Genotypic Homogeneity in Distinctive Transforming Growth Factor-Beta Induced (TGFBI) Protein Phenotypes
Abstract
1. Introduction
2. Patients and Methods
2.1. Patients and Blood Sample Collection
2.2. PCR Amplification Reaction and Sequencing
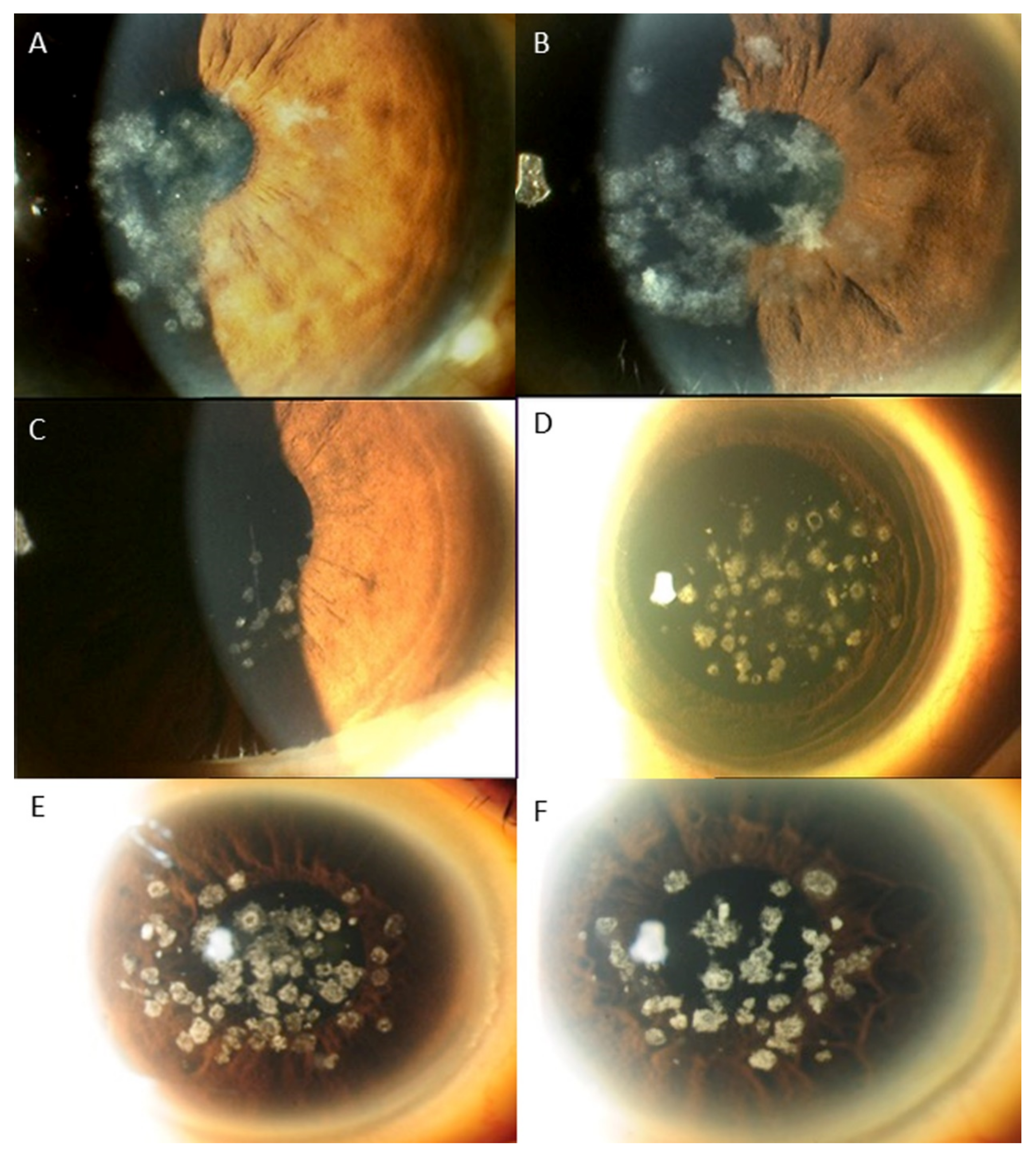
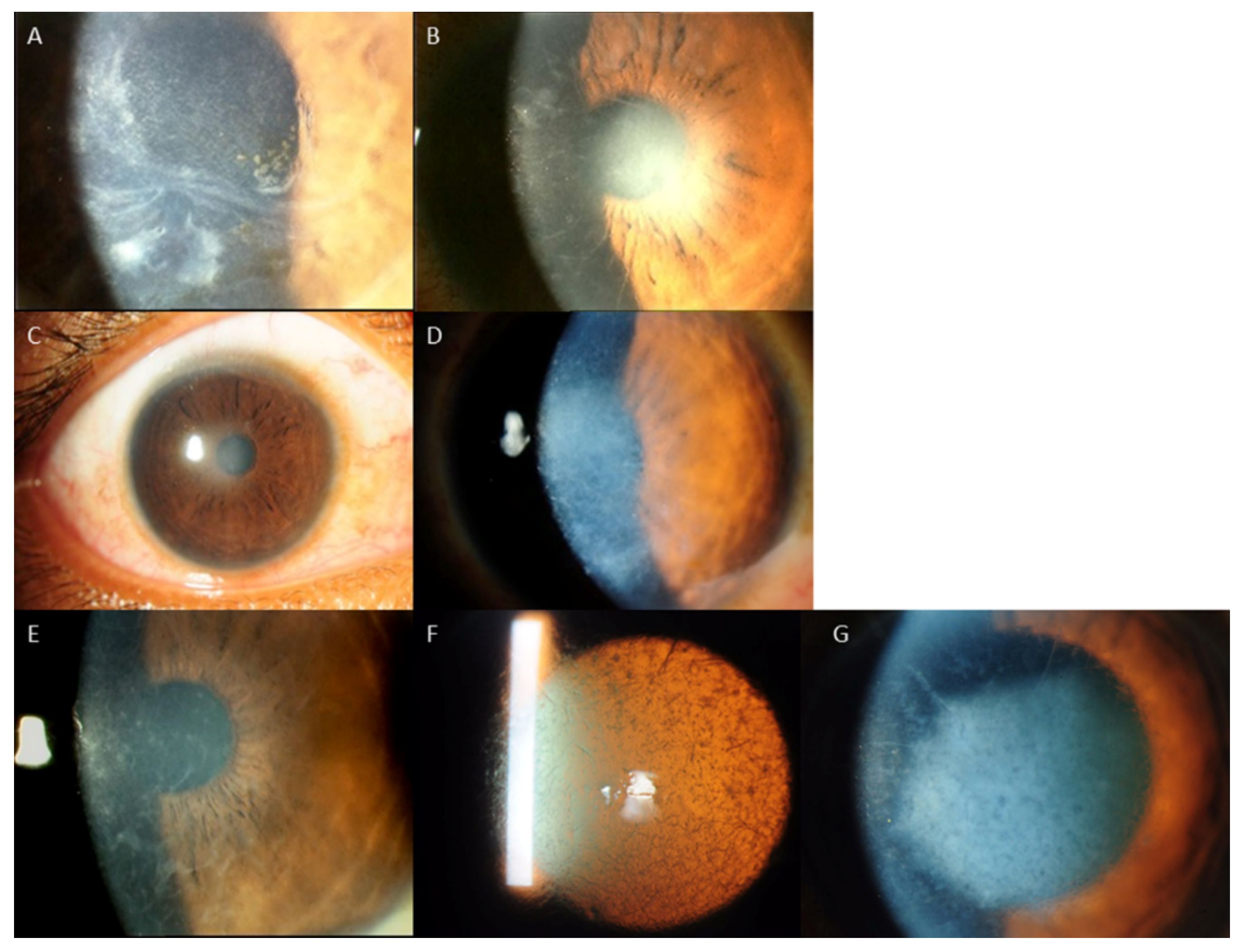
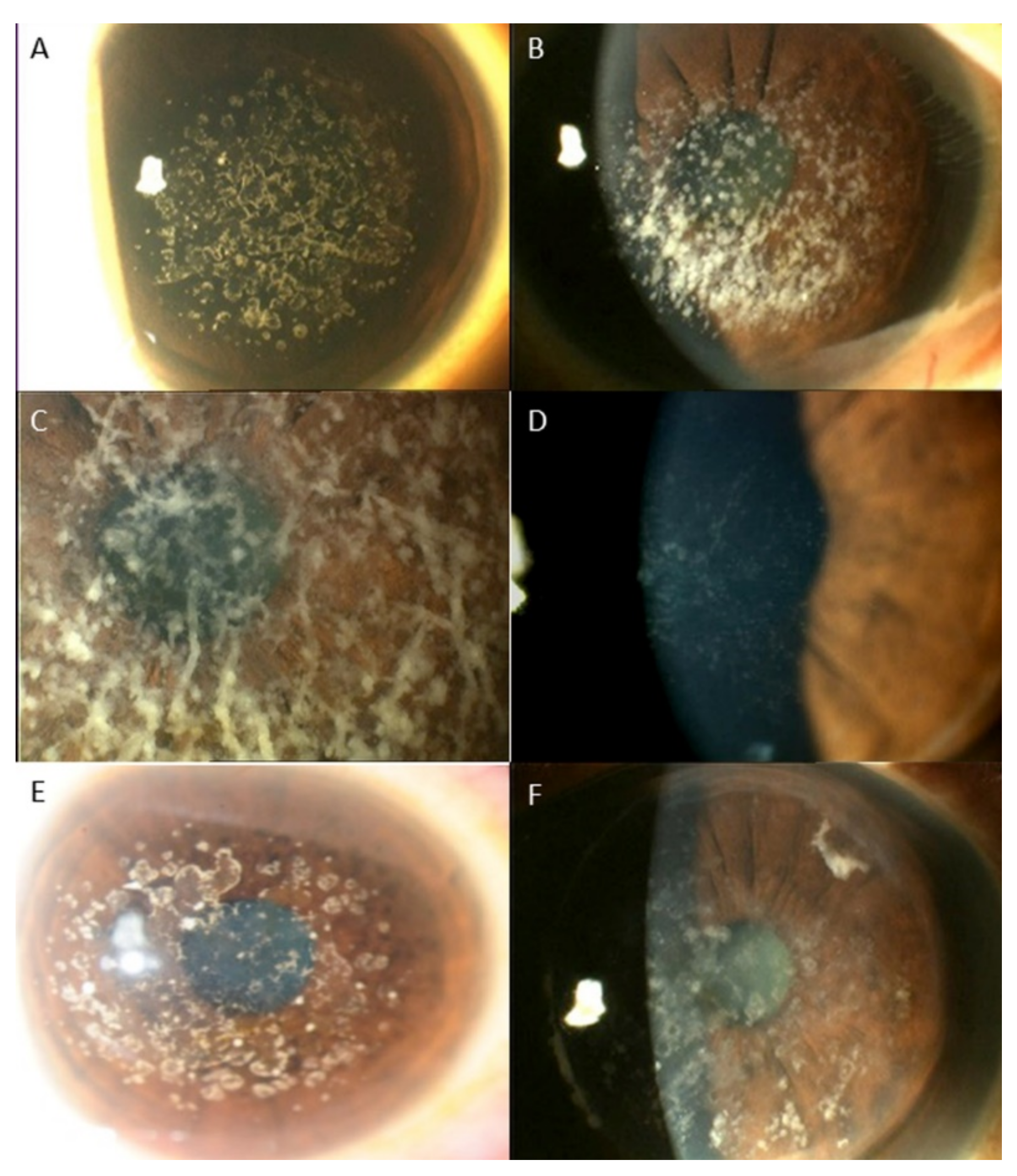
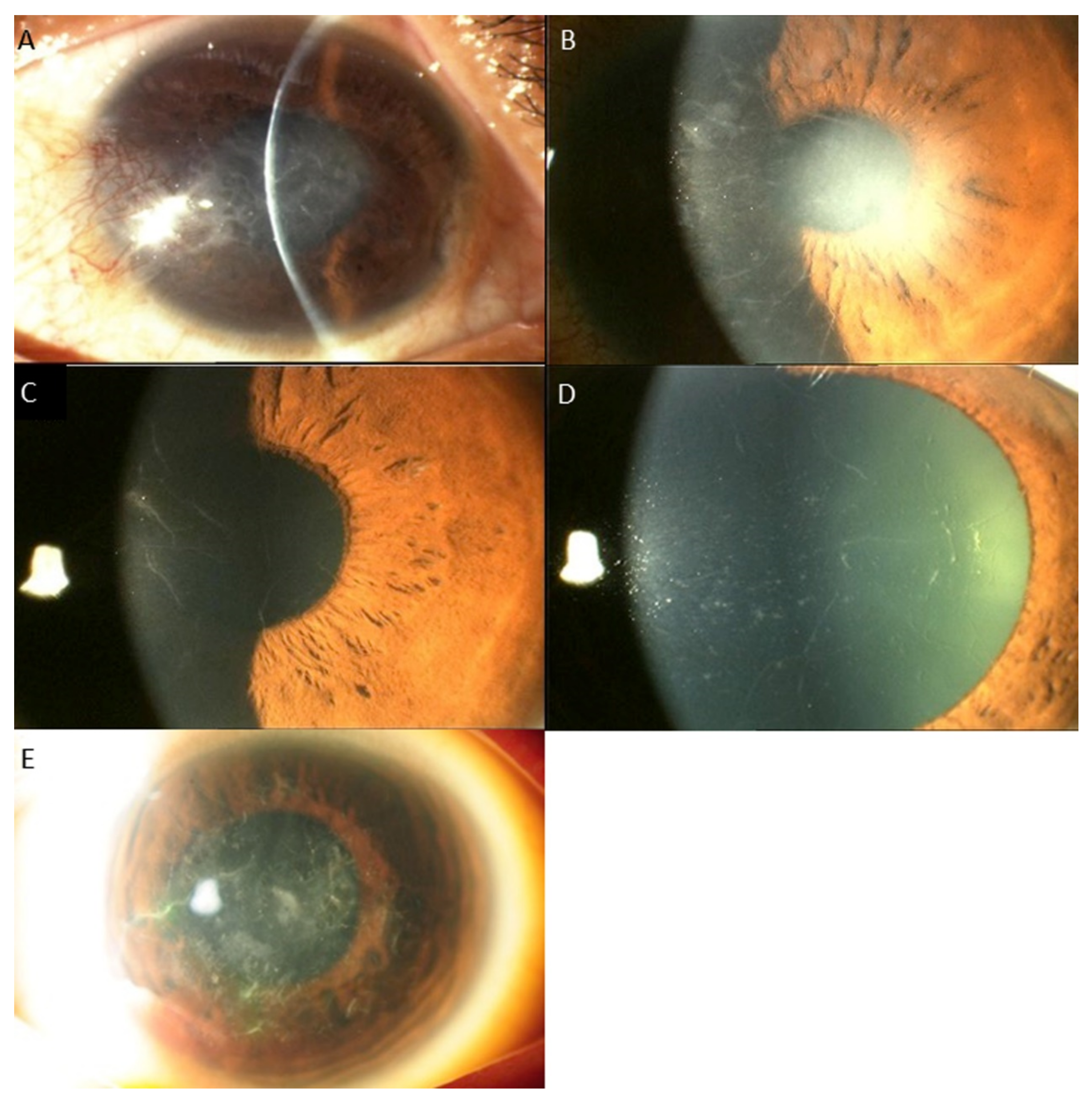
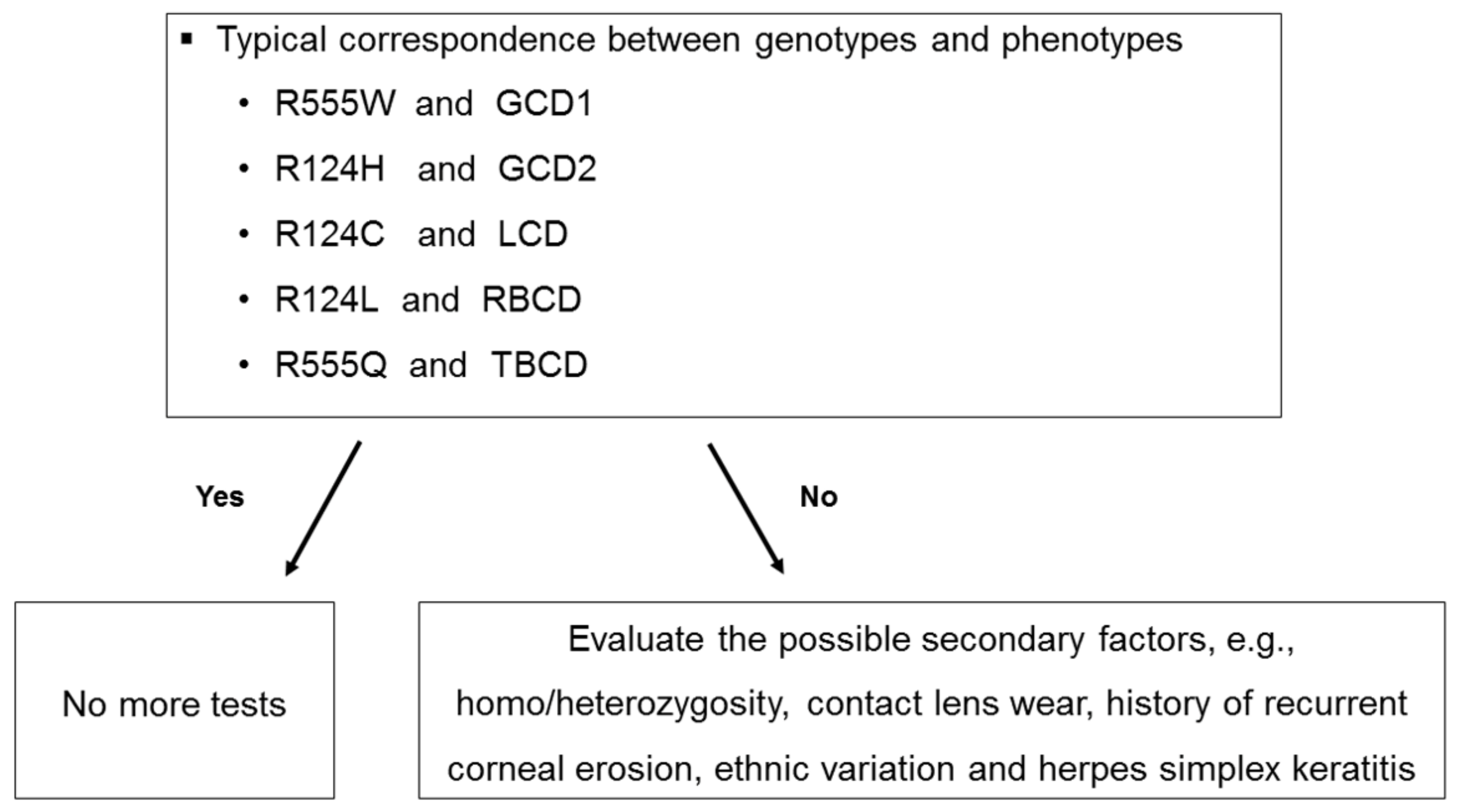
3. Results
Distribution of Genotypes and Phenotypes
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, T.; Yan, N.; Yu, W.; Liu, Y.; Liu, G.; Wu, X.; Lian, J.; Liu, X. Molecular genetics of Chinese families with TGFBI corneal dystrophies. Mol. Vis. 2011, 17, 380–387. [Google Scholar] [PubMed]
- Lakshminarayanan, R.; Chaurasia, S.S.; Anandalakshmi, V.; Chai, S.M.; Murugan, E.; Vithana, E.N.; Beuerman, R.W.; Mehta, J.S. Clinical and genetic aspects of the TGFBI-associated corneal dystrophies. Ocul. Surf. 2014, 12, 234–251. [Google Scholar] [CrossRef] [PubMed]
- Kheir, V.; Cortes-Gonzalez, V.; Zenteno, J.C.; Schorderet, D.F. Mutation update: TGFBI pathogenic and likely pathogenic variants in corneal dystrophies. Hum. Mutat. 2019, 40, 675–693. [Google Scholar] [CrossRef] [PubMed]
- Munier, F.L.; Korvatska, E.; Djemaï, A.; Le Paslier, D.; Zografos, L.; Pescia, G.; Schorderet, D.F. Kerato-epithelin mutations in four 5q31-linked corneal dystrophies. Nat. Genet. 1997, 15, 247–251. [Google Scholar] [CrossRef]
- Han, K.E.; Choi, S.I.; Kim, T.I.; Maeng, Y.S.; Stulting, R.D.; Ji, Y.W.; Kim, E.K. Pathogenesis and treatments of TGFBI corneal dystrophies. Prog. Retin. Eye Res. 2016, 50, 67–88. [Google Scholar] [CrossRef] [PubMed]
- Weiss, J.S.; Møller, H.U.; Lisch, W.; Kinoshita, S.; Aldave, A.J.; Belin, M.W.; Kivelä, T.; Busin, M.; Munier, F.L.; Seitz, B.; et al. The IC3D classification of the corneal dystrophies. Cornea 2008, 27 (Suppl. 2), S1–S83. [Google Scholar]
- Weiss, J.S.; Møller, H.U.; Aldave, A.J.; Seitz, B.; Bredrup, C.; Kivelä, T.; Munier, F.L.; Rapuano, C.J.; Nischal, K.K.; Kim, E.K.; et al. IC3D classification of corneal dystrophies—Edition 2. Cornea 2015, 34, 117–159. [Google Scholar] [CrossRef]
- Evans, C.J.; Davidson, A.E.; Carnt, N.; Rojas Lopez, K.E.; Veli, N.; Thaung, C.M.; Tuft, S.J.; Hardcastle, A.J. Genotype-Phenotype Correlation for TGFBI Corneal Dystrophies Identifies p.(G623D) as a Novel Cause of Epithelial Basement Membrane Dystrophy. Investig. Ophthalmol. Vis. Sci. 2016, 57, 5407–5414. [Google Scholar] [CrossRef]
- Nowinska, A.K.; Wylegala, E.; Janiszewska, D.A.; Dobrowolski, D.; Aragona, P.; Roszkowska, A.M.; Puzzolo, D. Genotype-phenotype correlation of TGFBI corneal dystrophies in Polish patients. Mol. Vis. 2011, 17, 2333–2342. [Google Scholar]
- Hou, Y.C.; Wang, I.J.; Hsiao, C.H.; Chen, W.L.; Hu, F.R. Phenotype-genotype correlations in patients with TGFBI-linked corneal dystrophies in Taiwan. Mol. Vis. 2012, 18, 362–371. [Google Scholar]
- Cao, W.; Ge, H.; Cui, X.; Zhang, L.; Bai, J.; Fu, S.; Liu, P. Reduced penetrance in familial Avellino corneal dystrophy associated with TGFBI mutations. Mol. Vis. 2009, 15, 70–75. [Google Scholar] [PubMed]
- Chang., L.; Zhiqun, W.; Shijing, D.; Chen, Z.; Qingfeng, L.; Li, L.; Xuguang, S. Arg124Cys mutation of the TGFBI gene in 2 Chinese families with Thiel-Behnke corneal dystrophy. Arch. Ophthalmol. 2009, 127, 641–644. [Google Scholar] [CrossRef] [PubMed]
- Patel, D.A.; Chang, S.-H.; Harocopos, G.J.; Vora, S.C.; Thang, D.H.; Huang, A.J.W. Granular and lattice deposits in corneal dystrophy caused by R124C mutation of TGFBIp. Cornea 2010, 29, 1215–1222. [Google Scholar] [CrossRef] [PubMed]
- Han, K.E.; Choi, S.I.; Chung, W.S.; Maeng, Y.S.; Stulting, R.D.; Ji, Y.W.; Kim, E.K. Extremely varied phenotypes in granular corneal dystrophy type 2 heterozygotes. Mol. Vis. 2012, 18, 1755–1762. [Google Scholar] [PubMed]
- Watanabe, H.; Hashida, Y.; Tsujikawa, K.; Tsujikawa, M.; Maeda, N.; Inoue, Y.; Yamamoto, S.; Tano, Y. Two patterns of opacity in corneal dystrophy caused by the homozygous BIG-H3 R124H mutation. Am. J. Ophthalmol. 2001, 132, 211–216. [Google Scholar] [CrossRef]
- Yang, Q.-N.; Zhao, Y.-W.; Guo, L.-H.; Yan, N.H.; Liu, X.Y.; Cai, S.P. Arg124Cys mutation of the TGFBI gene in a Chinese pedigree of Reis-Bücklers corneal dystrophy. Int. J. Ophthalmol. 2011, 4, 235–238. [Google Scholar]
- Ma, K.; Liu, G.; Yang, Y.; Yu, M.; Sui, R.; Yu, W.; Chen, X.; Deng, Y.; Yan, N.; Cao, G.; et al. TGFBI gene mutation analysis in a Chinese pedigree of Reis-Bücklers corneal dystrophy. Mol. Vis. 2010, 16, 556–561. [Google Scholar]
- Aldave, A.J.; Rayner, S.A.; Kim, B.T.; Prechanond, A.; Yellore, V.S. Unilateral lattice corneal dystrophy associated with the novel His572del mutation in the TGFBI gene. Mol. Vis. 2006, 12, 142–146. [Google Scholar]
- Fujiki, K.; Nakayasu, K.; Kanai, A. Corneal dystrophies in Japan. J. Hum. Genet. 2001, 46, 431–435. [Google Scholar] [CrossRef]
- Lee, J.H.; Cristol, S.M.; Kim, W.C.; Chung, E.S.; Tchah, H.; Kim, M.S.; Nam, C.M.; Cho, H.S.; Kim, E.K. Prevalence of granular corneal dystrophy type 2 (Avellino corneal dystrophy) in the Korean population. Ophthalmic Epidemiol. 2010, 17, 160–165. [Google Scholar] [CrossRef]
- Mashima, Y.; Yamamoto, S.; Inoue, Y.; Imamura, Y.; Yamada, M.; Ogata, T.; Kudoh, J.; Shimizu, N. Association of autosomal dominantly inherited corneal dystrophies with BIGH3 gene mutations in Japan. Am. J. Ophthalmol. 2000, 130, 516–517. [Google Scholar] [CrossRef]
- Chao-Shern, C.; DeDionisio, L.A.; Jang, J.H.; Chan, C.C.; Thompson, V.; Christie, K.; Nesbit, M.A.; McMullen, C.B.T. Evaluation of TGFBI corneal dystrophy and molecular diagnostic testing. Eye 2019, 33, 874–881. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Liu, Y.; Guan, T. Analysis of TGFBI Gene Mutations in Three Chinese Families with Corneal Dystrophy. J. Ophthalmol. 2019, 2019, 6769013. [Google Scholar] [CrossRef] [PubMed]
- Lakshminarayanan, R.; Vithana, E.N.; Chai, S.M.; Chaurasia, S.S.; Saraswathi, P.; Venkatraman, A.; Rojare, C.; Venkataraman, D.; Tan, D.; Aung, T.; et al. A novel mutation in transforming growth factor-beta induced protein (TGFβIp) reveals secondary structure perturbation in lattice corneal dystrophy. Br. J. Ophthalmol. 2011, 95, 1457–1462. [Google Scholar] [CrossRef]
- Fujiki, K.; Hotta, Y.; Nakayasu, K.; Yamaguchi, T.; Kato, T.; Uesugi, Y.; Ha, N.T.; Endo, S.; Ishida, N.; Lu, W.N.; et al. Six different mutations of TGFBI (betaig-h3, keratoepithelin) gene found in Japanese corneal dystrophies. Cornea 2000, 19, 842–845. [Google Scholar] [CrossRef]
- Yoshida, S.; Kumano, Y.; Yoshida, A.; Hisatomi, T.; Matsui, H.; Nishida, T.; Ishibashi, T.; Matsui, T. An analysis of BIGH3 mutations in patients with corneal dystrophies in the Kyushu district of Japan. Jpn. J. Ophthalmol. 2002, 46, 469–471. [Google Scholar] [CrossRef]
- Cho, K.J.; Mok, J.W.; Na, K.S.; Rho, C.R.; Byun, Y.S.; Hwang, H.S.; Hwang, K.Y.; Joo, C.K. TGFBI gene mutations in a Korean population with corneal dystrophy. Mol. Vis. 2012, 18, 2012–2021. [Google Scholar]
- Song, J.S.; Lim, D.H.; Chung, E.S.; Chung, T.Y.; Ki, C.S. Mutation Analysis of the TGFBI Gene in Consecutive Korean Patients with Corneal Dystrophies. Ann. Lab. Med. 2015, 35, 336–340. [Google Scholar] [CrossRef]
- Paliwal, P.; Sharma, A.; Tandon, R.; Sharma, A.; Vajpayee, R.B. TGFBI mutation screening and genotype-phenotype correlation in north Indian patients with corneal dystrophies. Mol. Vis. 2010, 16, 1429–1438. [Google Scholar]
- Chakravarthi, S.V.V.K.; Kannabiran, C.; Sridhar, M.S.; Vemuganti, G.K. TGFBI gene mutations causing lattice and granular corneal dystrophies in Indian patients. Investig. Ophthalmol. Vis. Sci. 2005, 46, 121–125. [Google Scholar] [CrossRef]
- Yang, J.; Han, X.; Huang, D.; Yu, L.; Zhu, Y.; Tong, Y.; Zhu, B.; Li, C.; Weng, M.; Ma, X.; et al. Analysis of TGFBI gene mutations in Chinese patients with corneal dystrophies and review of the literature. Mol. Vis. 2010, 16, 1186–1193. [Google Scholar]
- Wang, X.; Ying, M.; Fu, C.; Wang, Y.; Li, N. TGFBI gene mutations analysis in Chinese families with corneal dystrophies. Mol. Med. Rep. 2017, 15, 3198–3202. [Google Scholar] [CrossRef] [PubMed]
- Edelstein, S.L.; Huang, A.J.; Harocopos, G.J.; Waltman, S.R. Genotype of lattice corneal dystrophy (R124C mutation in TGFBI) in a patient presenting with features of avellino corneal dystrophy. Cornea 2010, 29, 698–700. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.W.; Kim, S.A.; Kang, E.M.; Kim, T.I.; Cho, H.S.; Kim, E.K. Lattice corneal dystrophy type IIIA with hyaline component from a novel A620P mutation and distinct surgical treatments. Cornea 2014, 33, 1324–1331. [Google Scholar] [CrossRef] [PubMed]
- Munier, F.L.; Frueh, B.E.; Othenin-Girard, P.; Uffer, S.; Cousin, P.; Wang, M.X.; Héon, E.; Black, G.C.; Blasi, M.A.; Balestrazzi, E.; et al. BIGH3 mutation spectrum in corneal dystrophies. Investig. Ophthalmol. Vis. Sci. 2002, 43, 949–954. [Google Scholar]
- Wheeldon, C.E.; de Karolyi, B.H.; Patel, D.V.; Sherwin, T.; McGhee, C.N.; Vincent, A.L. A novel phenotype-genotype relationship with a TGFBI exon 14 mutation in a pedigree with a unique corneal dystrophy of Bowman’s layer. Mol. Vis. 2008, 14, 1503–1512. [Google Scholar]
- Liskova, P.; Klintworth, G.K.; Bowling, B.L.; Filipec, M.; Jirsova, K.; Tuft, S.J.; Bhattacharya, S.S.; Hardcastle, A.J.; Ebenezer, N.D. Phenotype associated with the H626P mutation and other changes in the TGFBI gene in Czech families. Ophthalmic Res. 2008, 40, 105–108. [Google Scholar] [CrossRef]
- Solari, H.P.; Ventura, M.P.; Perez, A.B.; Sallum, J.M.; Burnier, M.N., Jr.; Belfort, R., Jr. TGFBI gene mutations in Brazilian patients with corneal dystrophy. Eye 2007, 21, 587–590. [Google Scholar] [CrossRef]
- Zeng, L.; Zhao, J.; Chen, Y.; Zhao, F.; Li, M.; Chao-Shern, C.; Moore, T.; Marshall, J.; Zhou, X. TGFBI Gene Mutation Analysis of Clinically Diagnosed Granular Corneal Dystrophy Patients Prior to PTK: A Pilot Study from Eastern China. Sci. Rep. 2017, 7, 596. [Google Scholar] [CrossRef]
- Roters, S.; Severin, M.; Konen, W.; Krieglstein, G.K. Treatment of granular dystrophy with soft contact lenses. Ophthalmologica 2004, 218, 70–72. [Google Scholar] [CrossRef]
- Okada, M.; Yamamoto, S.; Inoue, Y.; Watanabe, H.; Maeda, N.; Shimomura, Y.; Ishii, Y.; Tano, Y. Severe corneal dystrophy phenotype caused by homozygous R124H keratoepithelin mutations. Investig. Ophthalmol. Vis. Sci. 1998, 39, 1947–1953. [Google Scholar]
- Fujiki, K.; Hotta, Y.; Nakayasu, K.; Kanai, A. Homozygotic patient with betaig-h3 gene mutation in granular dystrophy. Cornea 1998, 17, 288–292. [Google Scholar] [CrossRef] [PubMed]
- Konishi, M.; Yamada, M.; Nakamura, Y.; Mashima, Y. Varied appearance of cornea of patients with corneal dystrophy associated with R124H mutation in the BIGH3 gene. Cornea 1999, 18, 424–429. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.W.; Kim, H.M.; Song, J.S. Phenotypic non-penetrance in granular corneal dystrophy type II. Graefes Arch. Clin. Exp. Ophthalmol. 2008, 246, 1629–1631. [Google Scholar] [CrossRef]
| Exon | Forward Primer | Reverse Primer | Product Size (bp) |
|---|---|---|---|
| 1 | CGGAGGCGCTCTCACTTCC | CGAGCCCCGACTACCTGACC | 265 |
| 2 | GGGAGTCATTAAAGTGGGGTGGA | AGCTTGGTCTCCTGGCTGGTTAC | 99 |
| 3 | CAACTTAGTGGAGAGGGGCCAGA | CTCTCTCCCACCATTCCCTTCC | 206 |
| 4 | GCCATCCCTCCTTCTGTCTTCTG | CCGGGCAGACGGAGGTCATC | 217 |
| 5 | ACTGACACCCTGTCCTTCCTCCT | AGCCCACACATGGAACAGAAATG | 261 |
| 6 | CTGCTCATCCTTGCTGCTTCTCT | AGAGTTCCTGCTAGGCCCCTCTT | 249 |
| 7 | TCTGTGGGGAGTGCCAGAGTC | CAAATGAGGCAGCAGCAGGA | 234 |
| 8 | TGGACCCTGACTTGACCTGAGTC | AAAGGATGGCAGAAGAGATGGTG | 311 |
| 9 | CCCTGGGGTGGATGAATGATAAA | GCCTCCAGGGACAATCTAACAGG | 251 |
| 10 | ATTGCAGGAGCACATCTCTCTGG | GCTTCCCAGGAGCATGATTTAGG | 230 |
| 11 | GCCCCTCGTGGAAGTATAACCAG | ATCCCACTCCAGCATGACCACT | 248 |
| 12 | TGACAGGTGACATTTTCTGTGTGTG | GGGCCCTGAGGGATCACTACTTT | 224 |
| 13 | TGACCAGGCTAATTACCATTCTTGG | CAGCCTTTGATTTGCAGGACACT | 210 |
| 14 | TGCTCTACTTTCAACCACTACTCTG | CCAACTGCCACATGAAGAAAAGG | 198 |
| 15 | TCACTCTGGTCAAACCTGCCTTT | CCTCTATGGCCCAAACAGAGGAC | 183 |
| 16 | GCCATTGTCATAAGCAGTTGCAG | ATACAGCAGATGGCAGGCTTGG | 176 |
| 17 | TGGGGAGATCTGCACCTATTTGA | GGTCAGCACACTGTACCATGCAC | 710 |
| Phenotype | Associated TGFBI Mutations (Genotypes) |
|---|---|
| Granular corneal dystrophy (GCD), type 1 (n = 11) | R124H (n = 3) |
| R555W (n = 8) | |
| Granular corneal dystrophy (GCD), type 2 (n = 6) | R124H (n = 5) |
| R124C (n = 1) | |
| Lattice corneal dystrophy (LCD) (n = 13) | R124C (n = 7) |
| H626R (n = 2) | |
| L550P (n = 2) | |
| A620D (n = 1) | |
| H572R (n = 1) | |
| Reis-Buckler dystrophy (RBCD) (n = 2) | R124L (n = 1) |
| R555Q (n = 1) |
| Ethnicity | Phenotype | Associated TGFBI Mutations |
|---|---|---|
| Chinese (n = 16) | GCD1 (n = 5) | R124H (n = 3) |
| R555W (n = 2) | ||
| GCD2 (n = 5) | R124H (n = 4) | |
| R124C (n = 1) | ||
| LCD (n = 4) | R124C (n = 1) | |
| A620D (n = 1) | ||
| H626R (n = 2) | ||
| RBCD (n = 2) | R124L (n = 1) | |
| R555Q (n = 1) | ||
| Malay (n = 6) | LCD (n = 6) | R124C (n = 6) |
| Indian (n = 5) | GCD1 (n = 5) | R555W (n = 5) |
| Others (n = 5) | GCD1 (n = 1) | R555W (n = 1) |
| GCD2 (n =1) | R124H (n = 1) | |
| LCD (n = 3) | L550P (n = 2) | |
| H572R (n = 1) |
| Locus | TGFBI Mutations | Phenotype |
|---|---|---|
| R124 (n = 17) | R124H (n = 8) | GCD1 (n = 3) |
| GCD 2 (n = 5) | ||
| R124C (n = 8) | GCD2 (n = 1) | |
| LCD (n = 7) | ||
| R124L (n = 1) | RBCD (n = 1) | |
| R555 (n = 9) | R555W (n = 8) | GCD1 (n = 8) |
| R555Q (n = 1) | RBCD (n = 1) | |
| Others (n = 6) | H626R (n = 2) | LCD |
| A620D (n = 1) | LCD | |
| L550P (n = 2) | LCD | |
| H572R (n = 1) | LCD |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, S.B.; Anandalakshmi, V.; Wong, C.W.; Ng, S.R.; Mehta, J.S. Genotypic Homogeneity in Distinctive Transforming Growth Factor-Beta Induced (TGFBI) Protein Phenotypes. Int. J. Mol. Sci. 2021, 22, 1230. https://doi.org/10.3390/ijms22031230
Han SB, Anandalakshmi V, Wong CW, Ng SR, Mehta JS. Genotypic Homogeneity in Distinctive Transforming Growth Factor-Beta Induced (TGFBI) Protein Phenotypes. International Journal of Molecular Sciences. 2021; 22(3):1230. https://doi.org/10.3390/ijms22031230
Chicago/Turabian StyleHan, Sang Beom, Venkatraman Anandalakshmi, Chee Wai Wong, Si Rui Ng, and Jodhbir S. Mehta. 2021. "Genotypic Homogeneity in Distinctive Transforming Growth Factor-Beta Induced (TGFBI) Protein Phenotypes" International Journal of Molecular Sciences 22, no. 3: 1230. https://doi.org/10.3390/ijms22031230
APA StyleHan, S. B., Anandalakshmi, V., Wong, C. W., Ng, S. R., & Mehta, J. S. (2021). Genotypic Homogeneity in Distinctive Transforming Growth Factor-Beta Induced (TGFBI) Protein Phenotypes. International Journal of Molecular Sciences, 22(3), 1230. https://doi.org/10.3390/ijms22031230








