Coxsackievirus B3 Infection of Human iPSC Lines and Derived Primary Germ-Layer Cells Regarding Receptor Expression
Abstract
1. Introduction
2. Results
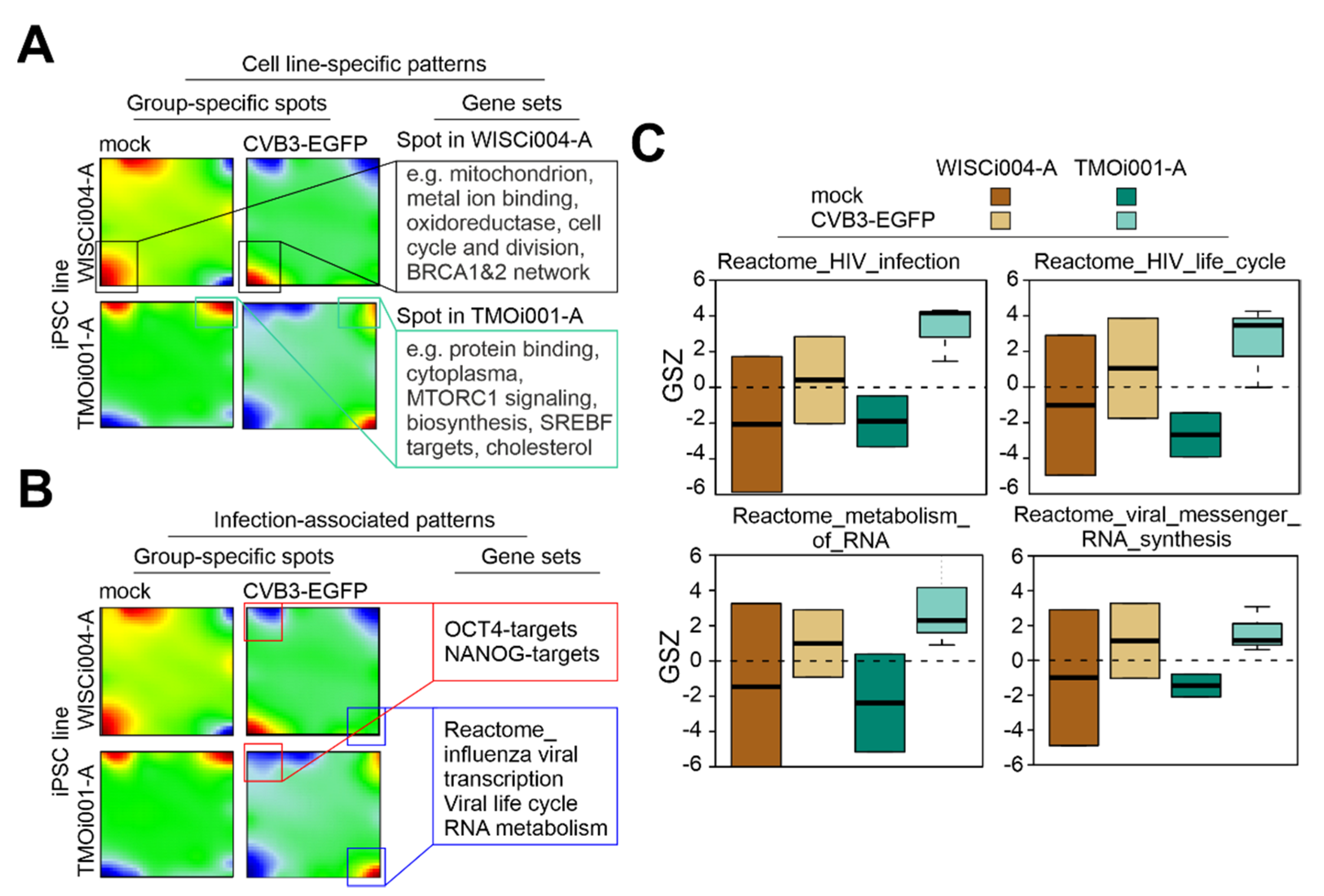
2.1. Transcriptomic Analysis Reveals Up-Regulation of RNA Metabolic Processes in iPSC Lines after CVB3-EGFP Infection
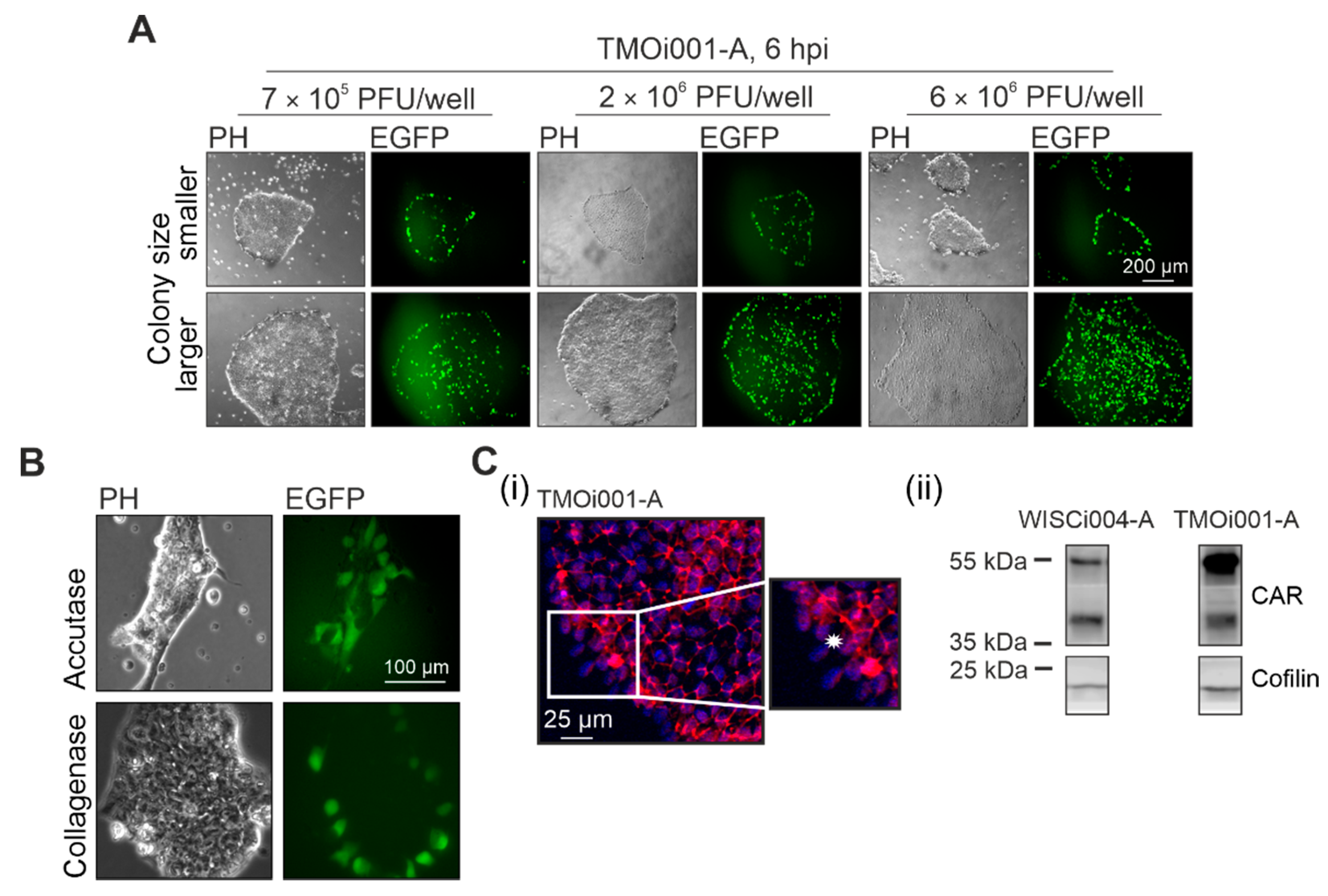
2.2. iPSC Colony Structure Determines Initial Infection Pattern of CVB3-EGFP
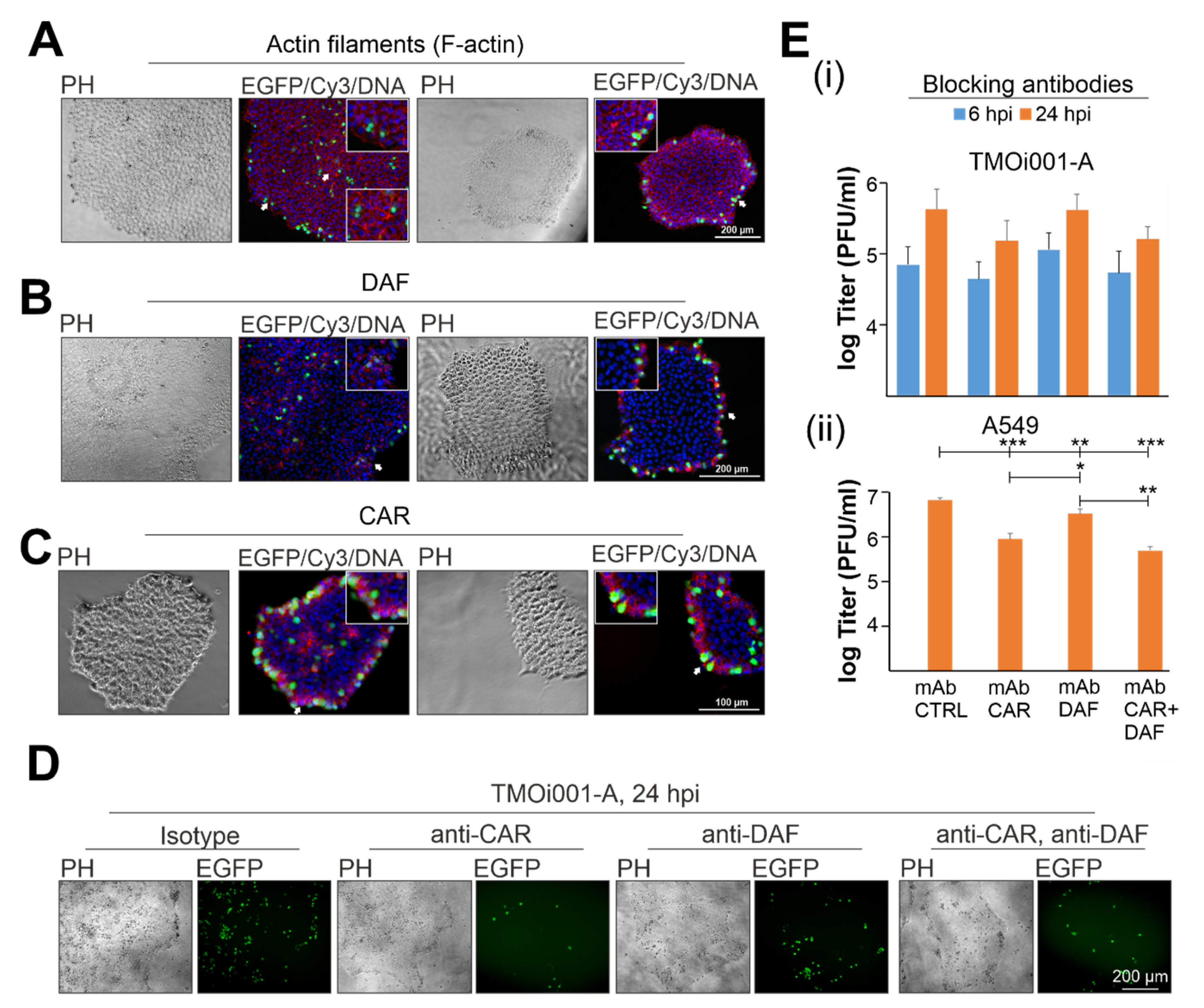
2.3. CAR Drives Initial Pattern of CVB3-EGFP Infection in iPSC Colonies
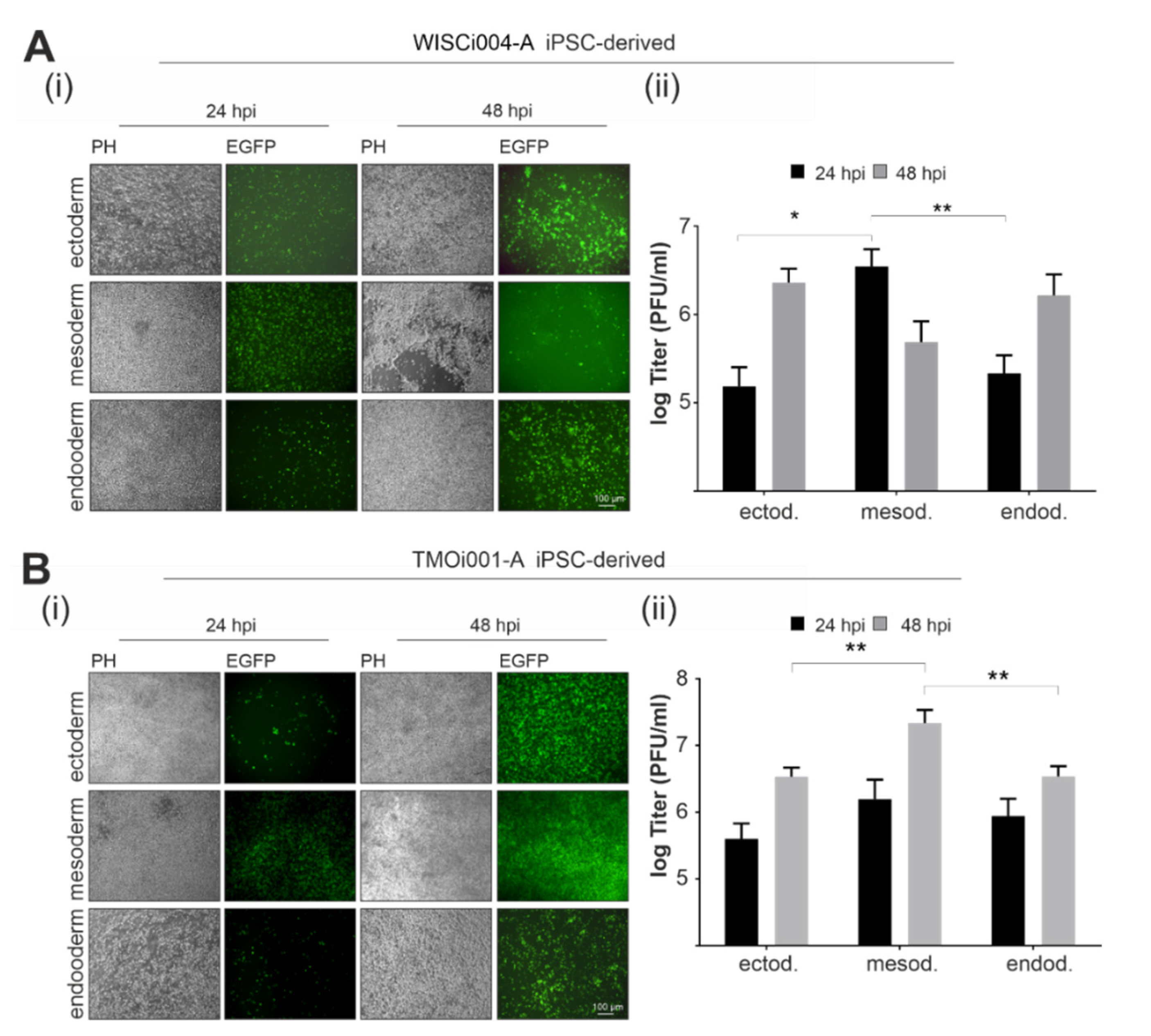
2.4. Primary Germ-Layer Cells Are Susceptible and Vulnerable to CVB3-EGFP Infection
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Cultivation
4.2. Directed Differentiation of iPSC Lines
4.3. Virus Infection
4.4. RNA Isolation, Microarray Gene Expression Analysis, and SOM Portrayal
4.5. Application of Blocking Antibodies
4.6. Western Blot Analysis
4.7. Immunofluorescence Analysis
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| iPSC | induced pluripotent stem cell |
| CVB3 | coxsackievirus B3 |
| EGFP | enhanced green fluorescent protein |
| IRES | internal ribosome entry site |
| CAR | coxsackie and adenovirus receptor |
| HFMD | hand, foot, and mouth disease |
References
- Coyne, C.B.; Lazear, H.M. Zika Virus—Reigniting the Torch. Nat. Rev. Microbiol. 2016, 14, 707–715. [Google Scholar] [CrossRef] [PubMed]
- Konstantinidou, A.; Anninos, H.; Spanakis, N.; Kotsiakis, X.; Syridou, G.; Tsakris, A.; Patsouris, E. Transplacental Infection of Coxsackievirus B3 Pathological Findings in the Fetus. J. Med. Virol. 2007, 79, 754–757. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.H.; Kim, J.W.; Hwang, J.Y.; Lee, K.M.; Shim, H.M.; Bae, Y.K.; Paik, S.S.; Park, H. Coxsackievirus B Infection Is Highly Related with Missed Abortion in Korea. Yonsei Med. J. 2014, 55, 1562–1567. [Google Scholar] [CrossRef] [PubMed]
- Al-Haddad, B.J.; Oler, E.; Armistead, B.; Elsayed, N.A.; Weinberger, D.R.; Bernier, R.; Burd, I.; Kapur, R.; Jacobsson, B.; Wang, C.; et al. The Fetal Origins of Mental Illness. Am. J. Obstet. Gynecol. 2019, 221, 549–562. [Google Scholar] [CrossRef]
- Tan, L.; Lacko, L.A.; Zhou, T.; Tomoiaga, D.; Hurtado, R.; Zhang, T.; Sevilla, A.; Zhong, A.; Mason, C.E.; Noggle, S.; et al. Pre- and Peri-Implantation Zika Virus Infection Impairs Fetal Development by Targeting Trophectoderm Cells. Nat. Commun. 2019, 10, 4155. [Google Scholar] [CrossRef]
- Khediri, Z.; Vauloup-Fellous, C.; Benachi, A.; Ayoubi, J.M.; Mandelbrot, L.; Picone, O. Adverse Effects of Maternal Enterovirus Infection on the Pregnancy Outcome: A Prospective and Retrospective Pilot Study. Virol. J. 2018, 15, 70. [Google Scholar] [CrossRef]
- Giachè, S.; Borchi, B.; Zammarchi, L.; Colao, M.G.; Ciccone, N.; Sterrantino, G.; Rossolini, G.M.; Bartoloni, A.; Trotta, M. Hand, Foot, and Mouth Disease in Pregnancy: 7 Years Tuscan Experience and Literature Review. J. Matern. Neonatal Med. 2019, 1–7. [Google Scholar] [CrossRef]
- Ouellet, A.; Sherlock, R.; Toye, B.; Fung, K.F.K. Antenatal Diagnosis of Intrauterine Infection with Coxsackievirus B3 Associated With Live Birth. Infect. Dis. Obstet. Gynecol. 2004, 12, 23–26. [Google Scholar] [CrossRef]
- Jaïdane, H.; Halouani, A.; Jmii, H.; Elmastour, F.; Mokni, M.; Aouni, M. Coxsackievirus B4 vertical transmission in a murine model. Virol. J. 2017, 14, 1–11. [Google Scholar] [CrossRef]
- Modlin, J.F.; Bowman, M. Perinatal Transmission of Coxsackievirus B3 in Mice. J. Infect. Dis. 1987, 156, 21–25. [Google Scholar] [CrossRef]
- Claus, C.; Jung, M.; Hübschen, J.M. Pluripotent Stem Cell-Based Models: A Peephole into Virus Infections during Early Pregnancy. Cells 2020, 9, 542. [Google Scholar] [CrossRef]
- Bowles, N.; Richardson, P.; Olsen, E.; Archard, L. Detection of Coxsackie-B-Virus-Specific Rna Sequences in Myocardial Biopsy Samples from Patients with Myocarditis and Dilated Cardiomyopathy. Lancet 1986, 1, 1120–1123. [Google Scholar] [CrossRef]
- Pauschinger, M.; Chandrasekharan, K.; Noutsias, M.; Schwimmbeck, L.P.; Schultheiss, H.-P. Viral heart disease: Molecular diagnosis, clinical prognosis, and treatment strategies. Med. Microbiol. Immunol. 2004, 193, 65–69. [Google Scholar] [CrossRef]
- Kindermann, I.; Kindermann, M.; Kandolf, R.; Klingel, K.; Bultmann, B.; Muller, T.; Lindinger, A.; Bohm, M. Predictors of Outcome in Patients with Suspected Myocarditis. Circulation 2008, 118, 639–648. [Google Scholar] [CrossRef] [PubMed]
- Daba, T.M.; Zhao, Y.; Pan, Z. Advancement of Mechanisms of Coxsackie Virus B3-Induced Myocarditis Pathogenesis and the Potential Therapeutic Targets. Curr. Drug Targets 2019, 20, 1461–1473. [Google Scholar] [CrossRef] [PubMed]
- Bergelson, J.M.; Cunningham, J.A.; Droguett, G.; Kurt-Jones, E.A.; Krithivas, A.; Hong, J.S.; Horwitz, M.S.; Crowell, R.L.; Finberg, R.W. Isolation of a Common Receptor for Coxsackie B Viruses and Adenoviruses 2 and 5. Science 1997, 275, 1320–1323. [Google Scholar] [CrossRef] [PubMed]
- Hafenstein, S.; Bowman, V.D.; Chipman, P.R.; Kelly, C.M.B.; Lin, F.; Medof, M.E.; Rossmann, M.G. Interaction of Decay-Accelerating Factor with Coxsackievirus B3. J. Virol. 2007, 81, 12927–12935. [Google Scholar] [CrossRef] [PubMed]
- Fechner, H.; Noutsias, M.; Tschoepe, C.; Hinze, K.; Wang, X.; Escher, F.; Pauschinger, M.; Dekkers, D.; Vetter, R.; Paul, M.; et al. Induction of Coxsackievirus-Adenovirus-Receptor Expression During Myocardial Tissue Formation and Remodeling: Identification of a Cell-to-Cell Contact-Dependent Regulatory Mechanism. Circulation 2003, 107, 876–882. [Google Scholar] [CrossRef]
- Honda, T.; Saitoh, H.; Masuko, M.; Katagiri-Abe, T.; Tominaga, K.; Kozakai, I.; Kobayashi, K.; Kumanishi, T.; Watanabe, Y.G.; Odani, S.; et al. The Coxsackievirus-Adenovirus Receptor Protein as a Cell Adhesion Molecule in the Developing Mouse Brain. Mol. Brain Res. 2000, 77, 19–28. [Google Scholar] [CrossRef]
- Hwang, J.Y.; Lee, K.M.; Kim, Y.H.; Shim, H.M.; Bae, Y.K.; Hwang, J.H.; Park, H. Pregnancy Loss Following Coxsackievirus B3 Infection in Mice During Early Gestation Due to High Expression of Coxsackievirus-Adenovirus Receptor (Car) in Uterus and Embryo. Exp. Anim. 2014, 63, 63–72. [Google Scholar] [CrossRef]
- Freiberg, F.; Sauter, M.; Pinkert, S.; Govindarajan, T.; Kaldrack, J.; Thakkar, M.; Fechner, H.; Klingel, K.; Gotthardt, M. Interspecies Differences in Virus Uptake Versus Cardiac Function of the Coxsackievirus and Adenovirus Receptor. J. Virol. 2014, 88, 7345–7356. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Krivega, M.; Geens, M.; Van de Velde, H. Car Expression in Human Embryos and Hesc Illustrates Its Role in Pluripotency and Tight Junctions. Reproduction 2014, 148, 531–544. [Google Scholar] [CrossRef] [PubMed]
- Hübner, D.; Jahn, K.; Pinkert, S.; Böhnke, J.; Jung, M.; Fechner, H.; Rujescu, D.; Liebert, U.G.; Claus, C. Infection of Ipsc Lines with Miscarriage-Associated Coxsackievirus and Measles Virus and Teratogenic Rubella Virus as a Model for Viral Impairment of Early Human Embryogenesis. ACS Infect. Dis 2017, 3, 886–897. [Google Scholar] [CrossRef]
- Bilz, N.C.; Willscher, E.; Binder, H.; Bohnke, J.; Stanifer, M.L.; Hubner, D.; Boulant, S.; Liebert, U.G.; Claus, C. Teratogenic Rubella Virus Alters the Endodermal Differentiation Capacity of Human Induced Pluripotent Stem Cells. Cells 2019, 8, 870. [Google Scholar] [CrossRef] [PubMed]
- Vissac, C.; De Latour, M.P.; Communal, Y.; Bignon, Y.-J.; Bernard-Gallon, D.J. Expression of Brca1 and Brca2 in Different Tumor Cell Lines with Various Growth Status. Clin. Chim. Acta 2002, 320, 101–110. [Google Scholar] [CrossRef]
- Hung, C.M.; Garcia-Haro, L.; Sparks, C.A.; Guertin, D.A. Mtor-Dependent Cell Survival Mechanisms. Cold Spring Harb. Perspect. Biol. 2012, 4, a008771. [Google Scholar] [CrossRef]
- Ben-Porath, I.; Thomson, M.W.; Carey, V.J.; Ge, R.; Bell, G.W.; Regev, A.; Weinberg, R.A. An Embryonic Stem Cell-Like Gene Expression Signature in Poorly Differentiated Aggressive Human Tumors. Nat. Genet. 2008, 40, 499–507. [Google Scholar] [CrossRef]
- Yudin, D.; Fainzilber, M. Ran on Tracks--Cytoplasmic Roles for a Nuclear Regulator. J. Cell Sci. 2009, 122, 587–593. [Google Scholar] [CrossRef]
- Tolbert, M.; Morgan, C.E.; Pollum, M.; Crespo-Hernández, C.E.; Li, M.-L.; Brewer, G.; Tolbert, B.S. Hnrnp A1 Alters the Structure of a Conserved Enterovirus Ires Domain to Stimulate Viral Translation. J. Mol. Biol. 2017, 429, 2841–2858. [Google Scholar] [CrossRef]
- Ortiz-Zapater, E.; Santis, G.; Parsons, M. CAR: A key regulator of adhesion and inflammation. Int. J. Biochem. Cell Biol. 2017, 89, 1–5. [Google Scholar] [CrossRef]
- Fechner, H.; Pinkert, S.; Wang, X.; Sipo, I.; Suckau, L.; Kurreck, J.; Dorner, A.; Sollerbrant, K.; Zeichhardt, H.; Grunert, H.-P.; et al. Coxsackievirus B3 and Adenovirus Infections of Cardiac Cells Are Efficiently Inhibited by Vector-Mediated Rna Interference Targeting Their Common Receptor. Gene Ther. 2007, 14, 960–971. [Google Scholar] [CrossRef] [PubMed]
- Pinkert, S.; Roger, C.; Kurreck, J.; Bergelson, J.M.; Fechner, H. The Coxsackievirus and Adenovirus Receptor: Glycosylation and the Extracellular D2 Domain Are Not Required for Coxsackievirus B3 Infection. J. Virol. 2016, 90, 5601–5610. [Google Scholar] [CrossRef] [PubMed]
- Hof, W.V.; Crystal, R.G. Fatty Acid Modification of the Coxsackievirus and Adenovirus Receptor. J. Virol. 2002, 76, 6382–6386. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Coyne, C.B.; Bergelson, J.M. Virus-Induced Abl and Fyn Kinase Signals Permit Coxsackievirus Entry through Epithelial Tight Junctions. Cell 2006, 124, 119–131. [Google Scholar] [CrossRef]
- Ullmann, U.; Veld, P.; Gilles, C.; Sermon, K.; De Rycke, M.; Van de Velde, H.; Van Steirteghem, A.; Liebaers, I. Epithelial-Mesenchymal Transition Process in Human Embryonic Stem Cells Cultured in Feeder-Free Conditions. Mol. Hum. Reprod. 2007, 13, 21–32. [Google Scholar] [CrossRef]
- Turco, M.Y.; Moffett, A. Development of the Human Placenta. Development 2019, 146, dev163428. [Google Scholar] [CrossRef]
- Pillet, S.; Billaud, G.; Omar, S.; Lina, B.; Pozzetto, B.; Schuffenecker, I. Multicenter Evaluation of the Enterovirus R-Gene Real-Time Rt-Pcr Assay for the Detection of Enteroviruses in Clinical Specimens. J. Clin. Virol. 2010, 47, 54–59. [Google Scholar] [CrossRef]
- Amstey, M.S.; Miller, R.K.; Menegus, M.A.; di Sant ‘Agnese, P.A. Enterovirus in Pregnant Women and the Perfused Placenta. Am. J. Obstet. Gynecol. 1988, 158, 775–782. [Google Scholar] [CrossRef]
- Chow, K.C.; Lee, C.C.; Lin, T.Y.; Shen, W.C.; Wang, J.H.; Peng, C.T.; Lee, C.C. Congenital Enterovirus 71 Infection: A Case Study with Virology and Immunohistochemistry. Clin. Infect. Dis 2000, 31, 509–512. [Google Scholar] [CrossRef]
- Chin, M.H.; Mason, M.J.; Xie, W.; Volinia, S.; Singer, M.; Peterson, C.; Ambartsumyan, G.; Aimiuwu, O.; Richter, L.; Zhang, J.; et al. Induced Pluripotent Stem Cells and Embryonic Stem Cells Are Distinguished by Gene Expression Signatures. Cell Stem Cell 2009, 5, 111–123. [Google Scholar] [CrossRef]
- Kaewseekhao, B.; Naranbhai, V.; Roytrakul, S.; Namwat, W.; Paemanee, A.; Lulitanond, V.; Chaiprasert, A.; Faksri, K. Comparative Proteomics of Activated Thp-1 Cells Infected with Mycobacterium Tuberculosis Identifies Putative Clearance Biomarkers for Tuberculosis Treatment. PLoS ONE 2015, 10, e0134168. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.-Y.; Li, M.-L.; Huang, P.-N.; Chien, K.-Y.; Horng, J.-T.; Shih, S.-R. Heterogeneous Nuclear Ribonuclear Protein K Interacts with the Enterovirus 71 5’ Untranslated Region and Participates in Virus Replication. J. Gen. Virol. 2008, 89, 2540–2549. [Google Scholar] [CrossRef] [PubMed]
- Warmflash, A.; Sorre, B.; Etoc, F.; Siggia, E.D.; Brivanlou, A.H. A Method to Recapitulate Early Embryonic Spatial Patterning in Human Embryonic Stem Cells. Nat. Methods 2014, 11, 847–854. [Google Scholar] [CrossRef] [PubMed]
- Cohen, C.J.; Shieh, J.T.C.; Pickles, R.J.; Okegawa, T.; Hsieh, J.-T.; Bergelson, J.M. The Coxsackievirus and Adenovirus Receptor Is a Transmembrane Component of the Tight Junction. Proc. Natl. Acad. Sci. USA 2001, 98, 15191–15196. [Google Scholar] [CrossRef] [PubMed]
- Cohen, C.J.; Gaetz, J.; Ohman, T.; Bergelson, J.M. Multiple Regions within the Coxsackievirus and Adenovirus Receptor Cytoplasmic Domain Are Required for Basolateral Sorting. J. Biol. Chem. 2001, 276, 25392–25398. [Google Scholar] [CrossRef]
- Excoffon, K.J.D.A.; Moninger, T.; Zabner, J. The Coxsackie B Virus and Adenovirus Receptor Resides in a Distinct Membrane Microdomain. J. Virol. 2003, 77, 2559–2567. [Google Scholar] [CrossRef]
- Bateman, A.; Martin, M.J.; O’Donovan, C.; Magrane, M.; Alpi, E.; Antunes, R.; Bely, B.; Bingley, M.; Bonilla, C.; Britto, R.; et al. Uniprot: The Universal Protein Knowledgebase. Nucleic Acids Res. 2017, 45, D158–D169. [Google Scholar]
- Wang, X.; Bergelson, J.M. Coxsackievirus and Adenovirus Receptor Cytoplasmic and Transmembrane Domains Are Not Essential for Coxsackievirus and Adenovirus Infection. J. Virol. 1999, 73, 2559–2562. [Google Scholar] [CrossRef]
- Dommergues, M.; Petitjean, J.; Aubry, M.C.; Delezoide, A.L.; Narcy, F.; Fallet-Bianco, C.; Freymuth, F.; Dumez, Y.; Lebon, P. Fetal Enteroviral Infection with Cerebral Ventriculomegaly and Cardiomyopathy. Fetal Diagn. Ther. 1994, 9, 77–78. [Google Scholar] [CrossRef]
- Verboon-Maciolek, M.A.; Groenendaal, F.; Cowan, F.; Govaert, P.; Van Loon, A.M.; De Vries, L.S. White Matter Damage in Neonatal Enterovirus Meningoencephalitis. Neurology 2006, 66, 1267–1269. [Google Scholar] [CrossRef]
- Hotta, Y.; Honda, T.; Naito, M.; Kuwano, R. Developmental Distribution of Coxsackie Virus and Adenovirus Receptor Localized in the Nervous System. Brain Res. Dev. Brain Res. 2003, 143, 1–13. [Google Scholar] [CrossRef]
- Wehbi, A.; Kremer, E.J.; Dopeso-Reyes, I.G. Location of the Cell Adhesion Molecule Coxsackievirus and Adenovirus Receptor in the Adult Mouse Brain. Front. Neuroanat. 2020, 14, 28. [Google Scholar] [CrossRef] [PubMed]
- Zussy, C.; Loustalot, F.; Junyent, F.; Gardoni, F.; Bories, C.; Valero, J.; Desarménien, M.G.; Bernex, F.; Henaff, D.; Bayo-Puxan, N.; et al. Coxsackievirus Adenovirus Receptor Loss Impairs Adult Neurogenesis, Synapse Content, and Hippocampus Plasticity. J. Neurosci. 2016, 36, 9558–9571. [Google Scholar] [CrossRef] [PubMed]
- Anasir, M.I.; Poh, C.L. Advances in Antigenic Peptide-Based Vaccine and Neutralizing Antibodies against Viruses Causing Hand, Foot, and Mouth Disease. Int. J. Mol. Sci. 2019, 20, 1256. [Google Scholar] [CrossRef] [PubMed]
- Du, R.; Mao, Q.; Hu, Y.; Lang, S.; Sun, S.; Li, K.; Gao, F.; Bian, L.; Yang, C.; Cui, B.; et al. A Potential Therapeutic Neutralization Monoclonal Antibody Specifically against Multi-Coxsackievirus A16 Strains Challenge. Hum. Vaccines Immunother. 2019, 15, 2343–2350. [Google Scholar] [CrossRef] [PubMed]
- Pinkert, S.; Dieringer, B.; Diedrich, S.; Zeichhardt, H.; Kurreck, J.; Fechner, H. Soluble Coxsackie- and Adenovirus Receptor (Scar-Fc); a Highly Efficient Compound against Laboratory and Clinical Strains of Coxsackie-B-Virus. Antiviral Res. 2016, 136, 1–8. [Google Scholar] [CrossRef]
- Chu, S.T.; Kobayashi, K.; Bi, X.; Ishizaki, A.; Tran, T.T.; Phung, T.T.B.; Pham, C.T.T.; Nguyen, L.V.; Ta, T.A.; Khu, D.T.K.; et al. Newly Emerged Enterovirus-A71 C4 Sublineage May Be More Virulent Than B5 in the 2015–2016 Hand-Foot-and-Mouth Disease Outbreak in Northern Vietnam. Sci. Rep. 2020, 10, 159. [Google Scholar] [CrossRef]
- Sarmirova, S.; Borsanyiova, M.; Benkoova, B.; Pospisilova, M.; Arumugam, R.; Berakova, K.; Gomolcak, P.; Reddy, J.; Bopegamage, S. Pancreas of Coxsackievirus-Infected Dams and Their Challenged Pups: A Complex Issue. Virulence 2019, 10, 207–221. [Google Scholar] [CrossRef]
- Paria, B.C.; Huet-Hudson, Y.M.; Dey, S.K. Blastocyst’s State of Activity Determines the Window of Implantation in the Receptive Mouse Uterus. Proc. Natl. Acad. Sci. USA 1993, 90, 10159–10162. [Google Scholar] [CrossRef]
- Deglincerti, A.; Etoc, F.; Guerra, M.C.; Martyn, I.; Metzger, J.; Ruzo, A.; Simunovic, M.; Yoney, A.; Brivanlou, A.H.; Siggia, E.D.; et al. Self-Organization of Human Embryonic Stem Cells on Micropatterns. Nat. Protoc. 2016, 11, 2223–2232. [Google Scholar] [CrossRef]
- Slifka, M.K.; Pagarigan, R.; Mena, I.; Feuer, R.; Whitton, J.L. Using Recombinant Coxsackievirus B3 to Evaluate the Induction and Protective Efficacy of Cd8+ T Cells During Picornavirus Infection. J. Virol. 2001, 75, 2377–2387. [Google Scholar] [CrossRef]
- Feuer, R.; Mena, I.; Pagarigan, R.; Slifka, M.K.; Whitton, J.L. Cell Cycle Status Affects Coxsackievirus Replication, Persistence, and Reactivation in Vitro. J. Virol. 2002, 76, 4430–4440. [Google Scholar] [CrossRef] [PubMed]
- Pinkert, S.; Klingel, K.; Lindig, V.; Dörner, A.; Zeichhardt, H.; Spiller, O.B.; Fechner, H. Virus-Host Coevolution in a Persistently Coxsackievirus B3-Infected Cardiomyocyte Cell Line. J. Virol. 2011, 85, 13409–13419. [Google Scholar] [CrossRef] [PubMed]
- Löffler-Wirth, H.; Kalcher, M.; Binder, H. Opossom: R-Package for High-Dimensional Portraying of Genome-Wide Expression Landscapes on Bioconductor. Bioinformatics 2015, 31, 3225–3227. [Google Scholar] [CrossRef] [PubMed]
- Wirth, H.; Von Bergen, M.; Binder, H. Mining Som Expression Portraits: Feature Selection and Integrating Concepts of Molecular Function. BioData Min. 2012, 5, 18. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Böhnke, J.; Pinkert, S.; Schmidt, M.; Binder, H.; Bilz, N.C.; Jung, M.; Reibetanz, U.; Beling, A.; Rujescu, D.; Claus, C. Coxsackievirus B3 Infection of Human iPSC Lines and Derived Primary Germ-Layer Cells Regarding Receptor Expression. Int. J. Mol. Sci. 2021, 22, 1220. https://doi.org/10.3390/ijms22031220
Böhnke J, Pinkert S, Schmidt M, Binder H, Bilz NC, Jung M, Reibetanz U, Beling A, Rujescu D, Claus C. Coxsackievirus B3 Infection of Human iPSC Lines and Derived Primary Germ-Layer Cells Regarding Receptor Expression. International Journal of Molecular Sciences. 2021; 22(3):1220. https://doi.org/10.3390/ijms22031220
Chicago/Turabian StyleBöhnke, Janik, Sandra Pinkert, Maria Schmidt, Hans Binder, Nicole Christin Bilz, Matthias Jung, Uta Reibetanz, Antje Beling, Dan Rujescu, and Claudia Claus. 2021. "Coxsackievirus B3 Infection of Human iPSC Lines and Derived Primary Germ-Layer Cells Regarding Receptor Expression" International Journal of Molecular Sciences 22, no. 3: 1220. https://doi.org/10.3390/ijms22031220
APA StyleBöhnke, J., Pinkert, S., Schmidt, M., Binder, H., Bilz, N. C., Jung, M., Reibetanz, U., Beling, A., Rujescu, D., & Claus, C. (2021). Coxsackievirus B3 Infection of Human iPSC Lines and Derived Primary Germ-Layer Cells Regarding Receptor Expression. International Journal of Molecular Sciences, 22(3), 1220. https://doi.org/10.3390/ijms22031220







