Molecular Mechanisms of Phosphate Sensing, Transport and Signalling in Streptomyces and Related Actinobacteria
Abstract
1. Introduction
2. High Affinity Phosphate Transport Systems
2.1. The High Affinity PstSCAB System
2.2. Sensing the Phosphate Limitation: The Seven Proteins Model of Wanner and the Intracellular Teichoic Acid Intermediate Signal Model
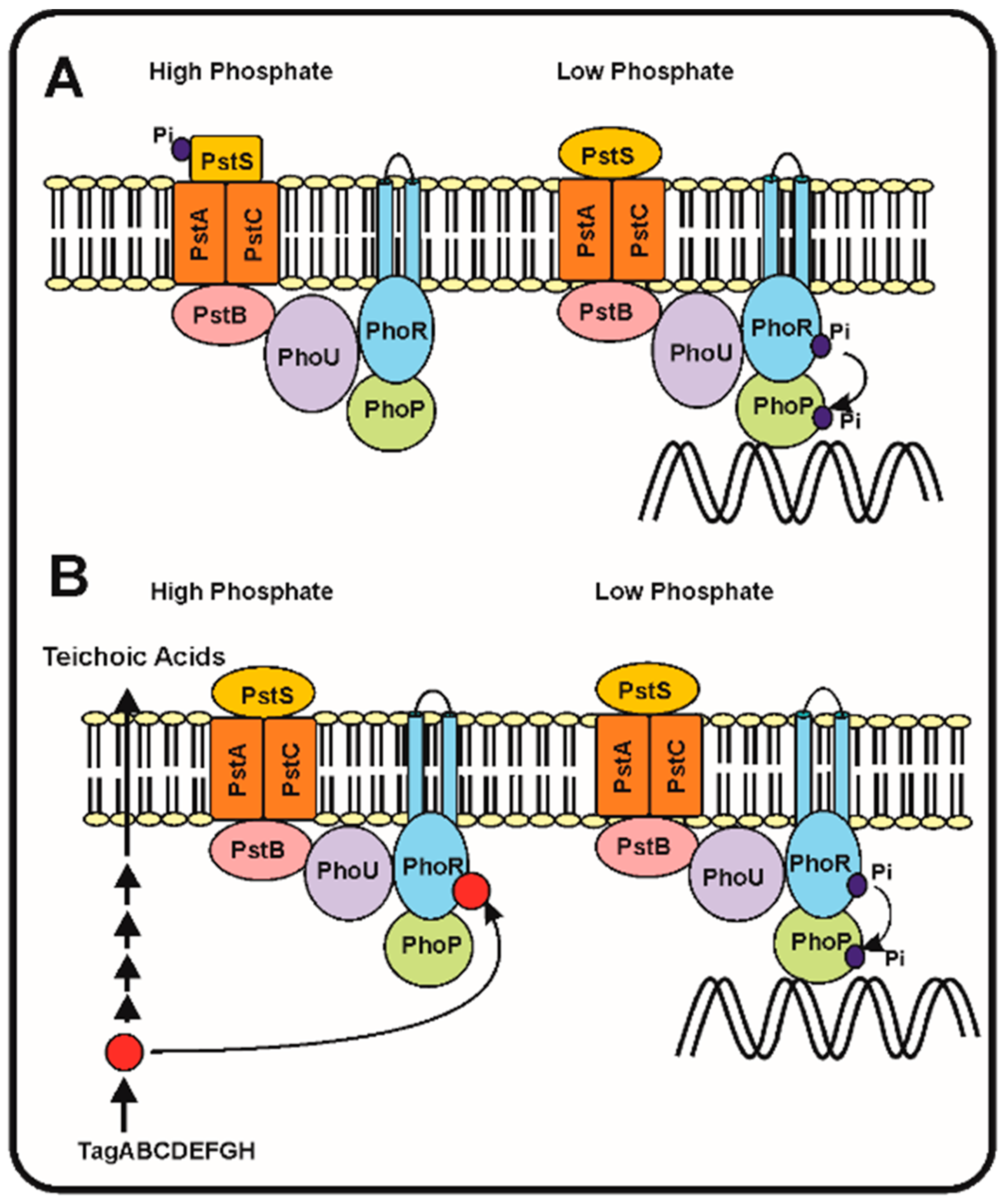
2.3. Discrimination between the Phosphate and Arsenate Anions by the Phosphate-Binding Protein PstS
2.4. Glycosylation and Release of PstS in Streptomyces Species: Which Is the Role of the Released PstS Protein?
2.5. A Second High Affinity Phosphate Transport System Exists in Mycobacterium smegmatis
3. Low Affinity Phosphate Transporters
3.1. Transport by the Inorganic Phosphate (Pit) System: Background Information in Gram-Negative Bacteria
3.2. The pitH1 and pitH2 Systems in Streptomyces and Corynebacterium glutamicum
3.3. PitH Genes in Other Streptomyces Species
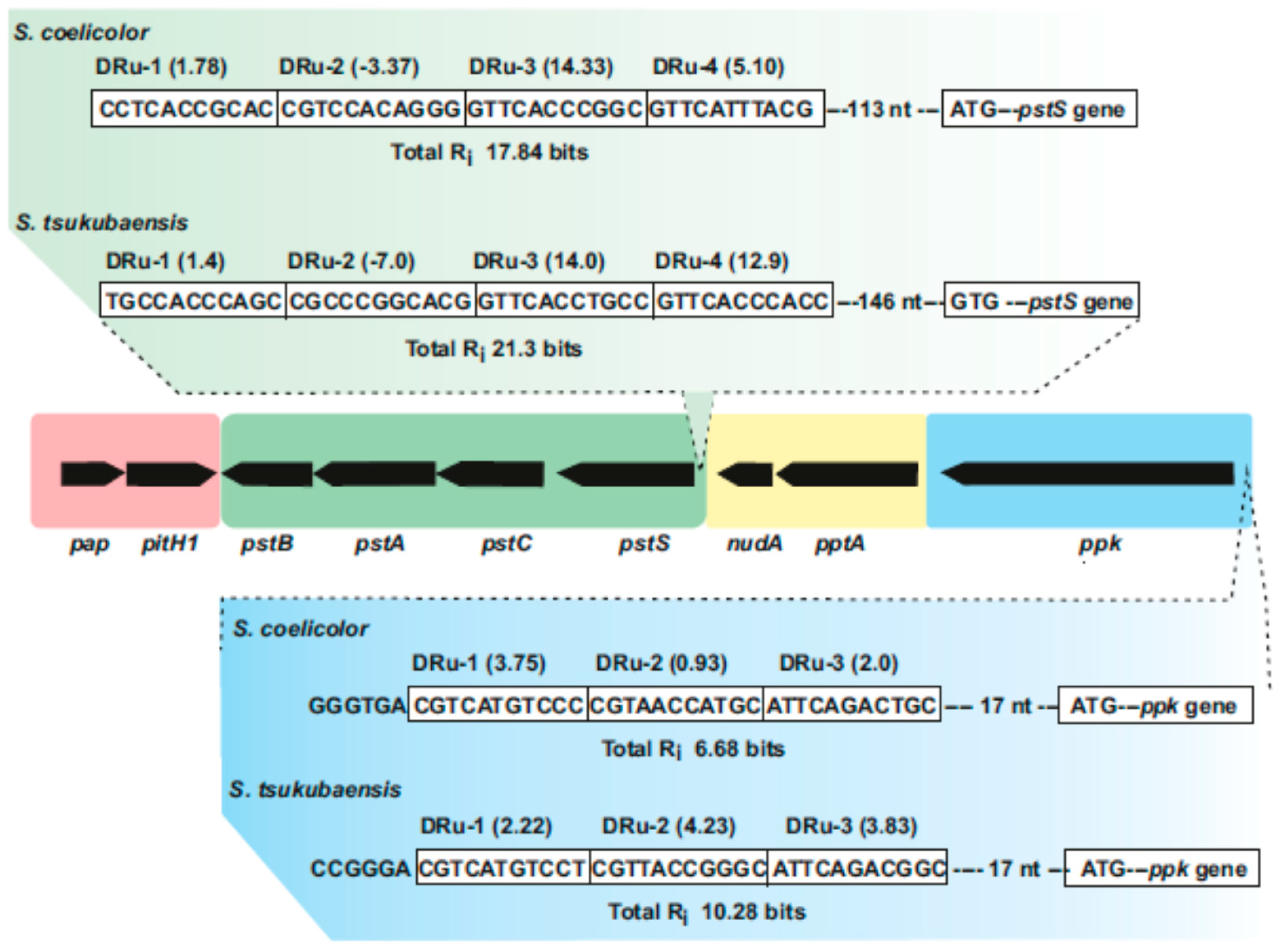
4. The pap-pitH1-pstSCAB-pptA-nudA-ppk Supercluster
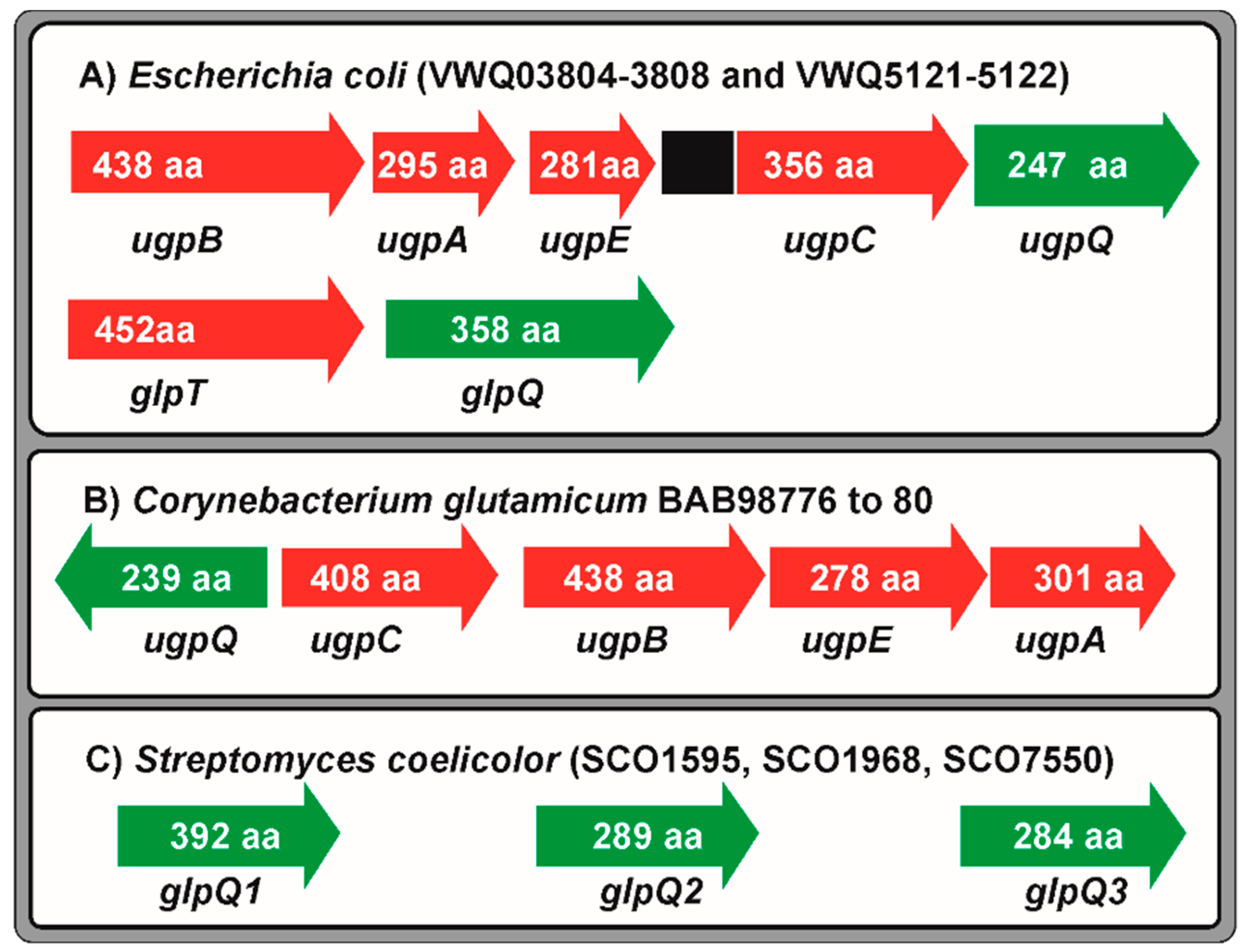
5. Transport and Utilization of Glycerol-3-Phosphate and Glycerophosphodiesters
Glycerophosphodiester Phosphodiesterases: Molecular Mechanisms of Binding of PhoP to the glpQ Promoters and Regulation by Phosphate and Carbon Sources
6. Utilization of Sugar Phosphates and Nucleotides: Dephosphorylation or Transport of Phosphorylated Compounds?
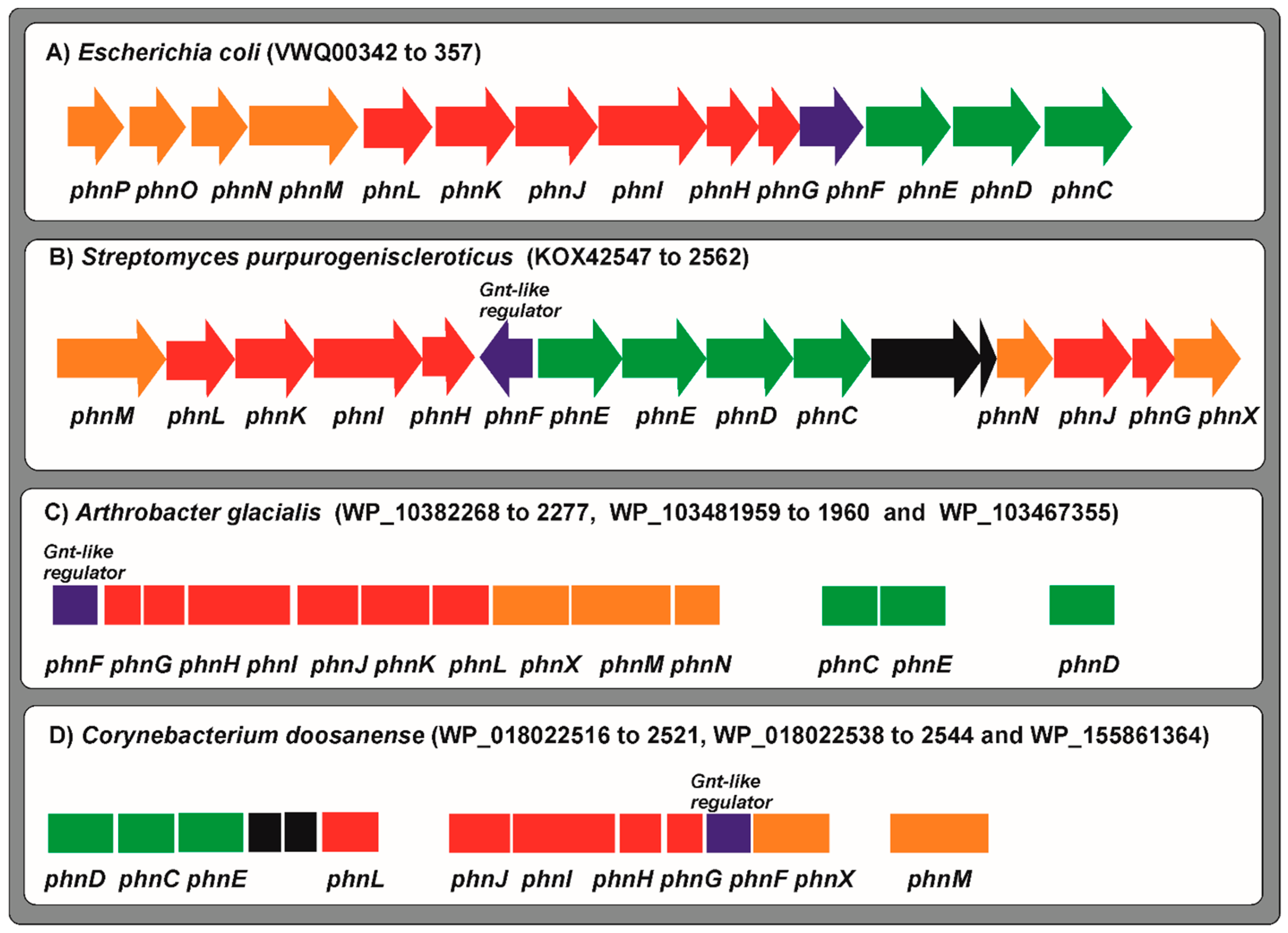
7. Transport and Utilization of Phosphonates
7.1. Phosphonate Transport and Utilization in Arthrobacter and Corynebacterium Species
7.2. The Ability to Utilize Phosphonates Is Restricted to Some Streptomyces sp.
8. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Santos-Beneit, F. The Pho regulon: A huge regulatory network in bacteria. Front. Microbiol. 2015, 6, 402. [Google Scholar] [CrossRef] [PubMed]
- Martín, J.F.; Liras, P. The Balance Metabolism Safety Net: Integration of Stress Signals by Interacting Transcriptional Factors in Streptomyces and Related Actinobacteria. Front. Microbiol. 2020, 10, 3120. [Google Scholar] [CrossRef] [PubMed]
- Jha, V.; Dafale, N.A.; Purohit, H.J. Regulatory rewiring through global gene regulations by PhoB and alarmone (p)ppGpp under various stress conditions. Microbiol. Res. 2019, 227, 126309. [Google Scholar] [CrossRef] [PubMed]
- Alori, E.T.; Glick, B.R.; Babalola, O.O. Microbial Phosphorus Solubilization and Its Potential for Use in Sustainable Agriculture. Front. Microbiol. 2017, 8, 971. [Google Scholar] [CrossRef]
- Demain, A.L. Antibiotics: Natural Products Essential to Human Health. Med. Res. Rev. 2009, 29, 821–842. [Google Scholar] [CrossRef]
- Robertsen, H.L.; Musiol-Kroll, E.M. Actinomycete-Derived Polyketides as a Source of Antibiotics and Lead Structures for the Development of New Antimicrobial Drugs. Antibiotics 2019, 8, 157. [Google Scholar] [CrossRef]
- Martín, J.F. Molecular mechanisms for the control by phosphate of the biosynthesis of antibiotics and other secondary metabolites. In Regulation of Secondary Metabolism in Actinomycetes; Shapiro, S., Ed.; CRC Press: Boca Ratón, FL, USA, 1989; pp. 213–237. [Google Scholar]
- Martín, J.F.; Liras, P. Organization and expression of genes involved in the biosynthesis of antibiotics and other secondary metabolites. Ann. Rev. Microbiol. 1989, 43, 173–206. [Google Scholar] [CrossRef]
- van Wezel, G.P.; McDowall, K.J. The regulation of the secondary metabolism of Streptomyces: New links and experimental advances. Nat. Prod. Rep. 2011, 28, 1311–1333. [Google Scholar] [CrossRef]
- Martín, J.F.; Liras, P. Novel Antimicrobial and other Bioactive Metabolites obtained from Silent Gene Clusters. In Antibiotics: Current Innovations and Future Trends; Demain, A.L., Sánchez, S., Eds.; Horizon Scientific Press and Caister Academic Press: Norfolk, UK, 2015; pp. 275–292. [Google Scholar]
- Moura, R.S.; Martín, J.F.; Martín, A.; Liras, P. Substrate analysis and molecular cloning of the extracellular alkaline phosphatase of Streptomyces griseus. Microbiology 2001, 147, 1525–1533. [Google Scholar] [CrossRef][Green Version]
- Apel, A.K.; Sola-Landa, A.; Rodríguez-García, A.; Martín, J.F. Phosphate control of phoA, phoC and phoD gene expression in Streptomyces coelicolor reveals significant differences in binding of PhoP to their promoter regions. Microbiology 2007, 153, 3527–3537. [Google Scholar] [CrossRef]
- Ordóñez-Robles, M.; Santos-Beneit, F.; Rodríguez-García, A.; Martín, J.F. Analysis of the Pho Regulon in Streptomyces tsukubaensis. Microbiol. Res. 2017, 205, 80–87. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Castro, M.; Barreiro, C.; Martín, J.F. Analysis and validation of the pho regulon in the tacrolimus-producer strain Streptomyces tsukubaensis: Differences with the model organism Streptomyces coelicolor. Appl. Microbiol. Biotechnol. 2018, 102, 7029–7045. [Google Scholar] [CrossRef] [PubMed]
- Ghorbel, S.; Smirnov, A.; Chouayekh, H.; Sperandio, B.; Esnault, C.; Kormanec, J.; Virolle, M.J. Regulation of ppk expression and in vivo function of Ppk in Streptomyces lividans TK24. J. Bacteriol. 2006, 188, 6269–6276. [Google Scholar] [CrossRef] [PubMed]
- Smirnov, A.; Esnault, C.; Prigent, M.; Holland, I.B.; Virolle, M.-J. Phosphate Homeostasis in Conditions of Phosphate Proficiency and Limitation in the Wild Type and the phoP Mutant of Streptomyces lividans. PLoS ONE 2015, 10, e0126221. [Google Scholar] [CrossRef]
- Rodríguez-García, A.; Barreiro, C.; Santos-Beneit, F.; Sola-Landa, A.; Martín, J.F. Genome-wide transcriptomic and proteomic analysis of the primary response to phosphate limitation in Streptomyces coelicolor M145 and in a ∆phoP mutant. Proteomics 2007, 7, 2410–2429. [Google Scholar] [CrossRef]
- Martín, J.F.; Santos-Beneit, F.; Rodríguez-García, A.; Sola-Landa, A.; Smith, M.C.; Ellingsen, T.E.; Wellington, E.M. Transcriptomic studies of phosphate control of primary and secondary metabolism in Streptomyces coelicolor. Appl. Microbiol. Biotechnol. 2012, 95, 61–75. [Google Scholar] [CrossRef]
- Martín, J.F.; Rodríguez-García, A.; Liras, P. The master regulator PhoP coordinates phosphate and nitrogen metabolism, respiration, cell differentiation and antibiotic biosynthesis: Comparison in Streptomyces coelicolor and Streptomyces avermitilis. J. Antibiot. 2017, 70, 534–541. [Google Scholar] [CrossRef]
- Sola-Landa, A.; Moura, R.S.; Martín, J.F. The two component PhoR-PhoP system controls both primary metabolism and secondary metabolite biosynthesis in Streptomyces lividans. Proc. Natl. Acad. Sci. USA 2003, 100, 6133–6138. [Google Scholar] [CrossRef]
- Sola-Landa, A.; Rodríguez-García, A.; Franco-Domínguez, E.; Martín, J.F. Binding of PhoP to promoters of phosphate regulated genes in Streptomyces coelicolor: Identification of PHO boxes. Mol. Microbiol. 2005, 56, 1373–1385. [Google Scholar] [CrossRef]
- Allenby, N.E.E.; Laing, E.; Bucca, G.; Kierzek, A.M.; Collin, C.P. Diverse control of metabolism and other cellular processes in Streptomyces coelicolor by the PhoP transcription factor: Genome-wide identification of in vivo targets. Nucleic Acid. Res. 2012, 40, 9543–9556. [Google Scholar] [CrossRef]
- Espitia, C.; Elinos, M.; Hernandez-Pando, R.; Mancilla, R. Phosphate starvation enhances expression of the immunodominant 38-kilodalton protein antigen of Mycobacterium tuberculosis: Demonstration by immunogold electron microscopy. Infect. Immun. 1992, 60, 2998–3001. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, J.L.; O’Gaora, P.; Gallagher, A.; Thole, J.E.; Young, D.B. Bacterial glycoproteins: A link between glycosylation and proteolytic cleavage of a 19 kDa antigen from Mycobacterium. EMBO J. 1996, 15, 3547–3554. [Google Scholar] [CrossRef] [PubMed]
- Lefevre, P.; Braibant, M.; de Wit, L.; Kalai, M.; Roeper, D.; Grotzinger, J.; Content, J. Three different putative phosphate transport receptors are encoded by the Mycobacterium tuberculosis genome and are present at the surface of Mycobacterium bovis BCG. J. Bacteriol. 1997, 179, 2900–2906. [Google Scholar] [CrossRef] [PubMed]
- Peirs, P.; Lefèvre, P.; Boarbi, S.; Wang, X.-M.; Denis, O.; Braibant, M.; Content, J. Mycobacterium tuberculosis with Disruption in Genes Encoding the Phosphate Binding Proteins PstS1 and PstS2 Is Deficient in Phosphate Uptake and Demonstrates Reduced In Vivo Virulence. Infect. Immun. 2005, 73, 1898–1902. [Google Scholar] [CrossRef] [PubMed]
- Ferraris, D.M.; Spallek, R.; Oehlmann, W.; Singh, M.; Rizzi, M. Crystal structure of the Mycobacterium tuberculosis phosphate binding protein PstS3. Proteins 2014, 82, 2268–2274. [Google Scholar] [CrossRef]
- Lichá, I.; Benes, I.; Janda, S.; Hostálek, Z.; Janácek, K. Characterization of phosphate transport in Streptomyces granaticolor. Biochem. Mol. Biol. Int. 1997, 41, 431–437. [Google Scholar]
- Wanner, B.L. Phosphorus assimilation and control of the phosphate regulon. In Escherichia coli and Salmonella: Cellular and Molecular Biology; Neidhart, F.C., Ed.; AASM Press: Washington, DC, USA, 1996; pp. 1357–1381. [Google Scholar]
- Allenby, N.E.; O’Connor, N.; Pragai, Z.; Carter, N.M.; Miethke, M.; Engelmann, S. Posttranscriptional regulation of the Bacillus subtilis pst operon encoding a phosphate specific ABC transporter. Microbiology 2004, 150, 2619–2628. [Google Scholar] [CrossRef]
- Wehmeier, S.; Varghese, A.S.; Gurcha, S.S.; Tissot, B.; Panico, M.; Hitchen, P.; Smith, M.C.M. Glycosylation of the phosphate binding protein, PstS, in Streptomyces coelicolor by a pathway that resembles protein O-mannosylation in eukaryotes. Mol. Microbiol. 2009, 71, 421–433. [Google Scholar] [CrossRef]
- Esteban, A.; Díaz, M.; Yepes, A.; Santamaría, R.I. Expression of the pstS gene of Streptomyces lividans is regulated by the carbon source and is partially independent of the PhoP regulator. BMC Microbiol. 2008, 8, 201. [Google Scholar] [CrossRef]
- Bentley, S.D.; Chater, K.F.; Cerdeño-Tárraga, A.M.; Challis, G.L.; Thomson, N.R.; James, K.D.; Hopwood, D.A. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 2002, 417, 141–147. [Google Scholar] [CrossRef]
- Díaz, M.; Esteban, A.; Fernández-Ábalos, J.M.; Santamaría, R.I. The high-affinity phosphate-binding protein PstS is accumulated under high fructose concentrations and mutation of the corresponding gene affects differentiation in Streptomyces lividans. Microbiology 2005, 151, 2583–2592. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chouayekh, H.; Virolle, M.J. The polyphosphate kinase plays a negative role in the control of antibiotic production in Streptomyces lividans. Mol. Microbiol. 2002, 43, 919–930. [Google Scholar] [CrossRef] [PubMed]
- Esnault, C.; Dulermo, T.; Smirnov, A.; Askora, A.; David, M.; Deniset-Besseau, A.; Virolle, M.J. Strong antibiotic production is correlated with highly active oxidative metabolism in Streptomyces coelicolor M145. Sci Rep. 2017, 7, 200. [Google Scholar] [CrossRef] [PubMed]
- Sola-Landa, A.; Rodríguez-García, A.; Apel, A.K.; Martín, J.F. Target genes and structure of the direct repeats in the DNA-binding sequences of the response regulator PhoP in Streptomyces coelicolor. Nucleic Acids Res. 2008, 36, 1358–1368. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Liao, C.-H.; Yao, L.; Xu, Y.; Liu, W.-B.; Zhou, Y.; Ye, B.-C. Nitrogen regulator GlnR controls uptake and utilization of non-phosphotransferase-system carbon sources in actinomycetes. Proc. Natl. Acad. Sci. USA 2015, 112, 15630–15635. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, Y.-J.; Wanner, B.L. Global regulation by the seven-component Pi signaling system. Curr. Op. Microbiol. 2010, 13, 198–203. [Google Scholar] [CrossRef] [PubMed]
- Martín-Martín, S.; Rodríguez-García, A.; Santos-Beneit, F.; Franco-Domínguez, E.; Sola-Landa, A.; Martín, J.F. Self-control of the PHO regulon: The PhoP-dependent protein PhoU controls negatively expression of genes of PHO regulon in Streptomyces coelicolor. J. Antibiotics 2018, 71, 113–122. [Google Scholar] [CrossRef]
- Botella, E.; Devine, S.K.; Hubner, S.; Salzberg, L.I.; Gale, R.T.; Brown, E.D.; Devine, K.M. PhoR autokinase activity is controlled by an intermediate in wall teichoic acid metabolism that is sensed by the intracellular PAS domain during the PhoPR-mediated phosphate limitation response of Bacillus subtilis. Mol. Microbiol. 2014, 94, 1242–1259. [Google Scholar] [CrossRef]
- 42 Kleinschnitz, E.M.; Latus, A.; Sigle, S.; Maldener, I.; Wohlleben, W.; Muth, G. Genetic analysis of SCO2997, encoding a TagF homologue, indicates a role for was teichoic acids in sporulation of Streptomyces coelicolor A3(2). J. Bacteriol. 2011, 193, 6080–6085. [Google Scholar] [CrossRef]
- Coyle, P.; Philcox, J.C.; Carey, L.C.; Rofe, A.M. Metallothionein: The multipurpose protein. Cell. Mol. Life Sci. 2002, 59, 627–647. [Google Scholar] [CrossRef]
- Moreau, P.L.; Gérard, F.; Lutz, N.W.; Cozzone, P. Non-growing Escherichia coli cells starved for glucose or phosphate use different mechanisms to survive oxidative stress. Mol. Microbiol. 2001, 39, 1048–1060. [Google Scholar] [CrossRef] [PubMed]
- Liras, P.; Martín, J.F.; Villanueva, J.R. Sequential expression of macromolecule biosynthesis and candicidin formation in Streptomyces griseus. J. Gen. Microbiol. 1977, 102, 269–277. [Google Scholar] [CrossRef] [PubMed]
- Martín, J.F.; Aparicio, J.F. Enzymology of the Polyenes Pimaricin and Candicidin Biosynthesis. Methods Enzymol. 2009, 459, 215–242. [Google Scholar] [PubMed]
- Martín, J.F.; Naharro, G.; Liras, P.; Villanueva, J.R. Isolation of mutants deregulated in phosphate control of candicidin biosynthesis. J. Antibiot 1979, 32, 600–606. [Google Scholar] [CrossRef]
- Elias, M.; Wellner, A.; Goldin-Azulay, K.; Chabriere, E.; Vorholt, J.A.; Erb, T.J.; Tawfik, D.S. The molecular basis of phosphate discrimination in arsenate-rich environments. Nature 2012, 491, 134–137. [Google Scholar] [CrossRef] [PubMed]
- González, D.; Richez, M.; Bergonzi, C.; Chabriere, E.; Elias, M. Crystal structure of the phosphate-binding protein (PBP-1) of an ABC-type phosphate transporter from Clostridium perfringens. Sci. Rep. 2014, 4, 6636. [Google Scholar] [CrossRef]
- Cowlishaw, D.A.; Smith, M.C. A gene encoding a homologue of dolichol phosphate-β-D mannose synthase is required for infection of Streptomyces coelicolor A3(2) by phage φC31. J. Bacteriol. 2002, 184, 6081–6083. [Google Scholar] [CrossRef]
- Cowlishaw, D.A.; Smith, M.C. Glycosylation of a Streptomyces coelicolor A3(2) cell envelope protein is required for infection by bacteriophage φC31. Mol. Microbiol. 2001, 41, 601–610. [Google Scholar] [CrossRef]
- Ong, E.; Kilburn, D.G.; Miller, R.C.; Warren, R.A. Streptomyces lividans glycosylates the linker region of a beta-1,4-glycanase from Cellulomonas fimi. J. Bacteriol. 1994, 176, 999–1008. [Google Scholar] [CrossRef][Green Version]
- Lara, M.; Servin-González, L.; Singh, M.; Moreno, C.; Cohen, I.; Nimtz, M.; Espitia, C. Expression, secretion, and glycosylation of the 45- and 47-kDa glycoprotein of Mycobacterium tuberculosis in Streptomyces lividans. Appl. Environ. Microbiol. 2004, 70, 679–685. [Google Scholar] [CrossRef]
- Zaborina, O.; Holbrook, C.; Chen, Y.; Long, J.; Zaborin, A.; Morozova, I.; Alverdy, J.C. Structure-Function Aspects of PstS in Multi-Drug-Resistant Pseudomonas aeruginosa. PLoS Pathog. 2008, 4, e43. [Google Scholar] [CrossRef] [PubMed]
- Tran, S.L.; Rao, M.; Simmers, C.; Gebhard, S.; Olsson, K.; Cook, G.M. Mutants of Mycobacterium smegmatis unable to grow at acidic pH in the presence of the protonophore carbonyl cyanide m-chlorophenylhydrazone. Microbiology 2005, 151, 665–672. [Google Scholar] [CrossRef] [PubMed]
- Gebhard, S.; Tran, S.L.; Cook, G.M. The Phn system of Mycobacterium smegmatis: A second high-affinity ABC-transporter for phosphate. Microbiology 2006, 152, 3453–3465. [Google Scholar] [CrossRef] [PubMed]
- Gebhard, S.; Busby, J.N.; Fritz, G.; Moreland, N.J.; Cook, G.M.; Lott, J.S.; Money, V.A. Crystal structure of PhnF, a GntR-family transcriptional regulator of phosphate transport in Mycobacterium smegmatis. J. Bacteriol. 2014, 196, 3472–3481. [Google Scholar] [CrossRef] [PubMed]
- Gebhard, S.; Cook, G.M. Differential regulation of high-affinity phosphate transport systems of Mycobacterium smegmatis: Identification of PhnF, a repressor of the phnDCE operon. J. Bacteriol. 2008, 190, 1335–1343. [Google Scholar] [CrossRef][Green Version]
- Kononova, S.V.; Nesmeyanova, M.A. Phosphonates and Their Degradation by Microorganisms. Biochemistry 2002, 67, 184–195. [Google Scholar] [PubMed]
- White, A.K.; Metcalf, W.W. Two C-P lyase operons in Pseudomonas stutzeri and their roles in the oxidation of phosphonates, phosphite, and hypophosphite. J. Bacteriol. 2004, 186, 4730–4739. [Google Scholar] [CrossRef]
- Bardin, S.; Dan, S.; Osteras, M.; Finan, T.M. A phosphate transport system is required for symbiotic nitrogen fixation by Rhizobium meliloti. J. Bacteriol. 1996, 178, 4540–4547. [Google Scholar] [CrossRef]
- Wanner, B.L.; Metcalf, W.W. Molecular genetic studies of a 10.9-kb operon in Escherichia coli for phosphonate uptake and biodegradation. FEMS Microbiol. Lett. 1992, 79, 133–139. [Google Scholar] [CrossRef]
- Hoffer, S.M.; van Uden, N.; Tommassen, J. Expression of the pho regulon interferes with induction of the uhpT gene in Escherichia coli K-12. Arch. Microbiol. 2001, 176, 370–376. [Google Scholar] [CrossRef]
- van Veen, H.W.; Abee, T.; Kortstee, G.J.; Konings, W.N.; Zehnder, A.J. Translocation of metal phosphate via the phosphate inorganic transport system of Escherichia coli. Biochemistry 1994, 33, 1766–1770. [Google Scholar] [CrossRef] [PubMed]
- van Veen, H.W. Phosphate transport in prokaryotes: Molecules, mediators and mechanisms. Ant. Van Leeuwen. 1997, 72, 299–315. [Google Scholar] [CrossRef] [PubMed]
- Voegele, R.T.; Bardin, S.; Finan, T.M. Characterization of the Rhizobium (Sinorhizobium) meliloti high- and low-affinity phosphate uptake systems. J. Bacteriol. 1997, 179, 7226–7232. [Google Scholar] [CrossRef]
- Russell, L.M.; Rosenberg, H. The Nature of the Link between Potassium Transport and Phosphate Transport in Escherichia coli. Biochem. J. 1980, 188, 715–723. [Google Scholar] [CrossRef] [PubMed]
- van Veen, H.W.; Abee, T.; Kortstee, G.J.; Konings, W.N.; Zehnder, A.J. Substrate specificity of the two phosphate transport systems of Acinetobacter johnsonii 210A in relation to phosphate speciation in its aquatic environment. J. Biol. Chem. 1994, 269, 16212–16216. [Google Scholar] [CrossRef]
- Beard, S.J.; Hashim, R.; Wu, G.; Binet, M.R.; Hughes, M.N.; Poole, R.K. Evidence for the transport of zinc(II) ions via the pit inorganic phosphate transport system in Escherichia coli. FEMS Microbiol. Lett. 2000, 184, 231–235. [Google Scholar] [CrossRef]
- Jackson, R.J.; Binet, M.R.B.; Lee, L.J.; Ma, R.; Graham, A.I.; McLeod, C.W.; Poole, R.K. Expression of the pitA phosphate/metal transporter of Escherichia coli is responsive to zinc and inorganic phosphate levels. FEMS Microbiol. Lett. 2008, 289, 219–224. [Google Scholar] [CrossRef]
- Graham, A.I.; Hunt, S.; Stokes, S.L.; Bramall, N.; Bunch, J.; Cox, A.G.; Poole, R.K. Severe zinc depletion of Escherichia coli: Roles for high affinity zinc binding by ZinT, zinc transport and zinc-independent proteins. J. Biol. Chem. 2009, 284, 18377–18389. [Google Scholar] [CrossRef]
- Keasling, J.D. Regulation of intracellular toxic metals and other cations by hydrolysis of polyphosphate. Ann. N. Y. Acad. Sci. 1997, 829, 242–249. [Google Scholar] [CrossRef]
- Harris, R.M.; Webb, D.C.; Howitt, M.; Cox, G.B. Characterization of PitA and PitB from Escherichia coli. J. Bacteriol. 2001, 183, 5008–5014. [Google Scholar] [CrossRef]
- Santos-Beneit, F.; Rodríguez-García, A.; Franco-Domínguez, E.; Martín, J.F. Phosphate-dependent regulation of the low- and high-affinity transport systems in the model actinomycete Streptomyces coelicolor. Microbiology 2008, 154, 56–2370. [Google Scholar] [CrossRef]
- Omura, S.; Ikeda, H.; Ishikawa, J.; Hanamoto, A.; Takahashi, C.; Shinose, M.; Hattori, M. Genome sequence of an industrial microorganism Streptomyces avermitilis deducing the ability of producing secondary metabolites. Proc. Natl. Acad. Sci. USA 2001, 98, 12215–12220. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, H.; Ishikawa, J.; Hanamoto, A.; Shinose, M.; Kikuchi, H.; Shiba, T.; Ōmura, S. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat. Biotechnol. 2003, 21, 526–531. [Google Scholar] [CrossRef] [PubMed]
- Bardin, S.D.; Voegele, R.T.; Finan, T.M. Phosphate assimilation in Rhizobium (Sinorhizobium) meliloti: Identification of a pit-like gene. J. Bacteriol. 1998, 180, 4219–4226. [Google Scholar] [CrossRef] [PubMed]
- Santos-Beneit, F.; Rodríguez-García, A.; Martín, J.F. Complex transcriptional control of the antibiotic regulator afsS in Streptomyces: PhoP and AfsR are overlapping, competitive activators. J. Bacteriol. 2011, 193, 2242–2251. [Google Scholar] [CrossRef]
- Schaaf, S.; Bott, M. Target Genes and DNA-Binding Sites of the Response Regulator PhoR from Corynebacterium glutamicum. J. Bacteriol. 2007, 189, 5002–5011. [Google Scholar] [CrossRef]
- Ishige, T.; Krause, M.; Bott, M.; Wendisch, V.F.; Sahm, H. The phosphate starvation stimulon of Corynebacterium glutamicum determined by DNA microarray analyses. J. Bacteriol. 2003, 185, 4519–4529. [Google Scholar] [CrossRef]
- Werten, S.; Rustmeier, N.H.; Gemmer, M.; Virolle, M.J.; Hinrichs, W. Structural and biochemical analysis of a phosin from Streptomyces chartreusis reveals a combined polyphosphate- and metal-binding fold. FEBS Lett. 2019, 593, 2019–2029. [Google Scholar] [CrossRef]
- Osbourn, A.E.; Field, B. Operons. Cell. Mol. Life Sci. 2009, 66, 3755–3775. [Google Scholar] [CrossRef]
- Morohoshi, T.; Maruo, T.; Shirai, Y.; Kato, J.; Ikeda, T.; Takiguchi, N.; Kuroda, A. Accumulation of inorganic polyphosphate in phoU mutants of Escherichia coli and Synechocystis sp. strain PCC6803. Appl. Environ. Microbiol. 2002, 68, 4107–4110. [Google Scholar] [CrossRef]
- Charaniya, S.; Mehra, S.; Lian, W.; Jayapal, K.P.; Karypis, G.; Hu, W.-S. Transcriptome dynamics-based operon prediction and verification in Streptomyces coelicolor. Nucleic Acids Res. 2007, 35, 7222–7236. [Google Scholar] [CrossRef] [PubMed]
- Jeong, Y.; Kim, J.N.; Kim, M.W.; Bucca, G.; Cho, S.; Yoon, Y.J.; Cho, B.K. The dynamic transcriptional and translational landscape of the model antibiotic producer Streptomyces coelicolor A3(2). Nat. Commun. 2016, 7, 11605. [Google Scholar] [CrossRef] [PubMed]
- Di Cenzo, G.C.; Sharthiya, H.; Nanda, A.; Zamani, M.; Finan, T.M. PhoU Allows Rapid Adaptation to High Phosphate Concentrations by Modulating PstSCAB Transport Rate in Sinorhizobium meliloti. J. Bacteriol. 2017, 199, e00143-17. [Google Scholar]
- Brzoska, P.; Boos, W. Characteristics of an ugp-encoded and phoB-dependent glycerophosphoryl diester phosphodiesterase which is physically dependent on the Ugp transport system of Escherichia coli. J. Bacteriol. 1988, 170, 4125–4135. [Google Scholar] [CrossRef] [PubMed]
- Brzoska, P.; Rimmele, M.; Brzostek, K.; Boos, W. The pho Regulon-Dependent Ugp Uptake System for Glycerol-3-Phosphate in Escherichia coli Is trans Inhibited by Pi. J. Bacteriol. 1994, 176, 15–20. [Google Scholar] [CrossRef]
- Larson, T.J.; Schumacher, G.; Boos, W. Identification of the glpT-encoded sn-glycerol-3-phosphate permease of Escherichia coli, an oligomeric integral membrane protein. J. Bacteriol. 1982, 152, 1008–1021. [Google Scholar]
- Larson, T.J.; Schumacher, G.; Boos, W. Periplasmic glycerophosphodiesterase of Escherichia coli, a new enzyme of the glp regulon. J. Biol. Chem. 1983, 258, 5428–5432. [Google Scholar] [CrossRef]
- Santos-Beneit, F.; Rodríguez-García, A.; Apel, A.K.; Martín, J.F. Phosphate and carbon source regulation of two PhoP-dependent glycerophosphodiester phosphodiesterase genes of Streptomyces coelicolor. Microbiology 2009, 155, 1800–1811. [Google Scholar] [CrossRef]
- Schneider, T.D. Information content of individual genetic sequences. J. Theor. Biol. 1997, 189, 427–441. [Google Scholar] [CrossRef]
- Claverys, J.P.; Havarstein, L.S. Cannibalism and fratricide: Mechanisms and raisons d´etre. Nat. Rev. Microbiol. 2007, 5, 219–229. [Google Scholar] [CrossRef]
- González-Pastor, J.E. Cannibalism: A social behavior in sporulating Bacillus subtilis. FEMS Microbiol. Rev. 2011, 35, 415–424. [Google Scholar] [CrossRef] [PubMed]
- Götz, A.; Goebel, W. Glucose and glucose 6-phosphate as carbon sources in extra- and intracellular growth of enteroinvasive Escherichia coli and Salmonella enterica. Microbiology 2010, 156, 1176–1187. [Google Scholar] [CrossRef] [PubMed]
- Merkel, T.J.; Nelson, D.M.; Brauer, C.L.; Kadner, R.J. Promoter elements required for positive control of transcription of the Escherichia coli uhpT gene. J. Bacteriol. 1992, 174, 2763–2770. [Google Scholar] [CrossRef]
- Verhamme, D.T.; Postma, P.W.; Crielaard, W.; Hellingwerf, K.J. Cooperativity in signal transfer through the Uhp system of Escherichia coli. J. Bacteriol. 2002, 184, 4205–4210. [Google Scholar] [CrossRef] [PubMed]
- Tenconi, E.; Jourdan, S.; Motte, P.; Virolle, M.J.; Rigali, S. Extracellular sugar phosphates are assimilated by Streptomyces in a PhoP-dependent manner. Antonie Van Leeuwenhoek 2012, 102, 425–433. [Google Scholar] [CrossRef]
- Martín, J.F.; Demain, A.L. Cleavage of adenosine 5′-monophosphate during uptake by Streptomyces griseus. J. Bacteriol. 1977, 132, 590–595. [Google Scholar] [CrossRef]
- Li, M.; Kim, T.-J.; Kwon, H.-J.; Shu, J.-W. Effects of extracellular ATP on the physiology of Streptomyces coelicolor A3(2). FEMS Microbiol. Lett. 2008, 286, 24–31. [Google Scholar] [CrossRef]
- Millan-Oropeza, A.; Henry, C.; Lejeune, C.; David, M.; Virolle, M.J. Expression of genes of the Pho regulon is altered in Streptomyces coelicolor. Sci Rep. 2020, 10, 8492. [Google Scholar] [CrossRef]
- Schowanek, D.; Verstraete, W. Phosphonate utilization by bacterial cultures and enrichments from environmental samples. Appl. Environ. Microbiol. 1990, 56, 895–903. [Google Scholar] [CrossRef]
- Chen, C.M.; Ye, Q.Z.; Zhu, Z.M.; Wanner, B.L.; Walsh, C.T. Molecular biology of carbon-phosphorus bond cleavage. Cloning and sequencing of the phn (psiD) genes involved in alkylphosphonate uptake and C-P lyase activity in Escherichia coli B. J. Biol Chem. 1990, 265, 4461–4471. [Google Scholar] [CrossRef]
- Metcalf, W.W.; Wanner, B.L. Evidence for a fourteen-gene, phnC to phnP locus for phosphonate metabolism in Escherichia coli. Gene 1993, 129, 27–32. [Google Scholar] [CrossRef]
- Imazu, K.; Tanaka, S.; Kuroda, A.; Anbe, Y.; Kato, J.; Ohtake, H. Enhanced utilization of phosphonate and phosphite by Klebsiella aerogenes. Appl. Environ. Microbiol. 1998, 64, 3754–3758. [Google Scholar] [CrossRef] [PubMed]
- Kertesz, M.; Elgorriaga, A.; Amrhein, N. Evidence for two distinct phosphonate-degrading enzymes (C-P lyases) in Arthrobacter sp. GLP-1. Biodegradation 1991, 2, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Krzyśko-Lupicka, T.; Strof, W.; Kubś, K.; Skorupa, M.; Wieczorek, P.; Lejczak, B.; Kafarski, P. The ability of soil-borne fungi to degrade organophosphonate carbon-to-phosphorus bonds. Appl. Micobiol. Biotechnol. 1997, 48, 549–552. [Google Scholar] [CrossRef] [PubMed]
- Bujacz, B.; Wieczorek, P.; Krzysko-Lupicka, T.; Golab, Z.; Lejczak, B.; Kavfarski, P. Organophosphonate Utilization by the Wild-Type Strain of Penicillium notatum. Appl. Microbiol. Biotechnol. 1995, 48, 549–552. [Google Scholar] [CrossRef] [PubMed]
- Surin, B.P.; Rosenberg, H.; Cox, G.B. Phosphate-specific transport system of Escherichia coli: Nucleotide sequence and gene-polypeptide relationships. J. Bacteriol. 1985, 161, 189–198. [Google Scholar] [CrossRef]
- Pipke, R.; Amrhein, N.; Jacob, G.S.; Schaefer, J.; Kishore, G.M. Metabolism of glyphosate in an Arthrobacter sp. GLP-1. Eur. J. Biochem. 1987, 165, 267–273. [Google Scholar] [CrossRef] [PubMed]
- Pipke, R.; Schulz, A.; Amrhein, N. Uptake of Glyphosate by an Arthrobacter sp. Appl. Environ. Microbiol. 1987, 53, 974–978. [Google Scholar] [CrossRef]
- Obojska, A.; Lejczak, B.; Kubrak, M. Degradation of phosphonates by streptomycete isolates. Appl. Microbiol. Biotechnol. 1999, 51, 872–876. [Google Scholar] [CrossRef]
- Thompson, C.J.; Seto, H. Bialaphos. Biotechnology 1995, 28, 197–222. [Google Scholar]
- Hendlin, D.; Stapley, E.O.; Jackson, M.; Wallick, H.; Miller, A.K.; Wolf, F.J.; Mochales, S. Phosphonomycin, a new antibiotic produced by strains of Streptomyces. Science 1969, 166, 122–123. [Google Scholar] [CrossRef] [PubMed]
- Cundliffe, E.; Demain, A.L. Avoidance of suicide in antibiotic-producing microbes. J. Ind. Microbiol. Biotechnol. 2010, 37, 643–672. [Google Scholar] [CrossRef] [PubMed]
- Martín, J.F.; Casqueiro, J.; Liras, P. Secretion systems for secondary metabolites: How producer cells send out messages of intercellular communication. Curr. Opin. Microbiol. 2005, 8, 282–293. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martín, J.F.; Liras, P. Molecular Mechanisms of Phosphate Sensing, Transport and Signalling in Streptomyces and Related Actinobacteria. Int. J. Mol. Sci. 2021, 22, 1129. https://doi.org/10.3390/ijms22031129
Martín JF, Liras P. Molecular Mechanisms of Phosphate Sensing, Transport and Signalling in Streptomyces and Related Actinobacteria. International Journal of Molecular Sciences. 2021; 22(3):1129. https://doi.org/10.3390/ijms22031129
Chicago/Turabian StyleMartín, Juan Francisco, and Paloma Liras. 2021. "Molecular Mechanisms of Phosphate Sensing, Transport and Signalling in Streptomyces and Related Actinobacteria" International Journal of Molecular Sciences 22, no. 3: 1129. https://doi.org/10.3390/ijms22031129
APA StyleMartín, J. F., & Liras, P. (2021). Molecular Mechanisms of Phosphate Sensing, Transport and Signalling in Streptomyces and Related Actinobacteria. International Journal of Molecular Sciences, 22(3), 1129. https://doi.org/10.3390/ijms22031129





