidiomycete Coprinus cinereus. Microbiol. Mol. Biol. Rev. 2000, 64, 316–353. [Google Scholar] [CrossRef]
- Eichlerová, I.; Homolka, L. Preparation and crossing of basidiospore-derived monokaryons—A useful tool for obtaining laccase and other ligninolytic enzyme higher-producing dikaryotic strains of Pleurotus ostreatus. Antonie Leeuwenhoek 1999, 75, 321–327. [Google Scholar] [CrossRef]
- Linke, D.; Omarini, A.B.; Takenberg, M.; Kelle, S.; Berger, R.G. Long-Term Monokaryotic Cultures of Pleurotus ostreatus var. florida Produce High and Stable Laccase Activity Capable to Degrade β-Carotene. Appl. Biochem. Biotechnol. 2019, 187, 894–912. [Google Scholar] [PubMed]
- L’opez, S.C.; Theelen, B.; Manserra, S.; Issak, T.Y.; Rytioja, J.; Mäkelä, M.R.; de Vries, R.P. Functional diversity in Dichomitus squalens monokaryons. IMA Fungus 2017, 8, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Omarini, A.B.; Plagemann, I.; Schimanski, S.; Krings, U.; Berger, R.G. Crosses between monokaryons of Pleurotus sapidus or Pleurotus florida show an improved biotransformation of (+)-valencene to (+)-nootkatone. Bioresour. Technol. 2014, 171, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Van der Nest, M.A.; Slippers, B.; Steenkamp, E.T.; De Vos, L.; Van Zyl, K.; Stenlid, J.; Wingfield, M.J.; Wingfield, B.D. Genetic linkage map for Amylostereum areolatum reveals an association between vegetative growth and sexual and self-recognition. Fungal Genet. Biol. 2009, 46, 632–641. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Postemsky, P.D.; Bidegain, M.A.; Lluberas, G.; Lopretti, M.I.; Bonifacino, S.; Inés Landache, M.; Zygadlo, J.A.; Fernández-Lahore, M.; Omarini, A.B. Biorefining via solid-state fermentation of rice and sunflower by-products employing novel monosporic strains from Pleurotus sapidus. Bioresour. Technol. 2019, 289, 121692. [Google Scholar] [CrossRef]
- Castanera, R.; Omarini, A.; Santoyo, F.; Pérez, G.; Pisabarro, A.G.; Ramírez, L. Non-Additive Transcriptional Profiles Underlie Dikaryotic Superiority in Pleurotus ostreatus Laccase Activity. PLoS ONE 2013, 8, e73282. [Google Scholar] [CrossRef]
- Linde, D.; Ruiz-Dueñas, F.J.; Fernández-Fueyo, E.; Guallar, V.; Hammel, K.E.; Pogni, R.; Martínez, A.T. Basidiomycete DyPs: Genomic diversity, structural–functional aspects, reaction mechanism and environmental significance. Arch. Biochem. Biophys. 2015, 574, 66–74. [Google Scholar] [CrossRef]
- Sugano, Y. DyP-type peroxidases comprise a novel heme peroxidase family. Cell. Mol. Life Sci. 2009, 66, 1387–1403. [Google Scholar] [CrossRef]
- Strittmatter, E.; Liers, C.; Ullrich, R.; Wachter, S.; Hofrichter, M.; Plattner, D.A.; Piontek, K. First Crystal Structure of a Fungal High-redox Potential Dye-decolorizing Peroxidase. J. Biol. Chem. 2013, 288, 4095–4102. [Google Scholar] [CrossRef]
- Linde, D.; Pogni, R.; Cañellas, M.; Lucas, F.; Guallar, V.; Baratto, M.C.; Sinicropi, A.; Sáez-Jiménez, V.; Coscolín, C.; Romero, A.; et al. Catalytic surface radical in dye-decolorizing peroxidase: A computational, spectroscopic and site-directed mutagenesis study. Biochem. J. 2015, 466, 253–262. [Google Scholar] [CrossRef]
- Fernández-Fueyo, E.; Linde, D.; Almendral, D.; López-Lucendo, M.F.; Ruiz-Dueñas, F.J.; Martínez, A.T. Description of the first fungal dye-decolorizing peroxidase oxidizing manganese(II). Appl. Microbiol. Biotechnol. 2015, 99, 8927–8942. [Google Scholar] [CrossRef] [PubMed]
- Krahe, N.-K.; Berger, R.G.; Ersoy, F. A DyP-Type Peroxidase of Pleurotus sapidus with Alkene Cleaving Activity. Molecules 2020, 25, 1536. [Google Scholar] [CrossRef]
- Behrens, C.J.; Zelena, K.; Berger, R.G. Comparative Cold Shock Expression and Characterization of Fungal Dye-Decolorizing Peroxidases. Appl. Biochem. Biotechnol. 2016, 179, 1404–1417. [Google Scholar] [CrossRef] [PubMed]
- Hofrichter, M.; Ullrich, R.; Pecyna, M.J.; Liers, C.; Lundell, T. New and classic families of secreted fungal heme peroxidases. Appl. Microbiol. Biotechnol. 2010, 87, 871–897. [Google Scholar] [CrossRef] [PubMed]
- Stahl, U.; Esser, K. Genetics of fruit body production in higher basidiomycetes. Mol. Gen. Genet. 1976, 148, 183–197. [Google Scholar] [CrossRef]
- Pringle, A.; Taylor, J.W. The fitness of filamentous fungi. Trends Microbiol. 2002, 10, 474–481. [Google Scholar] [CrossRef]
- Liu, S.-R.; Ke, B.-R.; Zhang, W.-R.; Liu, X.-R.; Wu, X.-P. Breeding of new Ganoderma lucidum strains simultaneously rich in polysaccharides and triterpenes by mating basidiospore-derived monokaryons of two commercial cultivars. Sci. Hortic. 2017, 216, 58–65. [Google Scholar] [CrossRef]
- Meissner, P.N.; Dailey, T.A.; Hift, R.J.; Ziman, M.; Corrigall, A.V.; Roberts, A.G.; Meissner, D.M.; Kirsch, R.E.; Dailey, H.A. A R59W mutation in human protoporphyrinogen oxidase results in decreased enzyme activity and is prevalent in South Africans with variegate porphyria. Nat. Genet. 1996, 13, 95–97. [Google Scholar] [CrossRef]
- De Jong, J.F.; Deelstra, H.J.; Wösten, H.A.B.; Lugones, L.G. RNA-mediated gene silencing in monokaryons and dikaryons of Schizophyllum commune. Appl. Environ. Microbiol. 2006, 72, 1267–1269. [Google Scholar] [CrossRef]
- Maurizi, M.R.; Rasulova, F. Degradation of l-glutamate dehydrogenase from Escherichia coli: Allosteric regulation of enzyme stability. Arch. Biochem. Biophys. 2002, 397, 206–216. [Google Scholar] [CrossRef]
- Castanera, R.; López-Varas, L.; Borgognone, A.; LaButti, K.; Lapidus, A.; Schmutz, J.; Grimwood, J.; Pérez, G.; Pisabarro, A.G.; Grigoriev, I.V.; et al. Transposable Elements versus the Fungal Genome: Impact on Whole-Genome Architecture and Transcriptional Profiles. PLoS Genet. 2016, 12, e1006108. [Google Scholar] [CrossRef] [PubMed]
- Castanera, R.; Borgognone, A.; Pisabarro, A.G.; Ramírez, L. Biology, dynamics, and applications of transposable elements in basidiomycete fungi. Appl. Microbiol. Biotechnol. 2017, 101, 1337–1350. [Google Scholar] [CrossRef] [PubMed]
- Clark, T.A.; Anderson, J.B. Dikaryons of the basidiomycete fungus Schizophyllum commune: Evolution in long-term culture. Genetics 2004, 167, 1663–1675. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ellingboe, A.H. Breeding for Mushroom Production in Lentinula edodes. In Genetics and Breeding of Edible Mushrooms; Chang, S.-T., Buswell, J.A., Miles, P.G., Eds.; Routledge: Philadelphia, PA, USA, 2018; pp. 111–123. [Google Scholar]
- Liu, T.; Li, H.; Ding, Y.; Qi, Y.; Gao, Y.; Song, A.; Shen, J.; Qiu, L. Genome-wide gene expression patterns in dikaryon of the basidiomycete fungus Pleurotus ostreatus. Braz. J. Microbiol. 2017, 48, 380–390. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Artimo, P.; Jonnalagedda, M.; Arnold, K.; Baratin, D.; Csardi, G.; de Castro, E.; Duvaud, S.; Flegel, V.; Fortier, A.; Gasteiger, E.; et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012, 40, W597–W603. [Google Scholar] [CrossRef]
- Brissos, V.; Tavares, D.; Sousa, A.C.; Robalo, M.P.; Martins, L.O. Engineering a Bacterial DyP-Type Peroxidase for Enhanced Oxidation of Lignin-Related Phenolics at Alkaline pH. ACS Catal. 2017, 7, 3454–3465. [Google Scholar] [CrossRef]
- Habib, M.H.; Rozeboom, H.J.; Fraaije, M.W. Characterization of a New DyP-Peroxidase from the Alkaliphilic Cellulomonad, Cellulomonas bogoriensis. Molecules 2019, 24, 1208. [Google Scholar] [CrossRef]
- Whittle, E.; Shanklin, J. Engineering Δ9-16:0-Acyl Carrier Protein (ACP) Desaturase Specificity Based on Combinatorial Saturation Mutagenesis and Logical Redesign of the Castor Δ9-18:0-ACP Desaturase. J. Biol. Chem. 2001, 276, 21500–21505. [Google Scholar] [CrossRef]
- Van den Heuvel, R.H.H.; van den Berg, W.A.M.; Rovida, S.; van Berkel, W.J.H. Laboratory-evolved Vanillyl-alcohol Oxidase Produces Natural Vanillin. J. Biol. Chem. 2004, 279, 33492–33500. [Google Scholar] [CrossRef] [PubMed]
- Morley, K.L.; Kazlauskas, R.J. Improving enzyme properties: When are closer mutations better? Trends Biotechnol. 2005, 23, 231–237. [Google Scholar] [CrossRef] [PubMed]
- Bross, P.; Jespersen, C.; Jensen, T.G.; Andresen, B.S.; Kristensen, M.J.; Winter, V.; Nandy, A.; Krautle, F.; Ghisla, S.; Bolund, L.; et al. Effects of two mutations detected in medium chain Acyl-CoA dehydrogenase (MCAD)-deficient patients on folding, oligomer assembly, and stability of MCAD enzyme. J. Biol. Chem. 1995, 270, 10284–10290. [Google Scholar] [CrossRef] [PubMed]
- Boer, H.; Koivula, A. The relationship between thermal stability and pH optimum studied with wild-type and mutant Trichoderma reesei cellobiohydrolase Cel7A. Eur. J. Biochem. 2003, 270, 841–848. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Ruiz, E.; Gonzalez-Perez, D.; Ruiz-Dueñas, F.J.; Martínez, A.T.; Alcalde, M. Directed evolution of a temperature-, peroxide- and alkaline pH-tolerant versatile peroxidase. Biochem. J. 2012, 441, 487–498. [Google Scholar] [CrossRef]
- Graf, E. Ullmann’s Encyclopedia of Industrial Chemistry; Fifth Compl. Revis. Edit. Vol.’s A 3 and A 4 Ed’s W. Gerhartz (executive), Y. St. Yamamoto (Senior Ed.), F. T. Campbell, R. Pfefferkorn und J. F. Rounsaville. VCH Verlagsges. Weinheim 1985, A 3 = 578 S., A 4. Pharm. Unserer Zeit 1987, 16, 63. [Google Scholar] [CrossRef]
- Sergeant, E.; Dempsey, B. Ionisation Constants of Organic Acids in Aqueous Solution; IUPAC Chemical Data Series 2; Pergamon Press: New York, NY, USA, 1979; ISBN 0080223397. [Google Scholar]
- Lauber, C.; Schwarz, T.; Nguyen, Q.K.; Lorenz, P.; Lochnit, G.; Zorn, H. Identification, heterologous expression and characterization of a dye-decolorizing peroxidase of Pleurotus sapidus. AMB Express 2017, 7, 164. [Google Scholar] [CrossRef]
- Salvachua, D.; Prieto, A.; Martinez, A.T.; Martinez, M.J. Characterization of a Novel Dye-Decolorizing Peroxidase (DyP)-Type Enzyme from Irpex lacteus and Its Application in Enzymatic Hydrolysis of Wheat Straw. Appl. Environ. Microbiol. 2013, 79, 4316–4324. [Google Scholar] [CrossRef]
- Arnao, M.B.; Acosta, M.; del Rio, J.A.; Varón, R.; García-Cánovas, F. A kinetic study on the suicide inactivation of peroxidase by hydrogen peroxide. Biochim. Biophys. Acta Protein Struct. Mol. Enzymol. 1990, 1041, 43–47. [Google Scholar] [CrossRef]
- Busse, N.; Wagner, D.; Kraume, M.; Czermak, P. Reaction Kinetics of Versatile Peroxidase for the Degradation of Lignin Compounds. Am. J. Biochem. Biotechnol. 2013, 9, 365–394. [Google Scholar] [CrossRef]
- Freihorst, D.; Brunsch, M.; Wirth, S.; Krause, K.; Kniemeyer, O.; Linde, J.; Kunert, M.; Boland, W.; Kothe, E. Smelling the difference: Transcriptome, proteome and volatilome changes after mating. Fungal Genet. Biol. 2018, 112, 2–11. [Google Scholar] [CrossRef] [PubMed]
- Madhavan, S.; Krause, K.; Jung, E.-M.; Kothe, E. Differential regulation of multi-copper oxidases in Schizophyllum commune during sexual development. Mycol. Prog. 2014, 13, 1009. [Google Scholar] [CrossRef]
- Borgognone, A.; Castanera, R.; Morselli, M.; López-Varas, L.; Rubbi, L.; Pisabarro, A.G.; Pellegrini, M.; Ramírez, L. Transposon-associated epigenetic silencing during Pleurotus ostreatus life cycle. DNA Res. 2018, 25, 451–464. [Google Scholar] [CrossRef] [PubMed]
- Linke, D.; Leonhardt, R.; Eisele, N.; Petersen, L.M.; Riemer, S.; Nimtz, M.; Berger, R.G. Carotene-degrading activities from Bjerkandera adusta possess an application in detergent industries. Bioprocess Biosyst. Eng. 2015, 38, 1191–1199. [Google Scholar] [CrossRef] [PubMed]
- Krings, U.; Lehnert, N.; Fraatz, M.A.; Hardebusch, B.; Zorn, H.; Berger, R.G. Autoxidation versus Biotransformation of α-Pinene to Flavors with Pleurotus sapidus: Regioselective Hydroperoxidation of α-Pinene and Stereoselective Dehydrogenation of Verbenol. J. Agric. Food Chem. 2009, 57, 9944–9950. [Google Scholar] [CrossRef]
- Aamir, S.; Sutar, S.; Singh, S.; Baghela, A. A rapid and efficient method of fungal genomic DNA extraction, suitable for PCR based molecular methods. Plant Pathol. Quar. 2015, 5, 74–81. [Google Scholar] [CrossRef]
- NetNGlyc 1.0 Server. Available online: http://www.cbs.dtu.dk/services/NetNGlyc/ (accessed on 14 November 2020).
- Nieter, A.; Kelle, S.; Takenberg, M.; Linke, D.; Bunzel, M.; Popper, L.; Berger, R.G. Heterologous production and characterization of a chlorogenic acid esterase from Ustilago maydis with a potential use in baking. Food Chem. 2016, 209, 1–9. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Britton, H.T.S.; Robinson, R.A. CXCVIII.—Universal buffer solutions and the dissociation constant of veronal. J. Chem. Soc. 1931, 1456–1462. [Google Scholar] [CrossRef]
- Galperin, I. Das Ligninolytische System von Pleurotus sapidus: Transkriptomanalyse und Heterologe Expression einer Arylalkoholoxidase; Justus-Liebig-Universität Gießen: Gießen, Germany, 2018. [Google Scholar]
- Teste, M.A.; Duquenne, M.; François, J.M.; Parrou, J.L. Validation of reference genes for quantitative expression analysis by real-time RT-PCR in Saccharomyces cerevisiae. BMC Mol. Biol. 2009, 10, 99. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, 45e. [Google Scholar] [CrossRef] [PubMed]
| PsaPOX Variant | Km (µM) | kcat (s−1) | kcat/Km (s−1 mM−1) |
|---|---|---|---|
| DK | 33.0 ± 2.5 | 6.8 ± 0.1 | 206 ± 4 |
| high | 25.8 ± 1.6 | 10.5 ± 0.1 | 408 ± 14 |
| low | 31.9 ± 2.3 | 3.8 ± 0.1 | 119 ± 2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krahe, N.-K.; Berger, R.G.; Witt, M.; Zorn, H.; Omarini, A.B.; Ersoy, F. Monokaryotic Pleurotus sapidus Strains with Intraspecific Variability of an Alkene Cleaving DyP-Type Peroxidase Activity as a Result of Gene Mutation and Differential Gene Expression. Int. J. Mol. Sci. 2021, 22, 1363. https://doi.org/10.3390/ijms22031363
Krahe N-K, Berger RG, Witt M, Zorn H, Omarini AB, Ersoy F. Monokaryotic Pleurotus sapidus Strains with Intraspecific Variability of an Alkene Cleaving DyP-Type Peroxidase Activity as a Result of Gene Mutation and Differential Gene Expression. International Journal of Molecular Sciences. 2021; 22(3):1363. https://doi.org/10.3390/ijms22031363
Chicago/Turabian StyleKrahe, Nina-Katharina, Ralf G. Berger, Martin Witt, Holger Zorn, Alejandra B. Omarini, and Franziska Ersoy. 2021. "Monokaryotic Pleurotus sapidus Strains with Intraspecific Variability of an Alkene Cleaving DyP-Type Peroxidase Activity as a Result of Gene Mutation and Differential Gene Expression" International Journal of Molecular Sciences 22, no. 3: 1363. https://doi.org/10.3390/ijms22031363
APA StyleKrahe, N.-K., Berger, R. G., Witt, M., Zorn, H., Omarini, A. B., & Ersoy, F. (2021). Monokaryotic Pleurotus sapidus Strains with Intraspecific Variability of an Alkene Cleaving DyP-Type Peroxidase Activity as a Result of Gene Mutation and Differential Gene Expression. International Journal of Molecular Sciences, 22(3), 1363. https://doi.org/10.3390/ijms22031363