Abstract
Na+/H+ exchange factor-1 (NHERF1), a multidomain PDZ scaffolding phosphoprotein, is required for the type II sodium-dependent phosphate cotransporter (NPT2A)-mediated renal phosphate absorption. Both PDZ1 and PDZ2 domains are involved in NPT2A-dependent phosphate uptake. Though harboring identical core-binding motifs, PDZ1 and PDZ2 play entirely different roles in hormone-regulated phosphate transport. PDZ1 is required for the interaction with the C-terminal PDZ-binding sequence of NPT2A (-TRL). Remarkably, phosphocycling at Ser290 distant from PDZ1, the penultimate step for both parathyroid hormone (PTH) and fibroblast growth factor-23 (FGF23) regulation, controls the association between NHERF1 and NPT2A. PDZ2 interacts with the C-terminal PDZ-recognition motif (-TRL) of G Protein-coupled Receptor Kinase 6A (GRK6A), and that promotes phosphorylation of Ser290. The compelling biological puzzle is how PDZ1 and PDZ2 with identical GYGF core-binding motifs specifically recognize distinct binding partners. Binding determinants distinct from the canonical PDZ-ligand interactions and located “outside the box” explain PDZ domain specificity. Phosphorylation of NHERF1 by diverse kinases and associated conformational changes in NHERF1 add more complexity to PDZ-binding diversity.
1. Introduction
Protein–protein interactions play a major role in regulation of intracellular phosphate bringing together different classes of proteins and stabilizing multiprotein complexes. The tight interaction between the type II sodium-dependent phosphate cotransporter (NPT2A) mediated renal phosphate uptake and Na+/H+ exchange regulatory factor-1 (NHERF1) is required for renal phosphate uptake. Two hormones parathyroid hormone (PTH) and Fibroblast growth factor 23 (FGF23) regulate phosphate homeostasis by controlling the NPT2A-NHERF1 association. Remarkably, PTH and FGF23 belong to different classes of receptors, a Family B G protein coupled receptor (PTHR, parathyroid hormone receptor) and receptor tyrosine kinase (FGFR1, fibroblast growth factor receptor 1), respectively, and stimulate two distinct signaling pathways involving activation of diverse kinases (Figure 1). The precise mechanism and modulators involved in NHERF1-dependent NPT2A-mediated hormone-sensitive phosphate uptake are not completely understood.
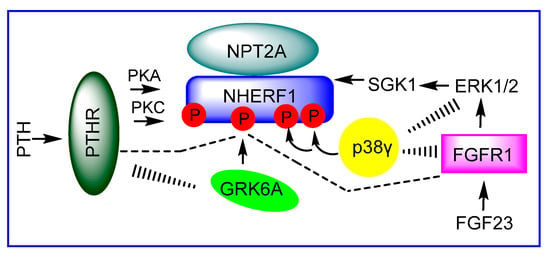
Figure 1.
NHERF1 brings together different classes of protein for NPT2A-dependent hormone-regulated phosphate transport in kidney cells. PTH and FGF23 hormones work through PTHR and FGFR1, respectively, and activate two distinct phosphorylation pathways leading to phosphorylation of NHERF1 by diverse kinases.
NHERF1 (EBP50) belonging to the NHERF family [] of PDZ adaptor proteins controls protein localization at the apical plasma membrane of polarized epithelial cells [,]. NHERF1 tethers binding partners through tandem PDZ domains named for the common structural domain shared by the postsynaptic density protein (PSD95), Drosophila disc large tumor suppressor (DlgA), and zonula occludens-1 protein (ZO-1), and a C-terminal ezrin-binding domain (EBD) associated with ezrin, radixin, and moesin through extreme C-terminal end representing a PDZ-ligand motif itself (-FSNL) (Figure 2). []. PDZ domains are approximately 90 aa in length and commonly represented as a box (Figure 2). Two flexible unstructured linkers connect PDZ1 and PDZ2, and PDZ2 and EBD. PDZ-dependent binding partners of NHERF1 include G-protein couple receptors (GPCR’s) (PTHR, CXCR2, β2AR), ion transporters (NPT2A, CFTR), transcriptional modifiers (TAZ/YAP65), and phosphoenzymes and posttranslational regulators (PKCα, GRK6A, SGK1, RGS14, MAPKs) (Table 1). These proteins may selectively bind PDZ1 or PDZ2, or both PDZ domains. The regulation of renal proximal tubular phosphate reabsorption requires PDZ-dependent interactions between NHERF1, NPT2A, and GRK6A.
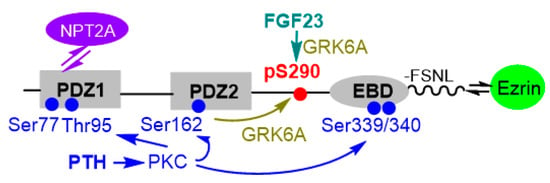
Figure 2.
Linear representation of NHERF1. NPT2A-dependent PTH- and FGF23-sensitive phosphate transport converge on Ser290 of NHERF1. PTH-stimulated phosphorylation employs PKC/PKCα. Both processes cause conformational changes in NHERF1 allowing GRK6A to phosphorylate Ser290 and disengage NPT2A from NHERF1-Ezrin complex, thereby arresting phosphate transport.

Table 1.
PDZ-selective NHERF1 interactions.
NPT2A (SLC34A1), the primary renal Na-dependent phosphate transporter, mediates hormone-regulated phosphate transport and is an essential binding partner of NHERF1 [,,,,]. NPT2A has eight putative transmembrane domains and several intra- and extracellular loops. The NPT2A intracellular C-terminus ends with a carboxy-terminal PDZ motif (-T−2-R-L0) that mediates its binding to NHERF1. Loss-of-function mutations in NHERF1 (PDZ1/L110V, PDZ2/R153Q and E225K) or NPT2A (R495H/C and S585P) disrupt phosphate metabolism and lead to hypophosphatemia [,,,]. Parathyroid hormone (PTH) and fibroblast growth factor 23 (FGF23) dissociate the NPT2A-NHERF1 binary complex by activating two distinct signaling pathways that converge at NHERF1 [,,,] resulting in NPT2A internalization and downregulation. PTH works through its cognate parathyroid hormone receptor (PTHR). PTHR contains seven transmembrane domains linked by three extracellular and three intracellular loops, and the intracellular C-terminus has a PDZ-binding sequence (-E−3-T−2-V-M0). PDZ-ligand interaction between NHERF1 and PTHR is critical for receptor signaling [,]. It is thought that NHERF1 homodimerization may play a key role in the formation of an extended multiprotein signaling complex that includes NHERF1, NPT2A, ezrin, PTHR and some kinases [,,,,]. The PDZ domains in NHERF1, like other such structures, are relatively rigid units that were crystallized and characterized [,]. Linkers between PDZ1 and PDZ2 and between PDZ2 and EBD are unstructured and flexible. This contributes to the challenge of solving the structure of full-length NHERF1. Both NHERF1 PDZ domains have a classical PDZ fold with two alpha-helixes and six beta-sheets and share 65% identity or sequence similarity. PDZ1 and PDZ2 are Class I PDZ domains with identical GYGF core-binding motifs, known also as carboxylate-binding sites. Available X-ray structures of PDZ1 or PDZ2 bound to the C-terminus of β2-AR (-DSLL), cystic fibrosis transmembrane conductance regulator (CFTR) (-DTRL) [,], or chemokine receptor-2 (CXCR2) (-STTL) [,] demonstrate that the preserved C-terminal X-S−2/T-X-Φ0 motif of peptide ligands (PDZ-binding motif) occupies the PDZ domain cavity as an antiparallel β-sheet. By convention, the sequential residues forming a PDZ-binding motif are numbered starting from the last position (Φ 0) and going upstream toward the N-terminus as –1, –2, –3, etc. Φ0 forms hydrophobic interactions with the GYGF motif, as well as hydrogen bonds through backbone atoms of PDZ1/PDZ2. Ser−2/Thr−2 of target C-terminal PDZ ligands establishes a direct side chain hydrogen bond with His72 or His212 from the top of the α2 helix of PDZ1 and PDZ2, respectively. These interactions are conserved, called canonical, and characterize Class I PDZ domains [,,]. Mutation of Tyr24/Tyr164 and Phe26/Phe166 to Ala in the GlyTyrGlyPhe [GYGF hereafter] motif (GlyAlaGlyAla sequence [GAGA hereafter]) of PDZ1/PDZ2 disrupts the hydrophobic network and blocks the association with ligands [,]. Remarkably, a substitution of the conserved Thr−2 by Cys in the C-terminus of PTHR (-ETVM) decreases but does not eliminate binding with His212 of PDZ2 and more essentially does not interfere with PTHR function [].
In addition to canonical interactions, NHERF1 PDZ1 and PDZ2 also form non-canonical interactions distal from the PDZ binding groove and located “outside the box”. The presence of such noncanonical interactions is illustrated by various domain-selective binding of different proteins that selectively engage NHERF1 PDZ1 or PDZ2 despite the identical canonical binding sites (Table 1). The non-canonical binding determinants are unique for individual PDZ domains []. There are several typical features making “outside the box” interactions specific. Frequently these interactions have an electrostatic nature []; some of them are regulated by phosphorylation [,,] or involved in a long-range allosteric network []. Further, the helical-turn-helical (ɑ3-turn-ɑ4) extension is an essential structural element common for many PDZ domains and represents a universal element stabilizing PDZ-ligand binding [,]. Whether the ɑ3-loop-ɑ4 extension allosterically affects the binding with target ligands or directly contacts upstream N-terminal residues of the bound target remains to be established.
The main feature making NHERF1 unique is its C-terminal tail corresponding to a PDZ-ligand motif (-FS−2NL0). It is believed that -FS−2NL0 may occupy PDZ2 rather than PDZ1 to form a self-inhibited conformation []. The physiological role of the self-inhibited conformer remains to be elucidated. The putative ability of NHERF1 to dimerize through its C-terminal PDZ-motif (-FS−2NL0) was extensively investigated [,,,,]. Based on dynamic light scattering experiments, NHERF1 in solution is monodispersed []. Curiously, the NHERF1-NHERF1 association decreased in the presence of okadaic acid suggesting that phosphorylation may regulate dimerization []. A comparable effect was observed in the presence of the C-terminal PDZ-ligand motif of the β2-AR (-DSLL) []. The biological importance of NHERF1 dimerization may relate to its ability to assemble large multi-protein complexes and serve as an adapter for different classes of proteins. However, this theory has not been analyzed in detail.
NHERF1 is a phosphoprotein harboring 31 Ser and 9 Thr residues. Notably, there is a high-density Ser cluster in the flexible hinge region of NHERF1 linking PDZ2 and the EBD with 17 Ser/Thr residues in this segment. Ser290 located in this region was identified as a phosphorylation site that allosterically regulates the interaction between NHERF1 and NPT2A through conformational changes near Glu43 [], a site that determines the binding specificity between PDZ1 and NPT2A []. Furthermore, dephosphorylation-phosphorylation cycling of Ser290 regulates NHERF1 self-assembly and NPT2A-dependent hormone-sensitive phosphate transport []. Remarkably, both PTH and FGF23 pathways contribute equally to the stabilization of the open state of NHERF1 via phosphorylation of Ser290 by G Protein-coupled Receptor Kinase 6A (GRK6A) (Figure 1) []. PTH-induced phosphorylation of Ser77 in conjunction with Thr95 has a similar effect on regulating phosphate uptake as does Ser290 phosphorylation. Ser77 and Thr95, located in PDZ1, are predicted PKC phosphorylation sites (Figure 1) [,]. Ser162 is a defined PKCɑ phosphorylation site [,]. Notably, PKCɑ is a major pathway of PTH signaling [,,] and is the only PKC isoform with a PDZ-recognition motif at its C-terminus (-SAV672) [,]. Two other PKC phosphorylation sites, Ser339 and Ser340, in the C-terminal region (Figure 2), promote conformational reorganization in NHERF1 and facilitate phosphorylation of Ser162 []. The role of this remarkable cooperativity on hormone-induced phosphorylation of NHERF1 has not yet been examined in vivo. Whether additional phospho-Ser/Thr sites regulate NHERF1-ligand interactions, conformational diversity, or phosphate transport is not known.
2. NHERF1 (PDZ1) Specifically Binds NPT2A for Hormone-Sensitive Phosphate Transport
2.1. NHERF1 PDZ1 and PDZ2 Domains Are Not Interchangeable
The association between NHERF1 PDZ1 and its natural ligand NPT2A (SLC34A1) is a perfect example representing NHERF1 PDZ-ligand specificity. The NHERF1 PDZ1-NPT2A complex is required for NPT2A-mediated hormone-sensitive renal phosphate transport [,,]. The interaction between PDZ1 and NPT2A occurs through the PDZ-ligand C-terminal -T−2R-1L0(639) sequence of NPT2A. The PDZ1-NPT2A complex is not assembled in the presence of NPT2A-Leu639Ala, a variant that has a defective C-terminal PDZ recognition motif (-T−2R−1L0/A) []. PDZ1 interacts with the C-terminal peptide ligand of NPT2A within the low micromolar range (3–5 µM), whereas the association between PDZ2 and NPT2A is insignificant [,]. The biological enigma is why and how PDZ1 and PDZ2 with identical conserved binding sites (GYGF and His) interact uniquely with NPT2A PDZ1. Analysis of possible binding determinants “outside the box” pointed to Glu43 located in the α1 helix of PDZ1. MD simulations applied in our study predict the formation of an ionic pair between Glu43 of PDZ1 and Arg−1 of the NPT2A C-terminal -T−2R−1L0 motif []. Long-range MD simulations (~ 100-ns) provide clear evidence that the negatively charged side chain of Glu43 and the side chain of His27 are involved in electrostatic interactions with the positively charged side chain of Arg−1 of NPT2A (-T−2-R−1-L0) and, more importantly, these interactions are persistent on the MD simulation time scale. In contrast to PDZ1, PDZ2 possesses Asp183 and Asn167 at the homologous positions. However, these residues do not form a stable interaction with Arg−1 []. We propose that the side chain of Asp183, which is relatively short compared to Glu43, is thereby unable to support direct interaction with the side chain of Arg−1. Our MD simulations [] and available NMR structure [] demonstrate that the sidechain of Asp183 is flexible and not involved in a stable interaction with Arg−1 and provide provisional support of this theory. Notably, the Glu43Asp mutation in PDZ1 leads to dramatic loss of affinity with a similar C-terminal -TRL motif of CFTR [], another biological partner of NHERF1 []. CFTR, like NPT2A, largely associates with PDZ1 but not with PDZ2 [,,]. Recently published X-ray structures and MD simulations of the PDZ2-CFTR complex suggest that the interaction between Asp183 and Arg−1 is underestimated and can be an important element of the complex []. However, how the association between Asp183 and Arg−1 regulates binding affinity between PDZ2 and CFTR remains unresolved.
To explore specificity of Glu43 and His27 on the interaction between PDZ1 and NPT2A we generated a PDZ1 variant where Glu43 and His27 were replaced by Asp and Asn, respectively. As anticipated, PDZ1 with the Glu43Asp/His27Asn mutations eliminates binding between PDZ1 and NPT2A. The relevance of Glu43 and His27 on NPT2A-dependent PTH-sensitive phosphate transport was validated by measuring phosphate uptake in OKH cells expressing the Glu43Asp/His27Asn-NHERF1 variant. As we presumed, Glu43Asp/His27Asn-NHERF1 blocks basal phosphate transport and is refractory to PTH []. Notably, when Asp183 and Asn167 in PDZ2 were replaced respectively by Glu and His, the corresponding residues in PDZ1, the Asp183Glu/Asn167His rescue variant bound the NPT2A C-terminal -TRL motif (Figure 3) []. However, Asp183Glu/Asn167His-NHERF1 did not support NPT2A-dependent PTH-sensitive phosphate uptake (unpublished observations). As expected, mutation of Glu43 to Gly in PDZ1 of NHERF1 blocked phosphate uptake (Figure 4) []. Experimental study confirms that PDZ1 is required for formation of the NPT2A complex, and regulation of NPT2A-mediated hormone-sensitive phosphate transport. This finding strongly supports the concept that the specificity of the PDZ1 domain is not determined by the conserved subset of residues (-23GYGF26- and His72) but rather by the “outside the box” determinant Glu43. Support for this conclusion also comes from thermodynamic parameters determined by isothermal microcalorimetry []. Substitution of Glu43 by Asp increases enthalpy (ΔHο) and provides a jump in entropy (ΔSο) making the interaction unfavorable. Mutation of His27 to Asn has a minor effect. A small change in the free energy (ΔΔGο), 0.8 and 0.4 kcal/mol for Glu43→Asp and His27→Asn, respectively, is attributed to the enthalpy-entropy compensation [] illustrated in Figure 5.
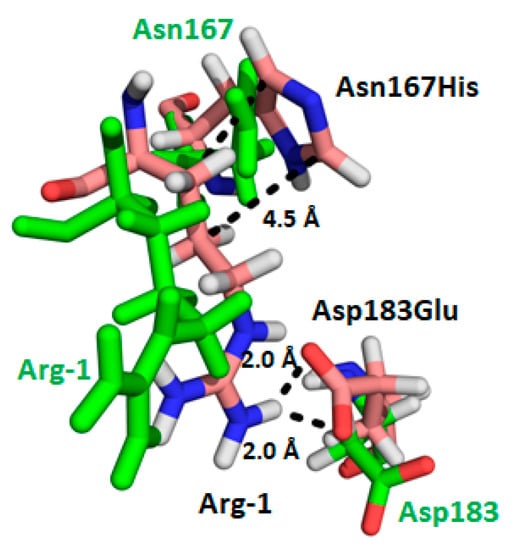
Figure 3.
Double mutation of Asp183 to Glu and Asn167 to His in PDZ2 favors the formation of electrostatic interactions with Arg-1 of the C-terminal -TRL motif of NPT2A shown in wheat sticks. The predicted interactions stabilizing the complex are shown as black dotted lines. Wild-type Asp183 and Asn167 of PDZ2 shown in green sticks do not interact with Arg-1 of NPT2A (green sticks) [].
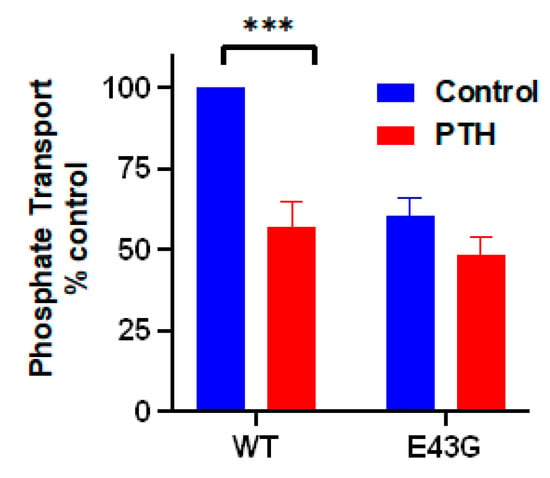
Figure 4.
Mutation of Glu43 to Gly in PDZ1 of NHERF1 inhibits NPT2A-dependent PTH-sensitive phosphate transport. Results report the mean ± SEM (n = 3, *** p < 0.001, ANOVA) [].
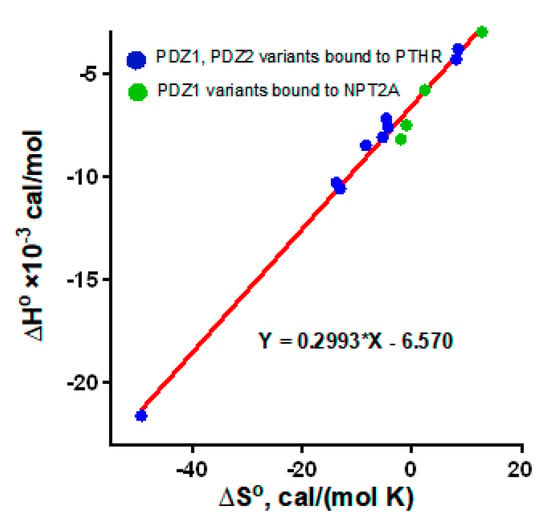
Figure 5.
Linear regression of ∆Hο vs. ∆Sο. Data for wild-type PDZ1, PDZ1-His27Asn, PDZ1-Glu43Asp, and PDZ1-His27Asn/Glu43Asp bound to the C-terminal motif of NPT2A are green (data from Table 2 []). Data for PDZ1 and PDZ2 bound to the ensemble of Ala variants of the C-terminal motif of PTHR are blue (data from Table 4 []).
2.2. The Role of the NPT2A Internal -TRL- Motif Remains to Be Explored
In addition to its canonical C-terminal -T−2RL0 PDZ-binding motif, NPT2A possess an internal PDZ -494T−2RL0- recognition sequence that has not been characterized. The C-terminal motif is critical for interaction with NHERF1 []. Whether both PDZ motifs contribute to or are required for proper localization and function of NPT2A and hormone action is unknown. Two disease-associated mutations (Arg495His, Arg495Cys) have recently been described in the putative internal PDZ motif (-494TRL496-), while a third (Ser585Pro) is located at the carboxy-terminal region. All are associated with elevated renal phosphate excretion and consequent hypophosphatemia [,,]. NPT2A-Arg495His and NPT2A-Ser585Pro display different cell localization compared to wild-type (WT) NPT2A []. The carboxy-terminal PDZ-binding motif (-T−2R-1L0) [,,] is required for cell membrane NPT2A localization and is necessary for hormone-regulated phosphate transport [,]. The involvement of the putative internal NPT2A PDZ motif in association with NHERF1 and its role in hormone action is not described. Pilot results indicate that mutation of the internal PDZ motif interferes with PTH and FGF23 action and inhibits regulated uptake (Figure 6) despite normal binding to NHERF1 (unpublished work).
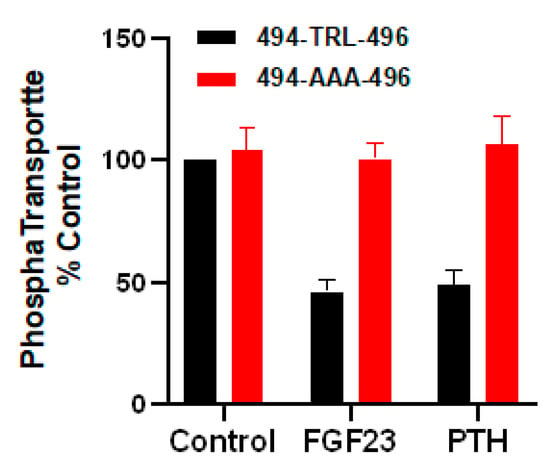
Figure 6.
Mutation of the WT internal PDZ-binding 494-TRL-496 motif of NPT2A to 494-AAA-496 blocks PTH- and FGF23-sensitive phosphate uptake. PTH- and FGF23-inhibitable phosphate uptake was measured in NPT2A CRISPR/Cas9 knockout OK cells transfected with a human NPT2A construct with carboxy-terminal -T−2R-1L0 → -AAA substitution. Results report the mean ± SEM (n = 5, p < 0.0001, ANOVA) (unpublished observation).
3. NPT2A-Dependent Hormone-Inhibitable Phosphate Transport Requires Association between PDZ2 and GRK6A
G protein-coupled receptor kinase 6A (GRK6A), a natural partner of NHERF1, possesses a canonical PDZ ligand (-T−2RL0) at its C-terminus. GRK6A, like NPT2A, associates with NHERF1 PDZ domains through its C-terminal motif (-T−2R−1L0). Knocking down Grk6a by siRNA blocks Npt2a-dependent phosphate uptake in response to PTH []. Thus, GRK6A is an essential regulatory component of NPT2A-dependent PTH-sensitive phosphate transport and corroborates previous findings that GRK6A pharmacological inhibitors abolish PTH action []. Binding affinities (3–5 µM) for the C-terminal PDZ ligand of GRK6A [] or NPT2A (22 aa) [] with NHERF1 are comparable and suggest that the binding mechanism is presumable identical. NPT2A and GRK6A interact with PDZ1 with a greater affinity than to PDZ2, thereby confirming that PDZ1 is naturally optimized to bind the -TRL sequence. Intriguingly, a minor interaction between GRK6A and PDZ2 is nonetheless critical for constitutive or PTH-induced phosphorylation of NHERF1 at Ser290 [,], and for PTH-sensitive phosphate transport []. This finding highlights a biological puzzle as to how PDZ2 becomes accessible to GRK6A and the role of PTH and FGF23 in this process. Again, canonical PDZ recognition sites (-GYGF- and His) required for the binding of the C-terminal Leu0 and Thr−2 of GRK6A cannot explain the observed specificity for PDZ2. Furthermore, mutation of the NHERF1 PDZ2 core-binding GYGF motif (N1P2-GAGA) decreased but did not abolish phosphate transport in response to PTH (Figure 7). This finding strongly suggests that the PDZ2 domain retains its ability to interact with the C-terminus of GRK6A in vivo. Clearly, binding determinants “outside the box” control the formation of the PDZ2-GRK6A complex. Analysis of residues that may contribute to the binding pointed to Ser162, known as a PKCα phosphorylation site in human NHERF1 [,]. Notably, NHERF1 homologs (mouse, rabbit) harbor Asn at the corresponding position. PKCα action is unique for PDZ2 inasmuch as PDZ1 has Asn22 at the homologous location. The published docking structure of PDZ2 interacting with its cognate C-terminal peptide demonstrates that Ser162 is masked by the C-terminal tail []. The in vivo significance of post-translational modification of Ser162 is unknown. We initiated studies to elucidate the role of NHERF1 Ser162 on NPT2A-dependent phosphate transport. Ala replacement at Ser162 (NHERF1 Ser162Ala) diminished PTH-inhibitable phosphate transport suggesting that Ser162 is an essential regulator of hormone-sensitive phosphate uptake. Unexpectedly, phosphomimic Ser162Asp mutation in NHERF1 disrupts basal phosphate transport and blocks PTH action on phosphate uptake (Figure 8) []. Clearly, the discrepancy in charge and size between phosphate and carboxylate groups of phosphorylated Ser and Asp, respectively explains the observed difference. Since the phosphomimic mutation is structurally remote and cannot directly mediate the interaction between PDZ1 and NPT2A we assume that NHERF1-Ser162Asp exerts a large conformational change that affects the interaction between PDZ1 and NPT2A. There is no high-resolution structure of full-length NHERF1. We speculate that incorporation of Asp162 with the negatively charge carboxylate group interrupts the self-inhibited NHERF1 conformation [], releases the C-terminal tail, and increases conformational dynamics of NHERF1. The structurally disordered C-terminal tail may adopt a variety of conformations and screen the association between PDZ1 and NPT2A. Another possibility is that the C-terminal tail engages PDZ1 and interferes with NPT2A binding. In contrast to NHERF1-Ser162Asp, the double phosphomimic mutation at Ser339Asp/Ser340Asp increases the binding affinity of both PDZ1 and PDZ2 for CFTR through conformational changes in the linker regions [] and long-range allosteric cooperativity in NHERF1 []. Of note, Ser339/Ser340 are PKC phosphorylation sites [,] like Ser162 []. It was suggested that Ser339/Ser340 phosphorylation promotes phosphorylation of Ser162 []. Whether this cooperativity exists in vivo remains to be established.
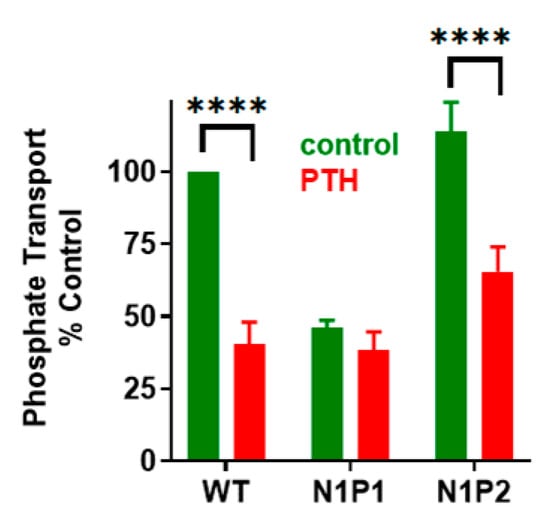
Figure 7.
Effect of NHERF1 PDZ mutants on basal and PTH-sensitive phosphate uptake. OKH cells were transiently transfected with WT-NHERF1, N1P1-GAGA, or N1P2-GAGA. Cells were treated with vehicle or with 100 nM PTH(1-34). N1P2-GAGA-NHERF1 and WT-NHERF1 support PTH-inhibitable phosphate uptake, whereas N1P1-GAGA does not. Results report the mean ± SEM (n = 4, **** p < 0.0001, ANOVA) [].
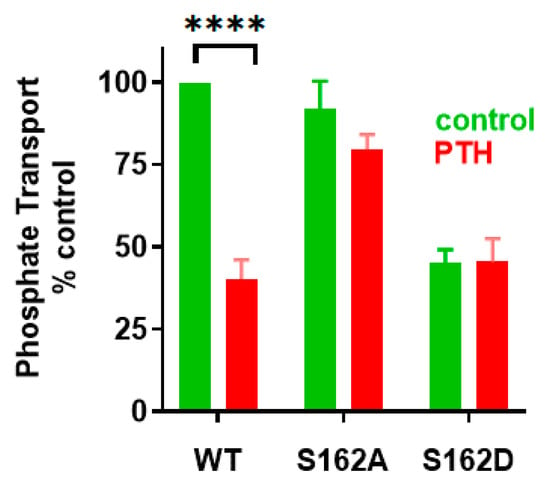
Figure 8.
Ser162 is essential for PTH-inhibitable phosphate uptake. OKH cells were transiently transfected with WT-NHERF1 or with Ser162Ala-NHERF1 or Ser162Asp- NHERF1. Cells were treated with vehicle or with 100 nM PTH(1-34). Results report the mean ± SEM (n = 4, **** p < 0.0001, ANOVA) [].
To explore the biochemical role of phospho-Ser162 (pSer) and its impact on the interaction between NHERF1 PDZ2 and GRK6A, pSer162 was genetically introduced in recombinant PDZ2 (133-300 aa) using amber codon suppression []. Previously, semi-synthesis was effectively applied to generate site-specific phosphorylated PDZ domains []. Here, we used amber codon suppression to genetically encode pSer at position 162 [], which in the future will allow to generate phosphorylated full-length proteins comprising both PDZ domains. Replacement of Ser162 by pSer162 in recombinant PDZ2 permits estimating the binding affinity between pSer162-PDZ2 and the C-terminal PDZ-ligand of GRK6A. Fluorescence anisotropy (FA) was applied to measure dissociation constants (KD’s) between pSer162-PDZ2, Ser162Ala-PDZ2, wild type PDZ2 (133–300 aa) and FITC-labeled GRK6A (22 aa). The results demonstrate that the KD values for the interaction between WT PDZ2 or Ser162Ala-PDZ2 and GRK6A were comparable (51.3 ± 0.4, 43.4 ± 0.4 µM) and double that for pSer162-PDZ2 (26.1 ± 0.8 µM), thus, demonstrating that the binding affinity of PDZ2 is regulated by phosphorylation of Ser162. Site-specific incorporation of pSer162 applied for NHERF1 PDZ2 introduces an analogous but not identical functional group compared to phosphomimetic mutagenesis. The phosphate group has a –2 negative charge compared to the single negative charge of the Asp carboxylate group. The increased size of the sidechain may importantly perturb the local protein conformation []. MD simulations provide structural details associated with incorporation of pSer162 (Figure 9). Three main observations come from the simulation studies. First, pSer162 forms an electrostatic interaction with the positively charged side chain of Arg−1 of the C-terminal TRL motif of GRK6A and therefore has a significant impact on dynamics and conformational flexibility of the C-terminus of GRK6A. Second, pSer162 promotes the formation of the electrostatic network (pSer162- Arg−1-Asp183) wherein the sidechain of Asp183 changes its orientation and moves toward the sidechain of Arg−1. Third, the GYGF/GAGA substitution in the carboxylate-binding site of pSer162-PDZ2 does not impede the interaction with GRK6A. Consequently, NHERF1 with the modified core-binding motif (N1P2-GAGA) supports PTH-sensitive phosphate uptake (Figure 7). Thus, a strong stabilizing influence of pSer162 underscores the limitation and potential hazard of using phosphomimetics to draw conclusions about phosphorylation and demonstrates the strength of introducing site-specific pSer using experimental and computational methods.
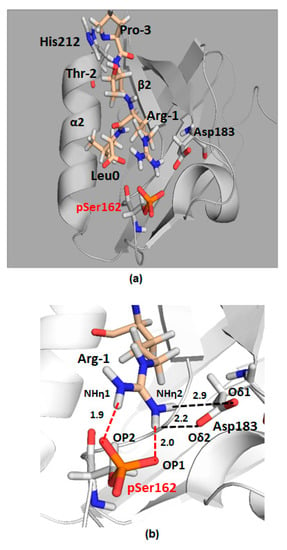
Figure 9.
Computational model of pSer162-PDZ2 bound to the C-terminal peptide of GRK6A. The PDZ2 domain is highlighted in grey and the GRK6A peptide is represented in wheat sticks. (a), The extreme C-terminus of GRK6A (-PTRL) is inserted in the binding pocket of pSer162-PDZ2 between the α2 helix and the β2 sheet. (b), electrostatic interactions between pSer162 or Asp183 of PDZ2 and Arg−1 of GRK6A are shown as red and black dotted lines, respectively; Average distances between OP1 of pSer162 and NHη1 of Arg−1 or between OP2 and NHη2 are 2.0 Å and 1.9 Å, respectively; between NHη1 of Arg−1 and Oδ1 or Oδ2 of Asp183 are 2.9 Å and 2.2 Å, respectively. Distances were calculated along the last 10-ns of MD simulation. Hydrogen atoms are white, oxygens are red, and nitrogens are blue.
4. Conformational Reorganization of NHERF1 Regulates NHERF1-NPT2A Interactions
4.1. Phosphorylation of Ser290 Controls NHERF1 Conformation and Interactions with NPT2A
Recent combined NMR and small-angle neutron scattering (SANS) experiments revealed that full-length NHERF1 cannot be characterized by a single conformation. Rather, NHERF1 represents an ensemble of diverse PDZ configurations connected by flexible linkers with the C-terminal unstructured tail []. Notably, the flexible linker connecting PDZ2 and the C-terminus has 17 Ser/Thr residues. Compared to structurally determined rigid domains, intrinsically disordered regions typically contain a high density of phosphorylation sites []. Site-specific phosphorylation within these regions promotes structurally relevant conformational transitions that affect protein function [,]. Reversible post-translational modification of Ser/Thr residues within this region may regulate NHERF1 activity and signaling. We showed that dephosphorylation-phosphorylation of Ser290 located in this flexible linker regulates the association between NHERF1 PDZ1 and NPT2A. Preventing Ser290 phosphorylation with a phosphoresistant mutation (Ser290Ala) or pharmacologically inhibiting the action of GRK6A kinase decreases the binding of NTP2A to NHERF1 and reduces PTH-sensitive phosphate transport []. Evidently, Ser290 determines regulated stability of the NHERF1-NPT2A complex through long range allosteric communication. Hydrogen deuterium exchange mass spectrometry (HDX-MS) analysis was used to analyze the region in NHERF1 undergoing conformational changes along the dephosphorylation-phosphorylation cycle of Ser290 []. In addition to the linker region between PDZ2 and EBD, substantial conformational changes were found in PDZ1 near Glu43, a critical determinant for NPT2A binding [,]. Thus, dephosphorylation-phosphorylation of Ser290 allosterically regulates the conformation of the side chain of Glu43 and switches the interaction on and off with Arg−1 of the C-terminal -TRL motif of NPT2A and thereby controls PTH-sensitive phosphate uptake.
4.2. NHERF1 Disease-Associated Mutations (Leu110Val, Arg153Gln and Glu225Lys) Affect Conformational Landscape and Interaction with NPT2A
Another example of allosteric regulation within NHERF1 relates to Leu110Val, Glu225Lys and Arg153Gln mutations identified in patients with hypophosphatemia []. The disease-associated mutations decrease or abolish the interaction between PDZ1 and NPT2A and block PTH-sensitive phosphate transport []. It was suggested that Arg153Gln and Glu225Lys, located in PDZ2, stabilize a self-inhibited conformation in which the NHERF1 C-terminus -FS−2NL0, a PDZ recognition motif itself, is engaged in the PDZ2 domain, masking PDZ1 and thereby disrupting the interaction with NPT2A []. Remarkably, when the C-terminal Leu0 of the NHERF1 PDZ ligand, required for engagement with PDZ2, was exchanged for Ala (NHERF1 Leu0Ala) and was paired with the Arg153Gln (NHERF1 Arg153Gln/Leu358Ala) disease mutation, the interaction with NPT2A was rescued and PTH-sensitive phosphate transport was restored and indistinguishable from that of wild-type NHERF1 [], notably in the ongoing presence of the naturally occurring Arg153Gln mutation. Leu110Val, a similar dysfunctional mutant, locates in the α4 helix of the carboxy-terminal helix-turn-helix extension (α3-loop-α4) of PDZ1 []. This subdomain of 30 residues allosterically regulates the binding affinity between NHERF1 PDZ domains and their cellular targets []. NMR studies suggested that an extensive hydrophobic network between PDZ1 and the α3-loop-α4 extension stabilizes the structure with Leu110 serving as a part of this hydrophobic network (64). It was proposed that mutation of Leu110 to Val could potentially disrupt the packing interactions and reduce the stability and affinity of the PDZ1 domain []. Replacement of Leu110, even with a shorter hydrophobic side chain of Val, will likely disturb the hydrophobic network inside the α3-loop-α4 extension and promote its rearrangement. In addition, conformational changes associated with such an Leu110Val mutation may limit phosphorylation of Thr95, a PKC phosphorylation site located near Leu110, and thereby disrupt cooperativity between Thr95 and Ser77 phosphorylation events required for normal NPT2A-dependent PTH-inhibitable phosphate transport []. These conjectures require experimental verification.
5. “Outside the Box” Determinants Are Involved in the Interaction between NHERF1 PDZ Domains and PTHR
PTHR, a Family B GPCR, plays a key role in mineral-ion metabolism and bone physiology [,]. The C-terminal sequence of PTHR represents a typical PDZ binding motif (-T−2-V-M0) with upstream Glu residues at positions -3, -5, and -6 (-E−6-E−5-W-E−3-T−2-V-M0). Although crystallographic or NMR structures for the complex between the PDZ domains of NHERF1 and PTHR have not been solved, molecular determinants of the interaction were characterized by different approaches. Biochemical studies [,] and Molecular Dynamics simulations [] predict that both PDZ domains engage in PDZ-ligand interactions with C-terminal Met0 and Thr−2 of PTHR. The promiscuous -T−2-V-M0 motif permits PTHR to bind PDZ1 or PDZ2, or both through canonical PDZ-ligand interactions established between the conserved GYGF motif of PDZ1/PDZ2 and the His72/His212 residue at the top of the α2 helix. Although these canonical interactions are essential components of the binding, determinants outside the PDZ-ligand pocket enhance formation of the binary [NHERF1-PTHR] complex [,]. MD simulations on a ~70-ns time scale provide clear evidence that Arg40 and His27 or Arg180 of PDZ1 and Asn169 in PDZ2 are involved in forming stable interactions with Glu−3 of PTHR [,]. Fluorescence anisotropy (FA), and isothermal titration calorimetry (ITC) measurements experimentally validated and extended computational predictions by showing that Glu−3 to Ala substitution destabilizes the complex []. This finding corroborates X-ray crystallographic [,] and NMR studies [] that demonstrated the formation of electrostatic interactions between Arg40/Arg180 of PDZ1/PDZ2 and Asp−3 of the C-terminal PDZ-binding motif of CFTR (-DTRL). Similar electrostatic interactions between NHERF1 PDZ1/PDZ2 and Asp−3 from the C-terminal PDZ-binding motif of the β-AR (-ESLL) were confirmed by X-ray crystallography [,]. The determined binding affinities in the nM–µM range for PDZ1 or PDZ2 domains bound the C-terminal peptide ligands of CFTR or PTHR support the formation of favorable electrostatic interactions [,,,].
In addition to the Arg40/Arg180-Glu−3 pair, residues from the β2-β3 loop of PDZ1/PDZ2 and the negatively charged Glu residues at positions –5 and –6 of the PTHR C-terminus are also involved in the interactions []. The β2-β3 loop is flexible. This explains why the impact of the residues from the loop on the binding with the upstream residues of the PTHR (E−6E−5-WETVM) is challenging to estimate by MD simulation []. The affinities determined by a plate-binding assay demonstrate that mutation of Glu585 (Glu−5) to Ala or double substitution of Glu585Ala/Glu586Ala decreases binding to NHERF1 []. Thermodynamic parameters of the binding between NHERF1 PDZ domains and the C-terminal peptide of PTHR (8 aa) with a single or double Ala substitution were evaluated by ITC []. The loss of the binding strength following Glu-to-Ala substitution is attributed to unfavorable changes in enthalpy (ΔHο) and entropy (ΔSο). Mutation of Glu−5 in combination with Glu−6 exerts a synergistic effect on ΔHο. Again, a corresponding change in the free energy of binding (ΔΔGο) is not observed or is only minor due to the enthalpy-entropy compensation and rather define the PDZ-ligand specificity []. The ΔSο values for PDZ1 and PDZ2 domains bound to the ensemble of Ala variants of the C-terminal motif of PTHR were plotted against ΔHο. A strong correlation between ΔSο and ΔHο is well-matched to PDZ1-NPT2A mutant variants (Figure 5). A linear plot for PDZ1-NPT2A and PDZ1/PDZ2-PTHR confirmed that the binding mechanism is similarly conserved.
6. Concluding Remarks
NHERF1 PDZ domain specificity for NPT2A and PTHR is modulated by residues outside the binding pocket and by allosteric long-range communication. Despite significant progress, crystallization or cryo-electron microscopy to solve the structure of NHERF1 PDZ domains with the C-terminal -TRL motif of NPT2A will help distinguish binding determinants and improve our understanding of the origins of NHERF1 and other PDZ protein functional specificity. The recent finding that an internal PDZ region in NPT2A may regulate phosphate transport raises new questions about the structural determinant of this interaction and its impact on NHERF1. The influence of post-translational, site-specific phosphorylation on NHERF1 binding specificity and regulation of NPT2A-mediated phosphate uptake in health and disease remains uncharacterized. A combination of structural analysis, protein engineering and cell biology will be required to address these gaps in our understanding.
Author Contributions
Conceptualization, T.M. and P.A.F.; writing—original draft preparation, T.M. and P.A.F.; writing—review and editing, T.M. and P.A.F.; visualization, T.M.; funding acquisition, P.A.F. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Institutes of Health (NIH), grant number DK105811-01A1.
Acknowledgments
We are especially grateful to Maria Vistrup-Parry, Kristian Strømgaard, Qiangmin Zhang, and W. Bruce Sneddon for their collegial collaboration.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Donowitz, M.; Cha, B.; Zachos, N.C.; Brett, C.L.; Sharma, A.; Tse, C.M.; Li, X. NHERF family and NHE3 regulation. J. Physiol. 2005, 567(Pt. 1), 3–11. [Google Scholar] [CrossRef] [PubMed]
- Ardura, J.A.; Friedman, P.A. Regulation of G protein-coupled receptor function by Na+/H+ exchange regulatory factors. Pharmacol. Rev. 2011, 63, 882–900. [Google Scholar] [CrossRef] [PubMed]
- Romero, G.; von Zastrow, M.; Friedman, P.A. Role of PDZ proteins in regulating trafficking, signaling, and function of GPCRs: Means, motif, and opportunity. Adv. Pharmacol. 2011, 62, 279–314. [Google Scholar] [PubMed]
- Wang, B.; Means, C.K.; Yang, Y.M.; Mamonova, T.; Bisello, A.; Altschuler, D.L.; Scott, J.D.; Friedman, P.A. Ezrin-anchored protein kinase A coordinates phosphorylation-dependent disassembly of a NHERF1 ternary complex to regulate hormone-sensitive phosphate transport. J. Biol. Chem. 2012, 287, 24148–24163. [Google Scholar] [CrossRef]
- Wagner, C.A.; Rubio-Aliaga, I.; Biber, J.; Hernando, N. Genetic diseases of renal phosphate handling. Nephrol. Dial. Transplant. 2014, 29 (Suppl. 4), iv45–iv54. [Google Scholar] [CrossRef]
- Hernando, N. NaPi-IIa interacting partners and their (un)known functional roles. Pflugers Arch. 2019, 471, 67–82. [Google Scholar] [CrossRef]
- Levi, M.; Gratton, E.; Forster, I.C.; Hernando, N.; Wagner, C.A.; Biber, J.; Sorribas, V.; Murer, H. Mechanisms of phosphate transport. Nat. Rev. Nephrol. 2019, 15, 482–500. [Google Scholar] [CrossRef]
- Hernando, N.; Gagnon, K.; Lederer, E. Phosphate Transport in Epithelial and Nonepithelial Tissue. Physiol. Rev. 2021, 101, 1–35. [Google Scholar] [CrossRef]
- Karim, Z.; Gerard, B.; Bakouh, N.; Alili, R.; Leroy, C.; Beck, L.; Silve, C.; Planelles, G.; Urena-Torres, P.; Grandchamp, B.; et al. NHERF1 mutations and responsiveness of renal parathyroid hormone. N. Engl. J. Med. 2008, 359, 1128–1135. [Google Scholar] [CrossRef]
- Gordon, R.J.; Li, D.; Doyle, D.; Zaritsky, J.; Levine, M.A. Digenic heterozygous mutations in SLC34A3 and SLC34A1 cause dominant hypophosphatemic rickets with hypercalciuria. J. Clin. Endocrinol. Metab. 2020, 105, 2392–2400. [Google Scholar] [CrossRef]
- Kang, S.J.; Lee, R.; Kim, H.S. Infantile hypercalcemia with novel compound heterozygous mutation in SLC34A1 encoding renal sodium-phosphate cotransporter 2a: A case report. Ann. Pediatr. Endocrinol. Metab. 2019, 24, 64–67. [Google Scholar] [CrossRef] [PubMed]
- Wagner, C.A.; Rubio-Aliaga, I.; Hernando, N. Renal phosphate handling and inherited disorders of phosphate reabsorption: An update. Pediatr. Nephrol. 2019, 34, 549–559. [Google Scholar] [CrossRef] [PubMed]
- Shenolikar, S.; Voltz, J.W.; Minkoff, C.M.; Wade, J.B.; Weinman, E.J. Targeted disruption of the mouse NHERF-1 gene promotes internalization of proximal tubule sodium-phosphate cotransporter type IIa and renal phosphate wasting. Proc. Natl. Acad. Sci. USA 2002, 99, 11470–11475. [Google Scholar] [CrossRef] [PubMed]
- Morales, F.C.; Takahashi, Y.; Kreimann, E.L.; Georgescu, M.M. Ezrin-radixin-moesin (ERM)-binding phosphoprotein 50 organizes ERM proteins at the apical membrane of polarized epithelia. Proc. Natl. Acad. Sci. USA 2004, 101, 17705–17710. [Google Scholar] [CrossRef]
- Hernando, N.; Deliot, N.; Gisler, S.M.; Lederer, E.; Weinman, E.J.; Biber, J.; Murer, H. PDZ-domain interactions and apical expression of type IIa Na/Pi cotransporters. Proc. Natl. Acad. Sci. USA 2002, 99, 11957–11962. [Google Scholar] [CrossRef]
- Weinman, E.J.; Steplock, D.; Shenolikar, S.; Biswas, R. Fibroblast growth factor-23-mediated inhibition of renal phosphate transport in mice requires sodium-hydrogen exchanger regulatory factor-1 (NHERF-1) and synergizes with parathyroid hormone. J. Biol. Chem. 2011, 286, 37216–37221. [Google Scholar] [CrossRef]
- Mahon, M.J.; Donowitz, M.; Yun, C.C.; Segre, G.V. Na+/H+ exchanger regulatory factor 2 directs parathyroid hormone 1 receptor signalling. Nature 2002, 417, 858–861. [Google Scholar] [CrossRef]
- Mahon, M.J.; Segre, G.V. Stimulation by parathyroid hormone of a NHERF-1-assembled complex consisting of the parathyroid hormone I receptor, phospholipase C beta, and actin increases intracellular calcium in opossum kidney cells. J. Biol. Chem. 2004, 279, 23550–23558. [Google Scholar] [CrossRef]
- Khundmiri, S.J.; Ahmad, A.; Bennett, R.E.; Weinman, E.J.; Steplock, D.; Cole, J.; Baumann, P.D.; Lewis, J.; Singh, S.; Clark, B.J.; et al. Novel regulatory function for NHERF-1 in Npt2a transcription. Am. J. Physiol. Renal Physiol. 2008, 294, F840–F849. [Google Scholar] [CrossRef]
- Reczek, D.; Berryman, M.; Bretscher, A. Identification of EBP50: A PDZ-containing phosphoprotein that associates with members of the ezrin-radixin-moesin family. J. Cell Biol. 1997, 139, 169–179. [Google Scholar] [CrossRef]
- Reczek, D.; Bretscher, A. The carboxyl-terminal region of EBP50 binds to a site in the amino-terminal domain of ezrin that is masked in the dormant molecule. J. Biol. Chem. 1998, 273, 18452–18458. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Bisello, A.; Yang, Y.; Romero, G.G.; Friedman, P.A. NHERF1 regulates parathyroid hormone receptor membrane retention without affecting recycling. J. Biol. Chem. 2007, 282, 36214–36222. [Google Scholar] [CrossRef] [PubMed]
- Karthikeyan, S.; Leung, T.; Ladias, J.A. Structural determinants of the Na+/H+ exchanger regulatory factor interaction with the β2 adrenergic and platelet-derived growth factor receptors. J. Biol. Chem. 2002, 277, 18973–18978. [Google Scholar] [CrossRef] [PubMed]
- Karthikeyan, S.; Leung, T.; Ladias, J.A.A. Structural basis of the Na+/H+ exchanger regulatory factor PDZ1 interaction with the carboxyl-terminal region of the cystic fibrosis transmembrane conductance regulator. J. Biol. Chem. 2001, 276, 19683–19686. [Google Scholar] [CrossRef] [PubMed]
- Lu, G.R.; Wu, Y.N.; Jiang, Y.Y.; Wang, S.; Hou, Y.N.; Guan, X.Q.; Brunzelle, J.; Sirinupong, N.; Sheng, S.J.; Li, C.Y.; et al. Structural insights into neutrophilic migration revealed by the crystal structure of the chemokine receptor CXCR2 in complex with the first PDZ domain of NHERF1. PLoS ONE 2013, 8. [Google Scholar] [CrossRef]
- Holcomb, J.; Jiang, Y.Y.; Guan, X.Q.; Trescott, L.; Lu, G.R.; Hou, Y.N.; Wang, S.; Brunzelle, J.; Sirinupong, N.; Li, C.Y.; et al. Crystal structure of the NHERF1 PDZ2 domain in complex with the chemokine receptor CXCR2 reveals probable modes of PDZ2 dimerization. Biochem. Biophys. Res. Commun. 2014, 448, 169–174. [Google Scholar] [CrossRef]
- Karthikeyan, S.; Leung, T.; Birrane, G.; Webster, G.; Ladias, J.A.A. Crystal structure of the PDZ1 domain of human Na+/H+ exchanger regulatory factor provides insights into the mechanism of carboxyl-terminal leucine recognition by class I PDZ domains. J. Mol. Biol. 2001, 308, 963–973. [Google Scholar] [CrossRef]
- Voltz, J.W.; Brush, M.; Sikes, S.; Steplock, D.; Weinman, E.J.; Shenolikar, S. Phosphorylation of PDZ1 domain attenuates NHERF-1 binding to cellular targets. J. Biol. Chem. 2007, 282, 33879–33887. [Google Scholar] [CrossRef]
- Mamonova, T.; Zhang, Q.; Chandra, M.; Collins, B.M.; Sarfo, E.; Bu, Z.; Xiao, K.; Bisello, A.; Friedman, P.A. Origins of PDZ binding specificity. A computational and experimental study using NHERF1 and the parathyroid hormone receptor. Biochemistry 2017, 56, 2584–2593. [Google Scholar] [CrossRef]
- Ernst, A.; Appleton, B.A.; Ivarsson, Y.; Zhang, Y.; Gfeller, D.; Wiesmann, C.; Sidhu, S.S. A structural portrait of the PDZ domain family. J. Mol. Biol. 2014, 426, 3509–3519. [Google Scholar] [CrossRef]
- McGarvey, J.C.; Xiao, K.; Bowman, S.L.; Mamonova, T.; Zhang, Q.; Bisello, A.; Sneddon, W.B.; Ardura, J.A.; Jean-Alphonse, F.; Vilardaga, J.P.; et al. Actin-Sorting Nexin 27 (SNX27)-Retromer Complex Mediates Rapid Parathyroid Hormone Receptor Recycling. J. Biol. Chem. 2016, 291, 10986–11002. [Google Scholar] [CrossRef] [PubMed]
- Clairfeuille, T.; Mas, C.; Chan, A.S.; Yang, Z.; Tello-Lafoz, M.; Chandra, M.; Widagdo, J.; Kerr, M.C.; Paul, B.; Merida, I.; et al. A molecular code for endosomal recycling of phosphorylated cargos by the SNX27-retromer complex. Nat. Struct. Mol. Biol. 2016, 23, 921–932. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, S.W.; Albertsen, L.; Moran, G.E.; Levesque, B.; Pedersen, S.B.; Bartels, L.; Wapenaar, H.; Ye, F.; Zhang, M.; Bowen, M.E.; et al. Site-specific phosphorylation of PSD-95 PDZ domains reveals fine-tuned regulation of protein-protein interactions. ACS Chem. Biol. 2017, 12, 2313–2323. [Google Scholar] [CrossRef] [PubMed]
- Gogl, G.; Biri-Kovacs, B.; Durbesson, F.; Jane, P.; Nomine, Y.; Kostmann, C.; Bilics, V.; Simon, M.; Remenyi, A.; Vincentelli, R.; et al. Rewiring of RSK-PDZ interactome by linear motif phosphorylation. J. Mol. Biol. 2019, 431, 1234–1249. [Google Scholar] [CrossRef]
- Amacher, J.F.; Brooks, L.; Hampton, T.H.; Madden, D.R. Specificity in PDZ-peptide interaction networks: Computational analysis and review. J. Struct Biol. X 2020, 4, 100022. [Google Scholar] [CrossRef]
- Bhattacharya, S.; Ju, J.H.; Orlova, N.; Khajeh, J.A.; Cowburn, D.; Bu, Z.M. Ligand-induced dynamic changes in extended PDZ domains from NHERF1. J. Mol. Biol. 2013, 425, 2509–2528. [Google Scholar] [CrossRef]
- Wang, C.K.; Pan, L.; Chen, J.; Zhang, M. Extensions of PDZ domains as important structural and functional elements. Protein Cell 2010, 1, 737–751. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, S.; Dai, Z.P.; Li, J.Q.; Baxter, S.; Callaway, D.J.E.; Cowburn, D.; Bu, Z.M. A conformational switch in the scaffolding protein NHERF1 controls autoinhibition and complex formation. J. Biol. Chem. 2010, 285, 9981–9994. [Google Scholar] [CrossRef]
- Morales, F.C.; Takahashi, Y.; Momin, S.; Adams, H.; Chen, X.; Georgescu, M.M. NHERF1/EBP50 head-to-tail intramolecular interaction masks association with PDZ domain ligands. Mol. Cell. Biol. 2007, 27, 2527–2537. [Google Scholar] [CrossRef]
- Shenolikar, S.; Minkoff, C.M.; Steplock, D.A.; Evangelista, C.; Liu, M.Z.; Weinman, E.J. N-terminal PDZ domain is required for NHERF dimerization. FEBS Lett. 2001, 489, 233–236. [Google Scholar] [CrossRef]
- Lau, A.G.; Hall, R.A. Oligomerization of NHERF-1 and NHERF-2 PDZ domains: Differential regulation by association with receptor carboxyl-termini and by phosphorylation. Biochemistry 2001, 40, 8572–8580. [Google Scholar] [CrossRef] [PubMed]
- Fouassier, L.; Nichols, M.T.; Gidey, E.; McWilliams, R.R.; Robin, H.; Finnigan, C.; Howell, K.E.; Housset, C.; Doctor, R.B. Protein kinase C regulates the phosphorylation and oligomerization of ERM binding phosphoprotein 50. Exp. Cell Res. 2005, 306, 264–273. [Google Scholar] [CrossRef]
- Li, J.Q.; Poulikakos, P.I.; Dai, Z.P.; Testa, J.R.; Callaway, D.J.E.; Bu, Z.M. Protein kinase c phosphorylation disrupts Na+/H+ exchanger regulatory factor 1 autoinhibition and promotes cystic fibrosis transmembrane conductance regulator macromolecular assembly. J. Biol. Chem. 2007, 282, 27086–27099. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Xiao, K.; Paredas, J.M.; Mamonova, T.; Sneddon, W.B.; Liu, H.; Wang, D.; Li, S.; McGarvey, J.C.; Uehling, D.; et al. Parathyroid hormone initiates dynamic NHERF1 phosphorylation cycling and conformational changes that regulate NPT2A-dependent phosphate transport. J. Biol. Chem. 2019, 294, 4546–4571. [Google Scholar] [CrossRef] [PubMed]
- Mamonova, T.; Zhang, Q.; Khajeh, J.A.; Bu, Z.; Bisello, A.; Friedman, P.A. Canonical and noncanonical sites determine NPT2A binding selectivity to NHERF1 PDZ1. PLoS ONE 2015, 10, e0129554. [Google Scholar] [CrossRef] [PubMed]
- Hall, R.A.; Spurney, R.F.; Premont, R.T.; Rahman, N.; Blitzer, J.T.; Pitcher, J.A.; Lefkowitz, R.J. G protein-coupled receptor kinase 6A phosphorylates the Na+/H+ exchanger regulatory factor via a PDZ domain-mediated interaction. J. Biol. Chem. 1999, 274, 24328–24334. [Google Scholar] [CrossRef]
- Cardone, R.A.; Bellizzi, A.; Busco, G.; Weinman, E.J.; Dell’aquila, M.E.; Casavola, V.; Azzariti, A.; Mangia, A.; Paradiso, A.; Reshkin, S.J. The NHERF1 PDZ2 domain regulates PKA-RhoA-p38-mediated NHE1 activation and invasion in breast tumor cells. Mol. Biol. Cell 2007, 18, 1768–1780. [Google Scholar] [CrossRef]
- Salyer, S.; Lesousky, N.; Weinman, E.J.; Clark, B.J.; Lederer, E.D.; Khundmiri, S.J. Dopamine regulation of Na+-K+-ATPase requires the PDZ-2 domain of sodium hydrogen regulatory factor-1 (NHERF-1) in opossum kidney cells. Am. J. Physiol. Cell Physiol. 2011, 300, C425–C434. [Google Scholar] [CrossRef]
- Raghuram, V.; Hormuth, H.; Foskett, J.K. A kinase-regulated mechanism controls CFTR channel gating by disrupting bivalent PDZ domain interactions. Proc. Natl. Acad. Sci. USA 2003, 100, 9620–9625. [Google Scholar] [CrossRef]
- Khundmiri, S.J.; Weinman, E.J.; Steplock, D.; Cole, J.; Ahmad, A.; Baumann, P.D.; Barati, M.; Rane, M.J.; Lederer, E. Parathyroid hormone regulation of NA+,K+-ATPase requires the PDZ 1 domain of sodium hydrogen exchanger regulatory factor-1 in opossum kidney cells. J. Am. Soc. Nephrol. 2005, 16, 2598–2607. [Google Scholar] [CrossRef]
- Weinman, E.J.; Steplock, D.; Shenolikar, S.; Blanpied, T.A. Dynamics of PTH-induced disassembly of Npt2a/NHERF-1 complexes in living OK cells. Am. J. Physiol. Renal Physiol. 2011, 300, F231–F235. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pouyssegur, J.; Sardet, C.; Franchi, A.; L’Allemain, G.; Paris, S. A specific mutation abolishing Na+/H+ antiport activity in hamster fibroblasts precludes growth at neutral and acidic pH. Proc. Natl. Acad. Sci. USA 1984, 81, 4833–4837. [Google Scholar] [CrossRef] [PubMed]
- Staudinger, J.; Lu, J.; Olson, E.N. Specific interaction of the PDZ domain protein PICK1 with the COOH terminus of protein kinase C-alpha. J. Biol. Chem. 1997, 272, 32019–32024. [Google Scholar] [CrossRef]
- Mahon, M.J. The parathyroid hormone 1 receptor directly binds to the FERM domain of ezrin, an interaction that supports apical receptor localization and signaling in LLC-PK1 cells. Mol. Endocrinol. 2009, 23, 1691–1701. [Google Scholar] [CrossRef]
- Cunningham, R.; XE, D.; Steplock, D.; Shenolikar, S.; Weinman, E.J. Defective PTH regulation of sodium-dependent phosphate transport in NHERF-1−/− renal proximal tubule cells and wild-type cells adapted to low-phosphate media. Am. J. Physiol. Renal Physiol. 2005, 289, F933–F9338. [Google Scholar] [CrossRef]
- Alshafie, W.; Chappe, F.G.; Li, M.; Anini, Y.; Chappe, V.M. VIP regulates CFTR membrane expression and function in Calu-3 cells by increasing its interaction with NHERF1 and P-ERM in a VPAC1- and PKCepsilon-dependent manner. Am. J. Physiol. Cell Physiol. 2014, 307, C107–C119. [Google Scholar] [CrossRef]
- Raghuram, V.; Mak, D.O.; Foskett, J.K. Regulation of cystic fibrosis transmembrane conductance regulator single-channel gating by bivalent PDZ-domain-mediated interaction. Proc. Natl. Acad. Sci. USA 2001, 98, 1300–1305. [Google Scholar] [CrossRef]
- Li, J.Q.; Dai, Z.P.; Jana, D.; Callaway, D.J.E.; Bu, Z.M. Ezrin controls the macromolecular complexes formed between an adapter protein Na+/H+ exchanger regulatory factor and the cystic fibrosis transmembrane conductance regulator. J. Biol. Chem. 2005, 280, 37634–37643. [Google Scholar] [CrossRef]
- Lee, J.H.; Richter, W.; Namkung, W.; Kim, K.H.; Kim, E.; Conti, M.; Lee, M.G. Dynamic regulation of cystic fibrosis transmembrane conductance regulator by competitive interactions of molecular adaptors. J. Biol. Chem. 2007, 282, 10414–10422. [Google Scholar] [CrossRef]
- Martin, E.R.; Barbieri, A.; Ford, R.C.; Robinson, R.C. In vivo crystals reveal critical features of the interaction between cystic fibrosis transmembrane conductance regulator (CFTR) and the PDZ2 domain of Na(+)/H(+) exchange cofactor NHERF1. J. Biol. Chem. 2020, 295, 4464–4476. [Google Scholar] [CrossRef]
- Rajagopal, A.; Braslavsky, D.; Lu, J.T.; Kleppe, S.; Clement, F.; Cassinelli, H.; Liu, D.S.; Liern, J.M.; Vallejo, G.; Bergada, I.; et al. Exome sequencing identifies a novel homozygous mutation in the phosphate transporter SLC34A1 in hypophosphatemia and nephrocalcinosis. J. Clin. Endocrinol. Metab. 2014, 99, E2451–E2456. [Google Scholar] [CrossRef] [PubMed]
- Mamonova, T.; Kurnikova, M.; Friedman, P.A. Structural basis for NHERF1 PDZ domain binding. Biochemistry 2012, 51, 3110–3120. [Google Scholar] [CrossRef] [PubMed]
- Vistrup-Parry, M.; Sneddon, W.B.; Bach, S.; Strømgaard, K.; Friedman, P.A.; Mamonova, T. Multisite NHERF1 phosphorylation controls GRK6A regulation of hormone-sensitive phosphate transport. J. Biol. Chem. under review.
- Rogerson, D.T.; Sachdeva, A.; Wang, K.; Haq, T.; Kazlauskaite, A.; Hancock, S.M.; Huguenin-Dezot, N.; Muqit, M.M.; Fry, A.M.; Bayliss, R.; et al. Efficient genetic encoding of phosphoserine and its nonhydrolyzable analog. Nat. Chem. Biol. 2015, 11, 496–503. [Google Scholar] [CrossRef]
- Toto, A.; Mattei, A.; Jemth, P.; Gianni, S. Understanding the role of phosphorylation in the binding mechanism of a PDZ domain. Protein Eng. Des. Sel. 2017, 30, 1–5. [Google Scholar] [CrossRef]
- Bhattacharya, S.; Stanley, C.B.; Heller, W.T.; Friedman, P.A.; Bu, Z. Dynamic structure of the full-length scaffolding protein NHERF1 influences signaling complex assembly. J. Biol. Chem. 2019, 294, 11297–11310. [Google Scholar] [CrossRef]
- Iakoucheva, L.M.; Radivojac, P.; Brown, C.J.; O’Connor, T.R.; Sikes, J.G.; Obradovic, Z.; Dunker, A.K. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 2004, 32, 1037–1049. [Google Scholar] [CrossRef]
- Darling, A.L.; Uversky, V.N. Intrinsic Disorder and Posttranslational Modifications: The Darker Side of the Biological Dark Matter. Front. Genet. 2018, 9, 158. [Google Scholar] [CrossRef]
- Bhattacherjee, A.; Wallin, S. Exploring protein-peptide binding specificity through computational peptide screening. PLoS Comput. Biol. 2013, 9, e1003277. [Google Scholar] [CrossRef]
- Weinman, E.J.; Steplock, D.; Zhang, Y.; Biswas, R.; Bloch, R.J.; Shenolikar, S. Cooperativity between the phosphorylation of Thr95 and Ser77 of NHERF-1 in the hormonal regulation of renal phosphate transport. J. Biol. Chem. 2010, 285, 25134–25138. [Google Scholar] [CrossRef]
- Sun, C.; Mierke, D.F. Characterization of interactions of Na+/H+ exchanger regulatory factor-1 with the parathyroid hormone receptor and phospholipase C. J. Pept. Res. 2005, 65, 411–417. [Google Scholar] [CrossRef] [PubMed]
- Cushing, P.R.; Fellows, A.; Villone, D.; Boisguerin, P.; Madden, D.R. The relative binding affinities of PDZ partners for CFTR: A biochemical basis for efficient endocytic recycling. Biochemistry 2008, 47, 10084–10098. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).