Brain-Derived Extracellular Vesicles in Health and Disease: A Methodological Perspective
Abstract
1. Introduction
1.1. Brief History of EVs
1.2. EVs in Central Nervous System (CNS) Physiology
1.3. EVs in CNS Pathologies
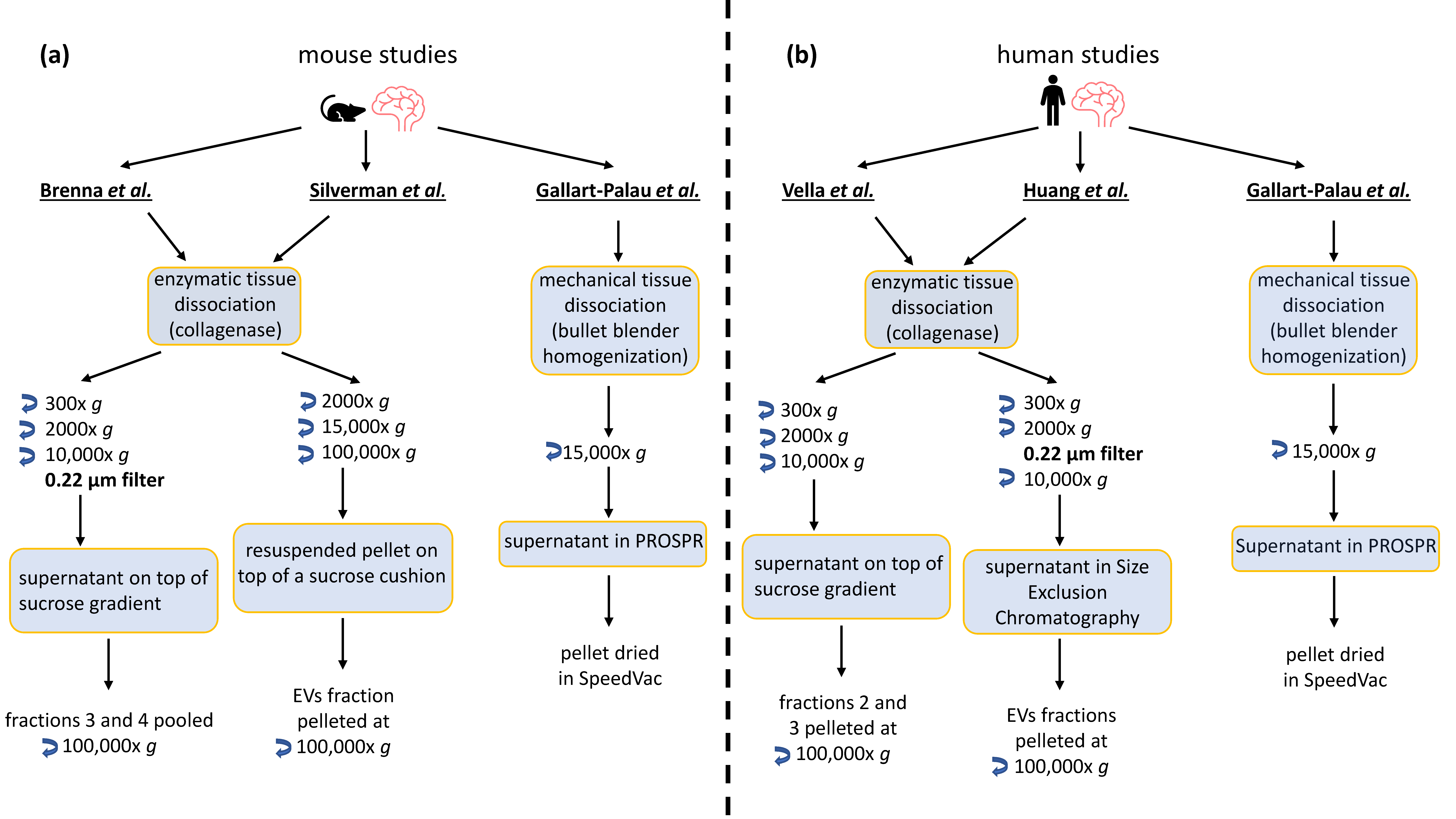
2. BDEVs: Comparison of Current Protocols
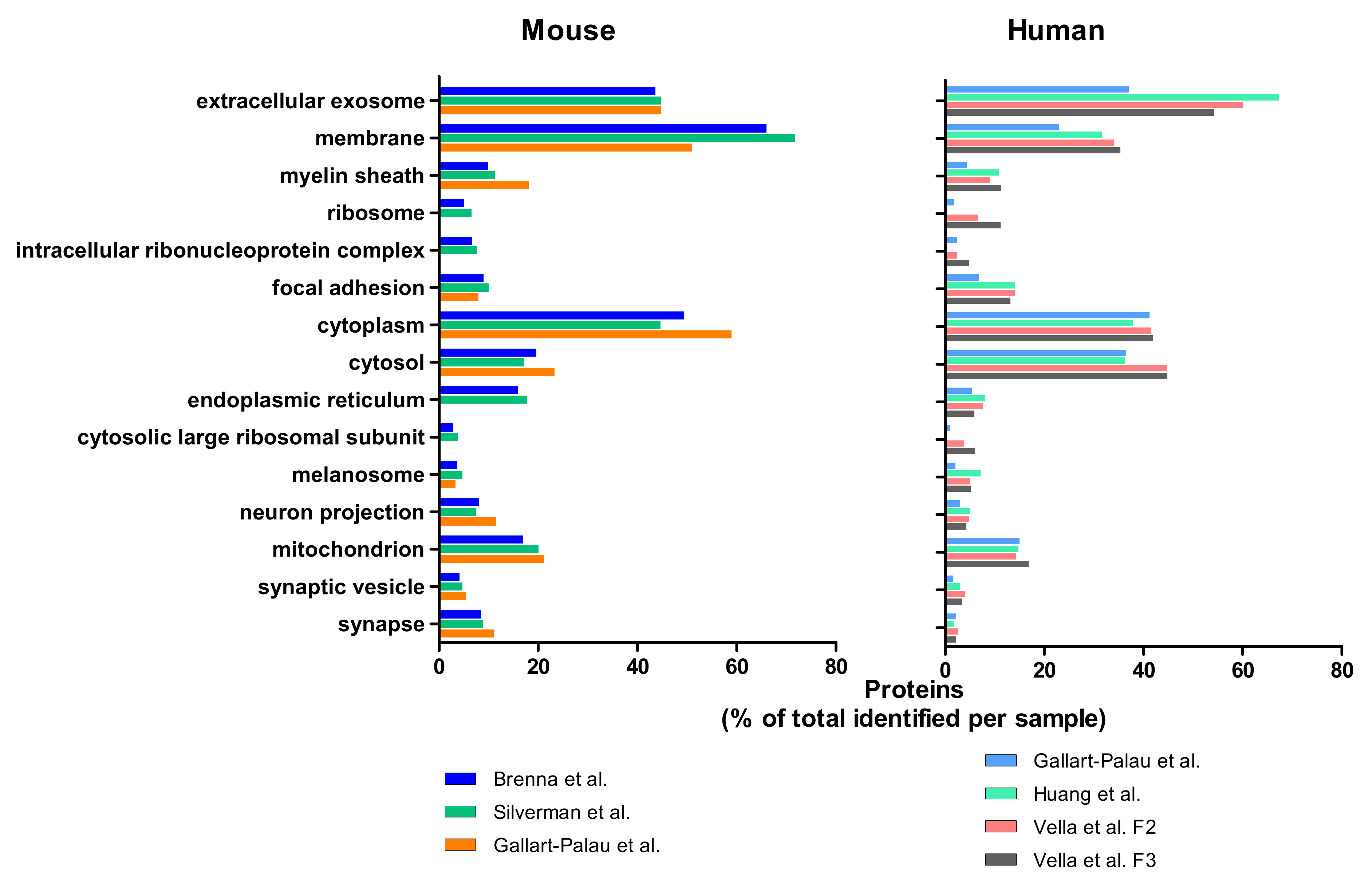
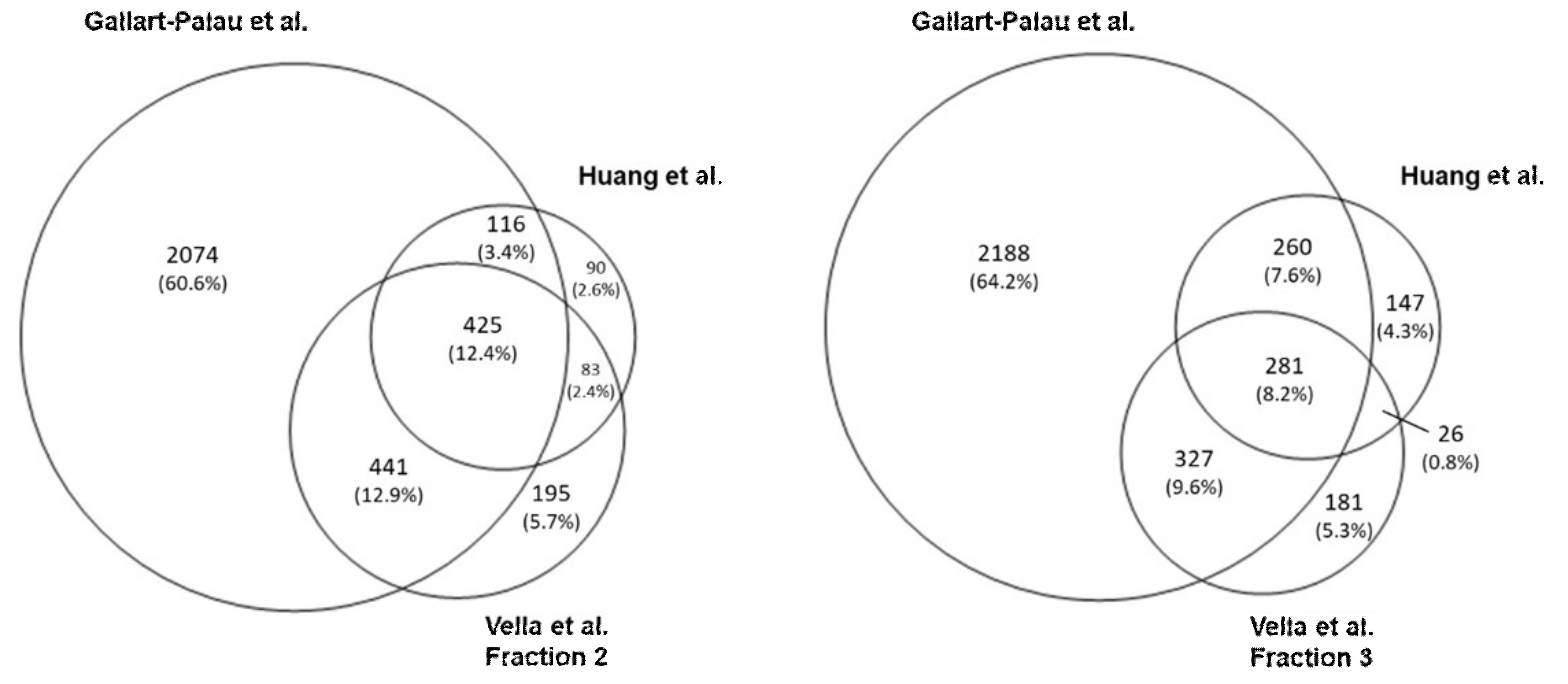
3. Mass Spectrometry Analysis-Based Comparison between Different BDEVs Isolation Protocols
3.1. Methods
3.2. Results
4. Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mercer, E.H.; Shaffer, B.M. Electron Microscopy of Solitary and Aggregated Slime Mould Cells. J. Biophys. Biochem. Cytol. 1960, 7, 353–356. [Google Scholar] [CrossRef] [PubMed]
- Vickerman, K. Patterns of cellular organisation in Limax amoebae. An electron microscope study. Exp. Cell Res. 1962, 26, 497–519. [Google Scholar] [PubMed]
- Sun, C.N. Lattice structures and osmiophilic bodies in the developing respiratory tissue of rats. J. Ultrastruct. Res. 1966, 15, 380–388. [Google Scholar] [CrossRef] [PubMed]
- Bonucci, E. Fine structure of early cartilage calcification. J. Ultrastruct. Res. 1967, 20, 33–50. [Google Scholar] [CrossRef]
- Anderson, H.C. Vesicles associated with calcification in the matrix of epiphyseal cartilage. J. Cell Biol. 1969, 41, 59–72. [Google Scholar] [CrossRef]
- Wolf, P. The nature and significance of platelet products in human plasma. Br. J. Haematol. 1967, 13, 269–288. [Google Scholar] [CrossRef]
- Chargaff, E.; West, R. The biological significance of the thromboplastic protein of blood. J. Biol. Chem. 1946, 166, 189–197. [Google Scholar] [CrossRef]
- Aaronson, S.; Behrens, U.; Orner, R.; Haines, T.H. Ultrastructure of intracellular and extracellular vesicles, membranes, and myelin figures produced by Ochromonas danica. J. Ultrastruct. Res. 1971, 35, 418–430. [Google Scholar] [CrossRef]
- Ronquist, G.; Brody, I. The prostasome: Its secretion and function in man. Biochim. Biophys. Acta 1985, 822, 203–218. [Google Scholar] [CrossRef]
- Ronquist, G.; Hedström, M. Restoration of detergent-inactivated adenosine triphosphatase activity of human prostatic fluid with concanavalin A. Biochim. Biophys. Acta 1977, 483, 483–486. [Google Scholar] [CrossRef]
- Trams, E.G.; Lauter, C.J.; Salem, N.; Heine, U. Exfoliation of membrane ecto-enzymes in the form of micro-vesicles. Biochim. Biophys. Acta 1981, 645, 63–70. [Google Scholar] [CrossRef]
- Poste, G.; Nicolson, G.L. Arrest and metastasis of blood-borne tumor cells are modified by fusion of plasma membrane vesicles from highly metastatic cells. Proc. Natl. Acad. Sci. USA 1980, 77, 399–403. [Google Scholar] [CrossRef] [PubMed]
- Pan, B.T.; Johnstone, R.M. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: Selective externalization of the receptor. Cell 1983, 33, 967–978. [Google Scholar] [CrossRef]
- Harding, C.; Heuser, J.; Stahl, P. Receptor-mediated endocytosis of transferrin and recycling of the transferrin receptor in rat reticulocytes. J. Cell Biol. 1983, 97, 329–339. [Google Scholar] [CrossRef]
- Harding, C.V.; Heuser, J.E.; Stahl, P.D. Exosomes: Looking back three decades and into the future. J. Cell Biol. 2013, 200, 367–371. [Google Scholar] [CrossRef]
- Johnstone, R.M.; Adam, M.; Hammond, J.R.; Orr, L.; Turbide, C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). J. Biol. Chem. 1987, 262, 9412–9420. [Google Scholar] [CrossRef]
- Johnstone, R.M.; Mathew, A.; Mason, A.B.; Teng, K. Exosome formation during maturation of mammalian and avian reticulocytes: Evidence that exosome release is a major route for externalization of obsolete membrane proteins. J. Cell. Physiol. 1991, 147, 27–36. [Google Scholar] [CrossRef]
- Raposo, G.; Nijman, H.W.; Stoorvogel, W.; Liejendekker, R.; Harding, C.V.; Melief, C.J.; Geuze, H.J. B lymphocytes secrete antigen-presenting vesicles. J. Exp. Med. 1996, 183, 1161–1172. [Google Scholar] [CrossRef]
- Zitvogel, L.; Regnault, A.; Lozier, A.; Wolfers, J.; Flament, C.; Tenza, D.; Ricciardi-Castagnoli, P.; Raposo, G.; Amigorena, S. Eradication of established murine tumors using a novel cell-free vaccine: Dendritic cell-derived exosomes. Nat. Med. 1998, 4, 594–600. [Google Scholar] [CrossRef]
- Raposo, G.; Tenza, D.; Mecheri, S.; Peronet, R.; Bonnerot, C.; Desaymard, C. Accumulation of major histocompatibility complex class II molecules in mast cell secretory granules and their release upon degranulation. Mol. Biol. Cell 1997, 8, 2631–2645. [Google Scholar] [CrossRef]
- Heijnen, H.F.; Schiel, A.E.; Fijnheer, R.; Geuze, H.J.; Sixma, J.J. Activated platelets release two types of membrane vesicles: Microvesicles by surface shedding and exosomes derived from exocytosis of multivesicular bodies and alpha-granules. Blood 1999, 94, 3791–3799. [Google Scholar] [CrossRef] [PubMed]
- Van Niel, G.; Raposo, G.; Candalh, C.; Boussac, M.; Hershberg, R.; Cerf-Bensussan, N.; Heyman, M. Intestinal epithelial cells secrete exosome-like vesicles. Gastroenterology 2001, 121, 337–349. [Google Scholar] [CrossRef] [PubMed]
- Colombo, M.; Raposo, G.; Théry, C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 2014, 30, 255–289. [Google Scholar] [CrossRef] [PubMed]
- Escola, J.-M.; Kleijmeer, M.J.; Stoorvogel, W.; Griffith, J.M.; Yoshie, O.; Geuze, H.J. Selective enrichment of tetraspan proteins on the internal vesicles of multivesicular endosomes and on exosomes secreted by human B-lymphocytes. J. Biol. Chem. 1998, 273, 20121–20127. [Google Scholar] [CrossRef]
- Théry, C.; Boussac, M.; Véron, P.; Ricciardi-Castagnoli, P.; Raposo, G.; Garin, J.; Amigorena, S. Proteomic analysis of dendritic cell-derived exosomes: A secreted subcellular compartment distinct from apoptotic vesicles. J. Immunol. 2001, 166, 7309–7318. [Google Scholar] [CrossRef]
- Stein, J.M.; Luzio, J.P. Ectocytosis caused by sublytic autologous complement attack on human neutrophils. The sorting of endogenous plasma-membrane proteins and lipids into shed vesicles. Biochem. J. 1991, 274 Pt 2, 381–386. [Google Scholar] [CrossRef]
- Hess, C.; Sadallah, S.; Hefti, A.; Landmann, R.; Schifferli, J.A. Ectosomes released by human neutrophils are specialized functional units. J. Immunol. 1999, 163, 4564–4573. [Google Scholar] [CrossRef]
- Mause, S.F.; Ritzel, E.; Liehn, E.A.; Hristov, M.; Bidzhekov, K.; Müller-Newen, G.; Soehnlein, O.; Weber, C. Platelet microparticles enhance the vasoregenerative potential of angiogenic early outgrowth cells after vascular injury. Circulation 2010, 122, 495–506. [Google Scholar] [CrossRef]
- Cocucci, E.; Racchetti, G.; Meldolesi, J. Shedding microvesicles: Artefacts no more. Trends Cell Biol. 2009, 19, 43–51. [Google Scholar] [CrossRef]
- Bastida, E.; Ordinas, A.; Escolar, G.; Jamieson, G.A. Tissue factor in microvesicles shed from U87MG human glioblastoma cells induces coagulation, platelet aggregation, and thrombogenesis. Blood 1984, 64, 177–184. [Google Scholar] [CrossRef]
- Lotvall, J.; Valadi, H. Cell to cell signalling via exosomes through esRNA. Cell Adhes. Migr. 2007, 1, 156–158. [Google Scholar] [CrossRef] [PubMed]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Tvall, J.O.L.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, S.; Palanisamy, V. Horizontal transfer of RNAs: Exosomes as mediators of intercellular communication. Wiley Interdiscip. Rev. RNA 2012, 3, 286–293. [Google Scholar] [PubMed]
- El Andaloussi, S.; Lakhal, S.; Mäger, I.; Wood, M.J. Exosomes for targeted siRNA delivery across biological barriers. Adv. Drug Deliv. Rev. 2013, 65, 391–397. [Google Scholar] [CrossRef] [PubMed]
- Hill, A.F. Extracellular Vesicles and Neurodegenerative Diseases. J. Neurosci. 2019, 39, 9269–9273. [Google Scholar] [CrossRef] [PubMed]
- Vassileff, N.; Cheng, L.; Hill, A.F. Extracellular vesicles-propagators of neuropathology and sources of potential biomarkers and therapeutics for neurodegenerative diseases. J. Cell Sci. 2020, 133. [Google Scholar] [CrossRef]
- Gould, S.J.; Raposo, G. As we wait: Coping with an imperfect nomenclature for extracellular vesicles. J. Extracell. Vesicles 2013, 2, 20389. [Google Scholar] [CrossRef]
- Lötvall, J.; Hill, A.F.; Hochberg, F.; Buzás, E.I.; Di Vizio, D.; Gardiner, C.; Gho, Y.S.; Kurochkin, I.V.; Mathivanan, S.; Quesenberry, P.; et al. Minimal experimental requirements for definition of extracellular vesicles and their functions: A position statement from the International Society for Extracellular Vesicles. J. Extracell. Vesicles 2014, 3, 26913. [Google Scholar] [CrossRef]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef]
- Bianco, F.; Pravettoni, E.; Colombo, A.; Schenk, U.; Möller, T.; Matteoli, M.; Verderio, C. Astrocyte-derived ATP induces vesicle shedding and IL-1 beta release from microglia. J. Immunol. 2005, 174, 7268–7277. [Google Scholar]
- Drago, F.; Lombardi, M.; Prada, I.; Gabrielli, M.; Joshi, P.; Cojoc, D.; Franck, J.; Fournier, I.; Vizioli, J.; Verderio, C. ATP Modifies the Proteome of Extracellular Vesicles Released by Microglia and Influences Their Action on Astrocytes. Front. Pharmacol. 2017, 8, 910. [Google Scholar] [CrossRef] [PubMed]
- Prada, I.; Gabrielli, M.; Turola, E.; Iorio, A.; D’Arrigo, G.; Parolisi, R.; De Luca, M.; Pacifici, M.; Bastoni, M.; Lombardi, M.; et al. Glia-to-neuron transfer of miRNAs via extracellular vesicles: A new mechanism underlying inflammation-induced synaptic alterations. Acta Neuropathol. 2018, 135, 529–550. [Google Scholar] [CrossRef] [PubMed]
- Bianco, F.; Perrotta, C.; Novellino, L.; Francolini, M.; Riganti, L.; Menna, E.; Saglietti, L.; Schuchman, E.H.; Furlan, R.; Clementi, E.; et al. Acid sphingomyelinase activity triggers microparticle release from glial cells. EMBO J. 2009, 28, 1043–1054. [Google Scholar] [CrossRef] [PubMed]
- Dugger, B.N.; Dickson, D.W. Pathology of Neurodegenerative Diseases. Cold Spring Harb. Perspect Biol. 2017, 9, a028035. [Google Scholar] [CrossRef] [PubMed]
- Rajendran, L.; Honsho, M.; Zahn, T.R.; Keller, P.; Geiger, K.D.; Verkade, P.; Simons, K. Alzheimer’s disease beta-amyloid peptides are released in association with exosomes. Proc. Natl. Acad. Sci. USA 2006, 103, 11172–11177. [Google Scholar] [CrossRef]
- Vingtdeux, V.; Hamdane, M.; Loyens, A.; Gelé, P.; Drobeck, H.; Bégard, S.; Galas, M.-C.; Delacourte, A.; Beauvillain, J.-C.; Buée, L.; et al. Alkalizing drugs induce accumulation of amyloid precursor protein by-products in luminal vesicles of multivesicular bodies. J. Biol. Chem. 2007, 282, 18197–18205. [Google Scholar] [CrossRef]
- Saman, S.; Kim, W.; Raya, M.; Visnick, Y.; Miro, S.; Saman, S.; Jackson, B.; McKee, A.C.; Alvarez, V.E.; Lee, N.C.Y.; et al. Exosome-associated tau is secreted in tauopathy models and is selectively phosphorylated in cerebrospinal fluid in early Alzheimer disease. J. Biol. Chem. 2012, 287, 3842–3849. [Google Scholar] [CrossRef]
- Baker, S.; Polanco, J.C.; Götz, J. Extracellular Vesicles Containing P301L Mutant Tau Accelerate Pathological Tau Phosphorylation and Oligomer Formation but Do Not Seed Mature Neurofibrillary Tangles in ALZ17 Mice. J. Alzheimer’s Dis. 2016, 54, 1207–1217. [Google Scholar] [CrossRef]
- Polanco, J.C.; Scicluna, B.J.; Hill, A.F.; Götz, J. Extracellular Vesicles Isolated from the Brains of rTg4510 Mice Seed Tau Protein Aggregation in a Threshold-dependent Manner. J. Biol. Chem. 2016, 291, 12445–12466. [Google Scholar] [CrossRef]
- Wang, Y.; Balaji, V.; Kaniyappan, S.; Krüger, L.; Irsen, S.; Tepper, K.; Chandupatla, R.; Maetzler, W.; Schneider, A.; Mandelkow, E.; et al. The release and trans-synaptic transmission of Tau via exosomes. Mol. Neurodegener. 2017, 12, 5. [Google Scholar] [CrossRef]
- Emmanouilidou, E.; Melachroinou, K.; Roumeliotis, T.; Garbis, SD.; Ntzouni, M.; Margaritis, LH.; Stefanis, L.; Vekrellis, K. Cell-produced alpha-synuclein is secreted in a calcium-dependent manner by exosomes and impacts neuronal survival. J. Neurosci. 2010, 30, 6838–6851. [Google Scholar] [CrossRef] [PubMed]
- Danzer, K.M.; Kranich, L.R.; Ruf, W.P.; Cagsal-Getkin, O.; Winslow, AR.; Zhu, L.; Vanderburg, CR.; McLean, PJ. Exosomal cell-to-cell transmission of alpha synuclein oligomers. Mol. Neurodegener. 2012, 7, 42. [Google Scholar] [CrossRef] [PubMed]
- Fevrier, B.; Vilette, D.; Archer, F.; Loew, D.; Faigle, W.; Vidal, M.; Laude, H.; Raposo, G. Cells release prions in association with exosomes. Proc. Natl. Acad. Sci. USA 2004, 101, 9683–9688. [Google Scholar] [CrossRef] [PubMed]
- Vella, L.J.; Sharples, R.A.; Lawson, V.A.; Masters, C.L.; Cappai, R.; Hill, A.F. Packaging of prions into exosomes is associated with a novel pathway of PrP processing. J. Pathol. 2007, 211, 582–590. [Google Scholar] [CrossRef]
- Falker, C.; Hartmann, A.; Guett, I.; Dohler, F.; Altmeppen, H.; Betzel, C.; Schubert, R.; Thurm, D.; Wegwitz, F.; Joshi, P.; et al. Exosomal cellular prion protein drives fibrillization of amyloid beta and counteracts amyloid beta-mediated neurotoxicity. J. Neurochem. 2015, 137, 88–100. [Google Scholar] [CrossRef]
- Soares Martins, T.; Trindade, D.; Vaz, M.; Campelo, I.; Almeida, M.; Trigo, G.; da Cruz E Silva, O.A.B.; Henriques, A.G. Diagnostic and therapeutic potential of exosomes in Alzheimer’s disease. J. Neurochem. 2020. [Google Scholar] [CrossRef]
- VandenDriessche, C.; Bruggeman, A.; Van Cauwenberghe, C.; Vandenbroucke, R.E. Extracellular Vesicles in Alzheimer’s and Parkinson’s Disease: Small Entities with Large Consequences. Cells 2020, 9, 2485. [Google Scholar] [CrossRef]
- Puig, B.; Brenna, S.; Magnus, T. Molecular Communication of a Dying Neuron in Stroke. Int. J. Mol. Sci. 2018, 19, 2834. [Google Scholar] [CrossRef]
- Wang, M.-M.; Feng, Y.-S.; Tan, Z.-X.; Xing, Y.; Dong, F.; Zhang, F. The role of exosomes in stroke. Mol. Biol. Rep. 2020, 47, 6217–6228. [Google Scholar] [CrossRef]
- Zhang, Z.G.; Chopp, M. Exosomes in stroke pathogenesis and therapy. J. Clin. Investig. 2016, 126, 1190–1197. [Google Scholar]
- Yates, A.G.; Anthony, D.C.; Ruitenberg, M.J.; Couch, Y. Systemic Immune Response to Traumatic CNS Injuries-Are Extracellular Vesicles the Missing Link? Front. Immunol. 2019, 10, 2723. [Google Scholar] [CrossRef] [PubMed]
- Osier, N.; Motamedi, V.; Edwards, K.; Puccio, A.; Diaz-Arrastia, R.; Kenney, K.; Gill, J. Exosomes in Acquired Neurological Disorders: New Insights into Pathophysiology and Treatment. Mol. Neurobiol. 2018, 55, 9280–9293. [Google Scholar] [CrossRef] [PubMed]
- SStenz, K.T.; Just, J.; Blauenfeldt, R.A.; Drasbek, K.R. Extracellular Vesicles in Acute Stroke Diagnostics. Biomedicines 2020, 8, 248. [Google Scholar] [CrossRef] [PubMed]
- Guedes, V.A.; Devoto, C.; Leete, J.; Sass, D.; Acott, JD.; Mithani, S.; Gill, JM. Extracellular Vesicle Proteins and MicroRNAs as Biomarkers for Traumatic Brain Injury. Front. Neurol. 2020, 11, 663. [Google Scholar] [CrossRef]
- Doeppner, T.R.; Herz, J.; Görgens, A.; Schlechter, J.; Ludwig, A.K.; Radtke, S.; de Miroschedji, K.; Horn, P.A.; Giebel, B.; Hermann, D.M. Extracellular Vesicles Improve Post-Stroke Neuroregeneration and Prevent Postischemic Immunosuppression. Stem Cells Transl. Med. 2015, 4, 1131–1143. [Google Scholar] [CrossRef]
- Doeppner, T.R.; Bähr, M.; Hermann, D.M.; Giebel, B. Concise Review: Extracellular Vesicles Overcoming Limitations of Cell Therapies in Ischemic Stroke. Stem Cells Transl. Med. 2017, 6, 2044–2052. [Google Scholar] [CrossRef]
- Zhang, Z.G.; Chopp, M. Promoting brain remodeling to aid in stroke recovery. Trends Mol. Med. 2015, 21, 543–548. [Google Scholar] [CrossRef]
- Zhang, H.; Wu, J.; Fan, Q.; Zhou, J.; Wu, J.; Liu, S.; Zang, J.; Ye, J.; Xiao, M.; Tian, T.; et al. Exosome-mediated targeted delivery of miR-210 for angiogenic therapy after cerebral ischemia in mice. J. Nanobiotechnol. 2019, 17, 1–13. [Google Scholar] [CrossRef]
- Yuan, J.; Botchway, B.O.A.; Zhang, Y.; Wang, X.; Liu, X. Combined bioscaffold with stem cells and exosomes can improve traumatic brain injury. Stem Cell Rev. Rep. 2019, 16, 323–334. [Google Scholar] [CrossRef]
- Verderio, C.; Muzio, L.; Turola, E.; Bergami, A.; Novellino, L.; Ruffini, F.; Riganti, L.; Corradini, I.; Francolini, M.; Garzetti, L.; et al. Myeloid microvesicles are a marker and therapeutic target for neuroinflammation. Ann. Neurol. 2012, 72, 610–624. [Google Scholar] [CrossRef]
- Jimenez, J.; Jy, W.; Mauro, L.M.; Horstman, LL.; Ahn, ER.; Ahn, YS.; Minagar, A. Elevated endothelial microparticle-monocyte complexes induced by multiple sclerosis plasma and the inhibitory effects of interferon-beta 1b on release of endothelial microparticles, formation and transendothelial migration of monocyte-endothelial microparticle complexes. Mult. Scler. J. 2005, 11, 310–315. [Google Scholar]
- Ciregia, F.; Urbani, A.; Palmisano, G. Extracellular Vesicles in Brain Tumors and Neurodegenerative Diseases. Front. Mol. Neurosci. 2017, 10, 276. [Google Scholar] [CrossRef] [PubMed]
- Harshyne, L.A.; Hooper, K.M.; Andrews, E.G.; Nasca, BJ.; Kenyon, LC.; Andrews, DW.; Hooper, DC. Glioblastoma exosomes and IGF-1R/AS-ODN are immunogenic stimuli in a translational research immunotherapy paradigm. Cancer Immunol. Immunother. 2014, 64, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Harshyne, L.A.; Nasca, B.J.; Kenyon, L.C.; Andrews, D.W.; Hooper, D.C. Serum exosomes and cytokines promote a T-helper cell type 2 environment in the peripheral blood of glioblastoma patients. Neuro Oncol. 2015, 18, 206–215. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, S.; Yao, J.; Lowery, F.J.; Zhang, Q.; Huang, W.C.; Li, P.; Li, M.; Wang, X.; Zhang, C.; et al. Microenvironment-induced PTEN loss by exosomal microRNA primes brain metastasis outgrowth. Nature 2015, 527, 100–104. [Google Scholar] [CrossRef]
- Perez-Gonzalez, R.; Gauthier, S.A.; Kumar, A.; Levy, E. The exosome secretory pathway transports amyloid precursor protein carboxyl-terminal fragments from the cell into the brain extracellular space. J. Biol. Chem. 2012, 287, 43108–43115. [Google Scholar] [CrossRef]
- Vella, L.J.; Scicluna, B.J.; Cheng, L.; Bawden, E.G.; Masters, C.L.; Ang, C.S.; Willamson, N.; McLean, C.; Barnham, KL.; Hill, AF. A rigorous method to enrich for exosomes from brain tissue. J. Extracell. Vesicles 2017, 6, 1348885. [Google Scholar] [CrossRef]
- Brenna, S.; Altmeppen, H.C.; Mohammadi, B.; Rissiek, B.; Schlink, F.; Ludewig, P.; Krisp, C.; Schlüter, H.; Failla, A.V.; Schneider, C.; et al. Characterization of brain-derived extracellular vesicles reveals changes in cellular origin after stroke and enrichment of the prion protein with a potential role in cellular uptake. J. Extracell. Vesicles 2020, 9, 1809065. [Google Scholar] [CrossRef]
- Jeppesen, D.K.; Fenix, A.M.; Franklin, J.L.; Higginbotham, J.N.; Zhang, Q.; Coffey, R.J. Reassessment of Exosome Composition. Tumor Biology 2019, 177, 428–445.e418. [Google Scholar] [CrossRef]
- Gallart-Palau, X.; Serra, A.; Sze, S.K. Enrichment of extracellular vesicles from tissues of the central nervous system by PROSPR. Mol. Neurodegener. 2016, 11, 1–13. [Google Scholar] [CrossRef]
- Huang, Y.; Cheng, L.; Turchinovich, A.; Mahairaki, V.; Troncoso, J.C.; Pletniková, O.; Haughey, N.J.; Vella, L.J.; Hill, A.F.; Zheng, L.; et al. Influence of species and processing parameters on recovery and content of brain tissue-derived extracellular vesicles. J. Extracell. Vesicles 2020, 9, 1785746. [Google Scholar] [CrossRef] [PubMed]
- Asai, H.; Ikezu, S.; Tsunoda, S.; Medalla, M.; Luebke, J.; Haydar, T.; Wolozin, B.; Butovsky, O.; Kügler, S.; Ikezu, T. Depletion of microglia and inhibition of exosome synthesis halt tau propagation. Nat. Neurosci. 2015, 18, 1584–1593. [Google Scholar] [CrossRef] [PubMed]
- Witwer, K.W.; Buzás, E.I.; Bemis, L.T.; Bora, A.; Lässer, C.; Lötvall, J.; Hoen, E.N.N.; Piper, M.G.; Sivaraman, S.; Skog, J.; et al. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J. Extracell. Vesicles 2013, 2. [Google Scholar] [CrossRef] [PubMed]
- Royo, F.; Théry, C.; Falcon-Perez, J.M.; Nieuwland, R.; Witwer, K.W. Methods for Separation and Characterization of Extracellular Vesicles: Results of a Worldwide Survey Performed by the ISEV Rigor and Standardization Subcommittee. Cells 2020, 9, 1955. [Google Scholar] [CrossRef] [PubMed]
- Cocozza, F.; Grisard, E.; Martin-Jaular, L.; Mathieu, M.; Théry, C. SnapShot: Extracellular Vesicles. Cell 2020, 182, 262–262.e1. [Google Scholar] [CrossRef] [PubMed]
- Brennan, K.; Martin, K.; FitzGerald, S.P.; O’Sullivan, J.; Wu, Y.; Blanco, A.; Richardson, C.; Mc Gee, MM. A comparison of methods for the isolation and separation of extracellular vesicles from protein and lipid particles in human serum. Sci. Rep. 2020, 10, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Dash, M.; Palaniyandi, K.; Ramalingam, S.; Sahabudeen, S.; Raja, N. Exosomes isolated from two different cell lines using three different isolation techniques show variation in physical and molecular characteristics. Biochim. Biophys. Acta BBA Biomembr. 2021, 1863, 183490. [Google Scholar] [CrossRef]
- Pérez-González, R.; Gauthier, S.A.; Kumar, A.; Saito, M.; Saito, M.; Levy, E. A Method for Isolation of Extracellular Vesicles and Characterization of Exosomes from Brain Extracellular Space. Methods Mol. Biol. 2017, 1545, 139–151. [Google Scholar] [CrossRef]
- Polanco, J.C.; Li, C.; Durisic, N.; Sullivan, R.; Götz, J. Exosomes taken up by neurons hijack the endosomal pathway to spread to interconnected neurons. Acta Neuropathol. Commun. 2018, 6, 1–14. [Google Scholar] [CrossRef]
- Dagur, R.S.; Liao, K.; Sil, S.; Niu, F.; Sun, Z.; Lyubchenko, YL.; Peeples, ES.; Hu, G.; Bucha, S. Neuronal-derived extracellular vesicles are enriched in the brain and serum of HIV-1 transgenic rats. J. Extracell. Vesicles 2020, 9, 1703249. [Google Scholar] [CrossRef]
- Gallart-Palau, X.; Serra, A.; Wong, A.S.; Sandin, S.; Lai, M.K.P.; Chen, C.P.; Kon, O.L.; Sze, S.K. Extracellular vesicles are rapidly purified from human plasma by PRotein Organic Solvent PRecipitation (PROSPR). Sci. Rep. 2015, 5, 14664. [Google Scholar] [CrossRef] [PubMed]
- Ruf, J.; Vairo, D.; Paganelli, F.; Guieu, R. Extracellular vesicles with ubiquitinated adenosine A. J. Cell. Mol. Med. 2019, 23, 6805–6811. [Google Scholar] [CrossRef]
- Silverman, J.M.; Christy, D.; Shyu, C.C.; Moon, K.-M.; Fernando, S.; Gidden, Z.; Cowan, C.M.; Ban, Y.; Stacey, R.G.; Grad, L.I.; et al. CNS-derived extracellular vesicles from superoxide dismutase 1 (SOD1). J. Biol. Chem. 2019, 294, 3744–3759. [Google Scholar] [CrossRef] [PubMed]
- Kowal, J.; Arras, G.; Colombo, M.; Jouve, M.; Morath, JP.; Primdal-Bengtson, B.; Dingli, F.; Loew, D.; Tkach, M.; Théry, C. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. USA 2016, 113, E968–E977. [Google Scholar] [CrossRef]
- Yelamanchili, S.V.; Lamberty, B.G.; Rennard, D.A.; Morsey, B.M.; Hochfelder, C.G.; Meays, B.M.; Levy, E.; Fox, H.S. MiR-21 in Extracellular Vesicles Leads to Neurotoxicity via TLR7 Signaling in SIV Neurological Disease. PLoS Pathog. 2015, 11, e1005032. [Google Scholar]
- Shahjin, F.; Guda, R.S.; Schaal, V.L.; Odegaard, K.; Clark, A.; Gowen, A.; Xiao, P.; Lisco, S.J.; Pendyala, G.; Yelamanchili, S.V. Brain-Derived Extracellular Vesicle microRNA Signatures Associated with In Utero and Postnatal Oxycodone Exposure. Cells 2019, 9, 21. [Google Scholar] [CrossRef]
- Volovitz, I.; Shapira, N.; Ezer, H.; Gafni, A.; Lustgarten, M.; Alter, T.; Ben-Horin, I.; Barzilai, O.; Shahar, T.; Kanner, A.; et al. A non-aggressive, highly efficient, enzymatic method for dissociation of human brain-tumors and brain-tissues to viable single-cells. BMC Neurosci. 2016, 17, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Levy, E. Exosomes in the Diseased Brain: First Insights from. Front. Neurosci. 2017, 11, 142. [Google Scholar] [CrossRef]
- Gallart-Palau, X.; Guo, X.; Serra, A.; Sze, S.K. Alzheimer’s disease progression characterized by alterations in the molecular profiles and biogenesis of brain extracellular vesicles. Alzheimer’s Res. Ther. 2020, 12, 54. [Google Scholar] [CrossRef]
- Pérez-González, R.; Kim, Y.; Miller, C.; Pacheco-Quinto, J.; Eckman, E.A.; Levy, E. Extracellular vesicles: Where the amyloid precursor protein carboxyl-terminal fragments accumulate and amyloid-β oligomerizes. FASEB J. 2020, 34, 12922–12931. [Google Scholar] [CrossRef]
- Gallart-Palau, X.; Serra, A.; Hase, Y.; Tan, C.F.; Chen, C.P.; Kalaria, R.N.; Sze, S.K. Brain-derived and circulating vesicle profiles indicate neurovascular unit dysfunction in early Alzheimer’s disease. Brain Pathol. 2019, 29, 593–605. [Google Scholar] [CrossRef] [PubMed]
- Papadopoulos, V.E.; Nikolopoulou, G.; Antoniadou, I.; Karachaliou, A.; Arianoglou, G.; Emmanouilidou, E.; Sardi, S.P.; Stefanis, L.; Vekrellis, K. Modulation of β-glucocerebrosidase increases α-synuclein secretion and exosome release in mouse models of Parkinson’s disease. Hum. Mol. Genet. 2018, 27, 1696–1710. [Google Scholar] [CrossRef] [PubMed]
- Ngolab, J.; Trinh, I.; Rockenstein, E.; Mante, M.; Florio, J.; Trejo, M.; Masliah, D.; Adame, A.; Masliah, E.; Rissman, R.A. Brain-derived exosomes from dementia with Lewy bodies propagate α-synuclein pathology. Acta Neuropathol. Commun. 2017, 5, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Vassileff, N.; Vella, L.J.; Rajapaksha, H.; Shambrook, M.; Kenari, A.N.; McLean, C.; Hill, A.F.; Cheng, L. Revealing the Proteome of Motor Cortex Derived Extracellular Vesicles Isolated from Amyotrophic Lateral Sclerosis Human Postmortem Tissues. Cells 2020, 9, 1709. [Google Scholar] [CrossRef] [PubMed]
- Chand, S.; Jo, A.; Vellichirammal, N.N.; Gowen, A.; Guda, C.; Schaal, V.; Odegaard, K.; Lee, H.; Pendyala, G.; Yelamanchili, SV. Comprehensive Characterization of Nanosized Extracellular Vesicles from Central and Peripheral Organs: Implications for Preclinical and Clinical Applications. ACS Appl. Nano Mater. 2020, 3, 8906–8919. [Google Scholar] [CrossRef] [PubMed]

| Human | Mouse | ||
|---|---|---|---|
| Study | Proteins | Study | Proteins |
| Gallart-Palau et al. | 3056 | Silverman et al. | 1191 |
| Huang et al. | 714 | Gallart-Palau et al. | 444 |
| Vella et al. F2 | 1144 | Brenna et al. | 1518 |
| Vella et al. F3 | 815 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brenna, S.; Krisp, C.; Altmeppen, H.C.; Magnus, T.; Puig, B. Brain-Derived Extracellular Vesicles in Health and Disease: A Methodological Perspective. Int. J. Mol. Sci. 2021, 22, 1365. https://doi.org/10.3390/ijms22031365
Brenna S, Krisp C, Altmeppen HC, Magnus T, Puig B. Brain-Derived Extracellular Vesicles in Health and Disease: A Methodological Perspective. International Journal of Molecular Sciences. 2021; 22(3):1365. https://doi.org/10.3390/ijms22031365
Chicago/Turabian StyleBrenna, Santra, Christoph Krisp, Hermann Clemens Altmeppen, Tim Magnus, and Berta Puig. 2021. "Brain-Derived Extracellular Vesicles in Health and Disease: A Methodological Perspective" International Journal of Molecular Sciences 22, no. 3: 1365. https://doi.org/10.3390/ijms22031365
APA StyleBrenna, S., Krisp, C., Altmeppen, H. C., Magnus, T., & Puig, B. (2021). Brain-Derived Extracellular Vesicles in Health and Disease: A Methodological Perspective. International Journal of Molecular Sciences, 22(3), 1365. https://doi.org/10.3390/ijms22031365







