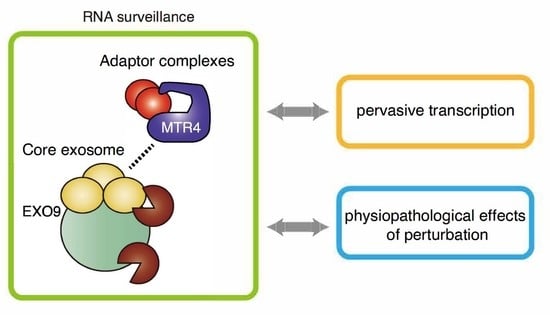
Nuclear RNA Exosome and Pervasive Transcription: Dual Sculptors of Genome Function
Abstract
1. Introduction
2. Molecular Mechanisms of RNA Surveillance by RNA Exosome
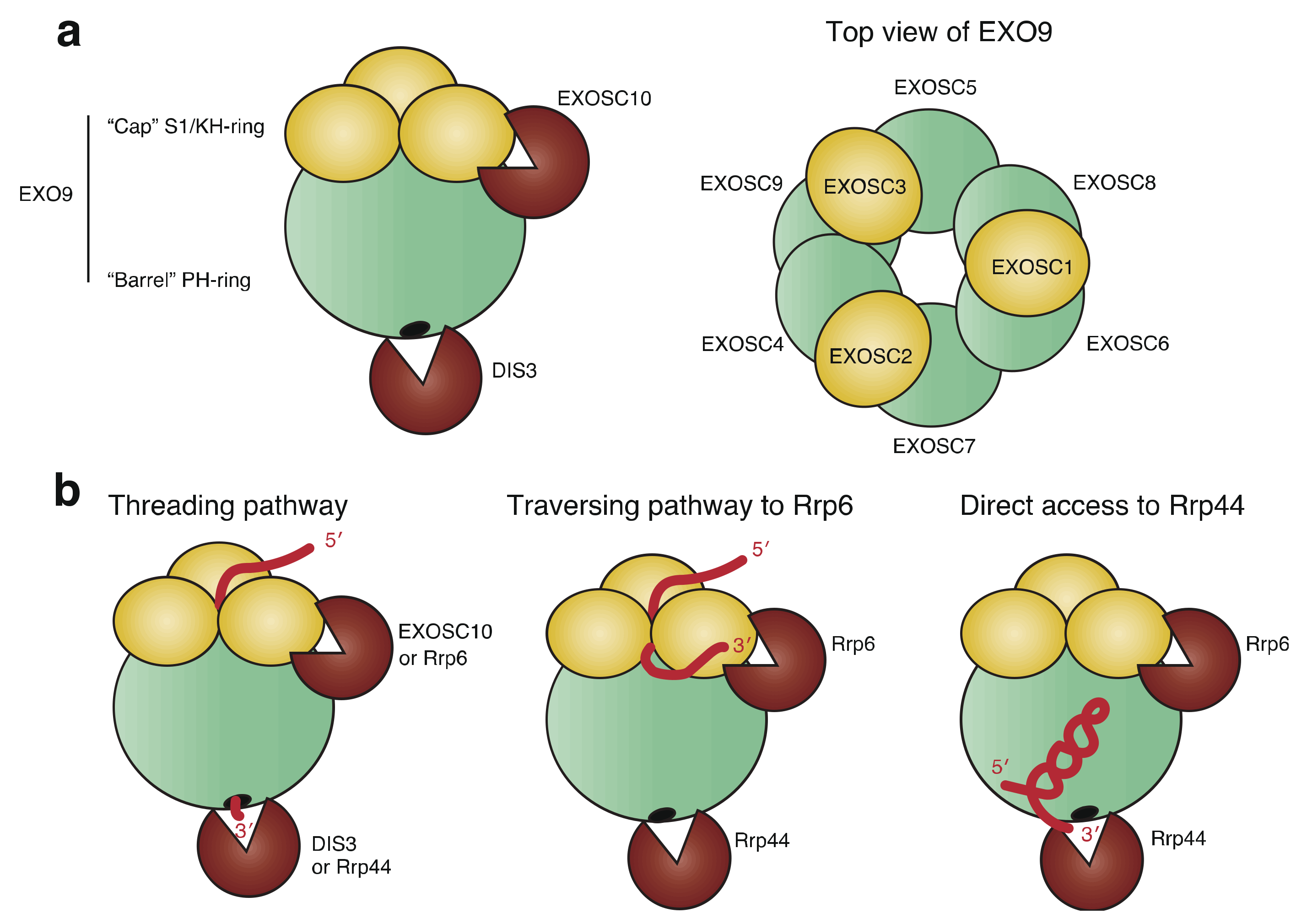
2.1. Structure of RNA Exosome
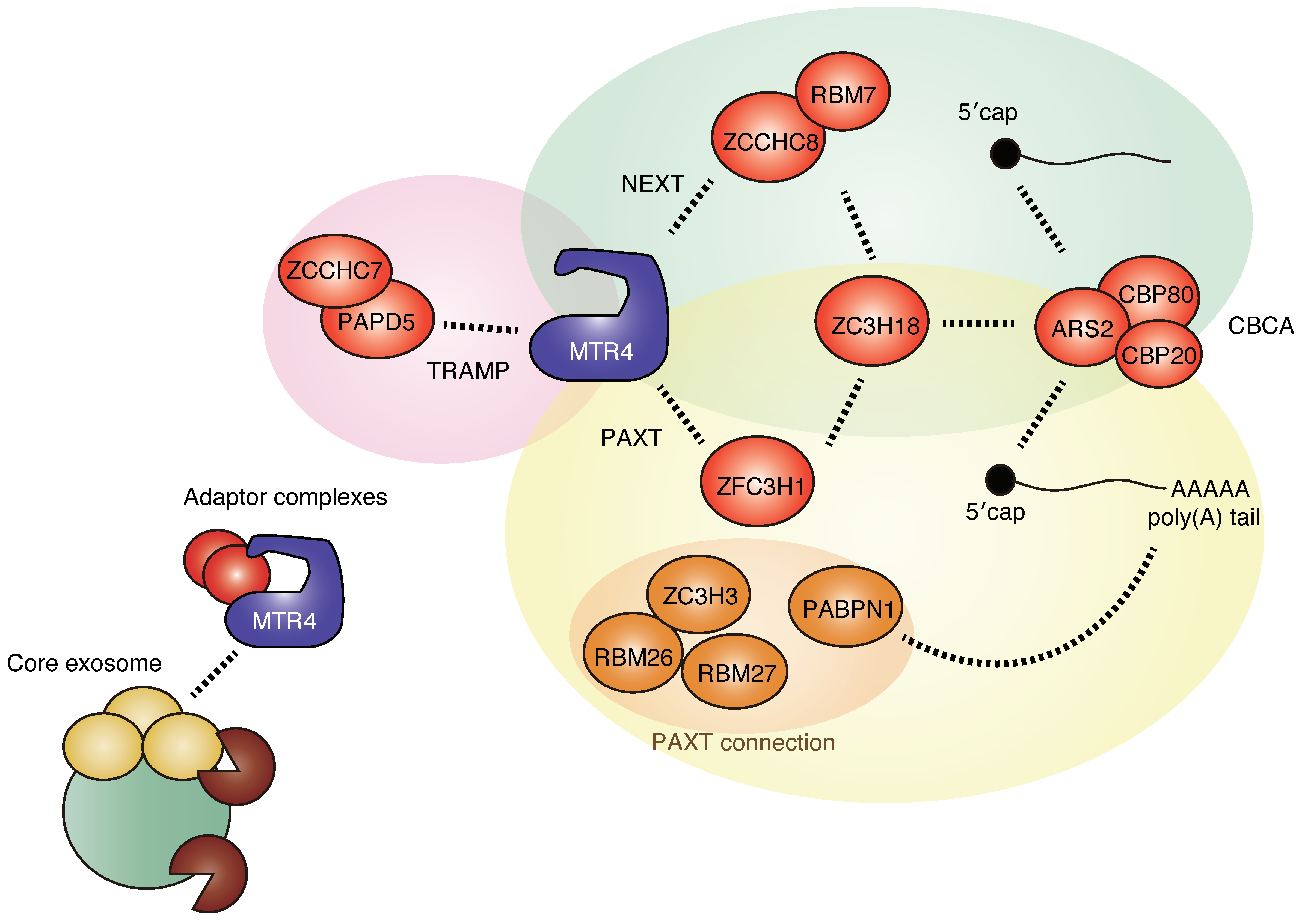
2.2. Sorting of Target RNAs by Adaptors and Cofactors of RNA Exosome
2.3. Balance between RNA Export and Degradation
3. Evolutionary Interplays between RNA Exosome and Pervasive Transcription
4. Biological Significance of Human Nuclear RNA Exosome
4.1. Regulation of Genome Stability, Chromatin Structure, and DNA Damage Response
4.2. Gene Regulation and Differentiation
5. Pathological Significance of Human Nuclear RNA Exosome
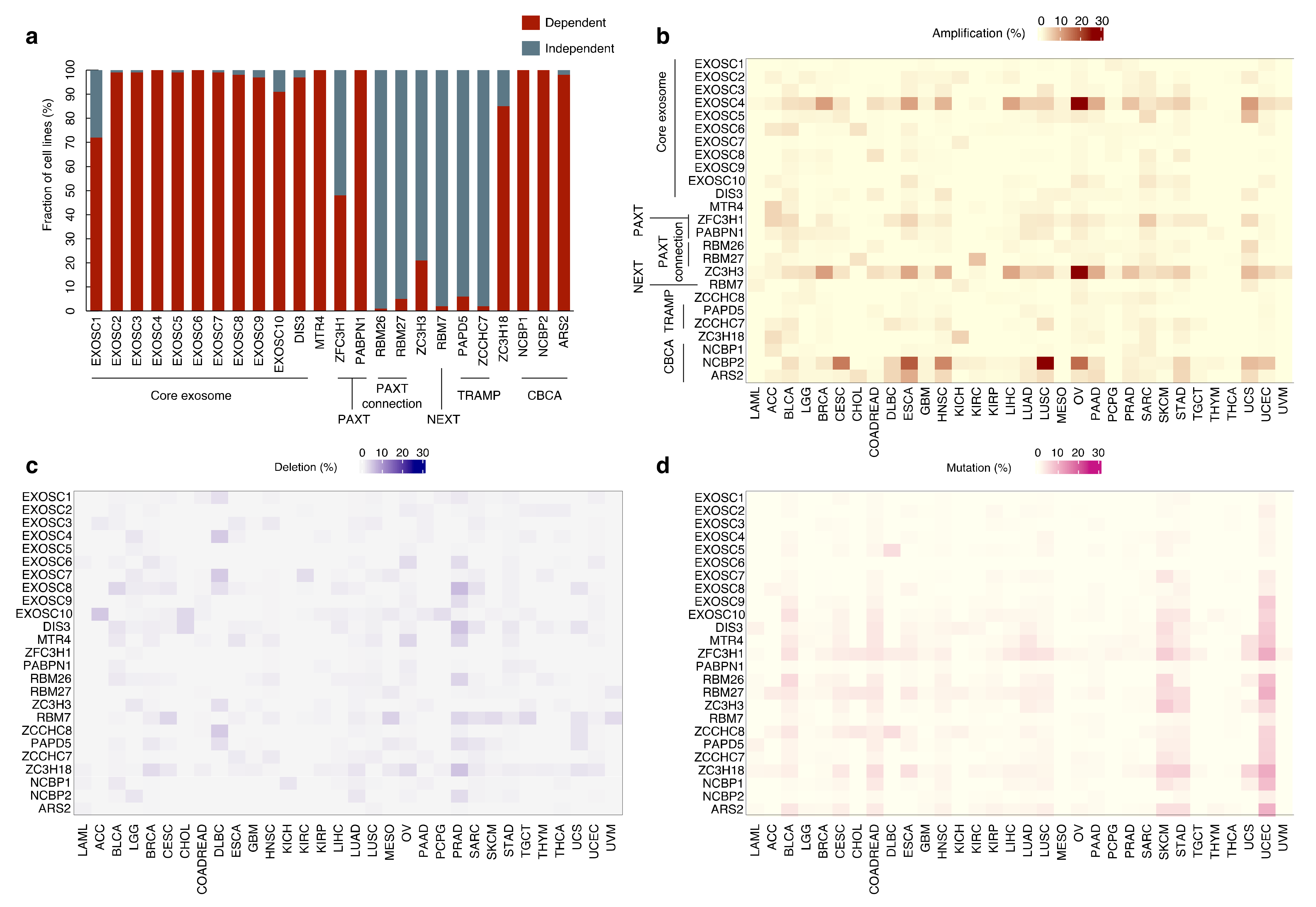
5.1. Mutations in RNA Exosome Components and Rare Diseases
5.2. RNA Exosome and Cancer
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Haberle, V.; Stark, A. Eukaryotic core promoters and the functional basis of transcription initiation. Nat. Rev. Mol. Cell Biol. 2018, 19, 621–637. [Google Scholar] [CrossRef]
- Cramer, P. Organization and regulation of gene transcription. Nature 2019, 573, 45–54. [Google Scholar] [CrossRef]
- Ogami, K.; Chen, Y.; Manley, J.L. RNA surveillance by the nuclear RNA exosome: Mechanisms and significance. Non-Coding RNA 2018, 4, 8. [Google Scholar] [CrossRef] [PubMed]
- Schmid, M.; Jensen, T.H. Controlling nuclear RNA levels. Nat. Rev. Genet. 2018, 19, 518–529. [Google Scholar] [CrossRef]
- Garland, W.; Jensen, T.H. Nuclear sorting of RNA. Wiley Interdiscip. Rev. RNA 2020, 11, e1572. [Google Scholar] [CrossRef]
- Januszyk, K.; Lima, C.D. The eukaryotic RNA exosome. Curr. Opin. Struct. Biol. 2014, 24, 132–140. [Google Scholar] [CrossRef] [PubMed]
- Zinder, J.C.; Lima, C.D. Targeting RNA for processing or destruction by the eukaryotic RNA exosome and its cofactors. Genes Dev. 2017, 31, 88–100. [Google Scholar] [CrossRef]
- Makino, D.L.; Conti, E. Structure determination of an 11-subunit exosome in complex with RNA by molecular replacement. Acta Crystallogr. Sect. D Biol. Crystallogr. 2013, 69, 2226–2235. [Google Scholar] [CrossRef] [PubMed]
- Wasmuth, E.V.; Januszyk, K.; Lima, C.D. Structure of an Rrp6-RNA exosome complex bound to poly(A) RNA. Nature 2014, 511, 435–439. [Google Scholar] [CrossRef] [PubMed]
- Makino, D.L.; Schuch, B.; Stegmann, E.; Baumgärtner, M.; Basquin, C.; Conti, E. RNA degradation paths in a 12-subunit nuclear exosome complex. Nature 2015, 524, 54–58. [Google Scholar] [CrossRef]
- Bonneau, F.; Basquin, J.; Ebert, J.; Lorentzen, E.; Conti, E. The yeast exosome functions as a macromolecular cage to channel RNA substrates for degradation. Cell 2009, 139, 547–559. [Google Scholar] [CrossRef]
- Liu, J.J.; Bratkowski, M.A.; Liu, X.; Niu, C.Y.; Ke, A.; Wang, H.W. Visualization of distinct substrate-recruitment pathways in the yeast exosome by EM. Nat. Struct. Mol. Biol. 2014, 21, 95–102. [Google Scholar] [CrossRef]
- Han, J.; van Hoof, A. The RNA Exosome Channeling and Direct Access Conformations Have Distinct In Vivo Functions. Cell Rep. 2016, 16, 3348–3358. [Google Scholar] [CrossRef] [PubMed]
- Zinder, J.C.; Wasmuth, E.V.; Lima, C.D. Nuclear RNA Exosome at 3.1 Å Reveals Substrate Specificities, RNA Paths, and Allosteric Inhibition of Rrp44/Dis3. Mol. Cell 2016, 64, 734–745. [Google Scholar] [CrossRef]
- Gerlach, P.; Schuller, J.M.; Bonneau, F.; Basquin, J.; Reichelt, P.; Falk, S.; Conti, E. Distinct and evolutionary conserved structural features of the human nuclear exosome complex. eLife 2018, 7, e38686. [Google Scholar] [CrossRef] [PubMed]
- Weick, E.M.; Puno, M.R.; Januszyk, K.; Zinder, J.C.; DiMattia, M.A.; Lima, C.D. Helicase-Dependent RNA Decay Illuminated by a Cryo-EM Structure of a Human Nuclear RNA Exosome-MTR4 Complex. Cell 2018, 173, 1663–1677.e1621. [Google Scholar] [CrossRef] [PubMed]
- Dai, L.; Zhao, T.; Bisteau, X.; Sun, W.; Prabhu, N.; Lim, Y.T.; Sobota, R.M.; Kaldis, P.; Nordlund, P. Modulation of Protein-Interaction States through the Cell Cycle. Cell 2018, 173, 1481–1494.e1413. [Google Scholar] [CrossRef]
- Fox, M.J.; Mosley, A.L. Rrp6: Integrated roles in nuclear RNA metabolism and transcription termination. Wiley Interdiscip. Rev. RNA 2016, 7, 91–104. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Xiao, Q.; Sun, Z.; Wang, B.; Wang, L.; Wang, N.; Wang, K.; Song, C.; Yang, Q. Exosome component 1 cleaves single-stranded DNA and sensitizes human kidney renal clear cell carcinoma cells to poly(ADP-ribose) polymerase inhibitor. eLife 2021, 10, e69454. [Google Scholar] [CrossRef]
- Lubas, M.; Christensen, M.S.; Kristiansen, M.S.; Domanski, M.; Falkenby, L.G.; Lykke-Andersen, S.; Andersen, J.S.; Dziembowski, A.; Jensen, T.H. Interaction profiling identifies the human nuclear exosome targeting complex. Mol. Cell 2011, 43, 624–637. [Google Scholar] [CrossRef]
- Wu, Y.; Liu, W.; Chen, J.; Liu, S.; Wang, M.; Yang, L.; Chen, C.; Qi, M.; Xu, Y.; Qiao, Z.; et al. Nuclear Exosome Targeting Complex Core Factor Zcchc8 Regulates the Degradation of LINE1 RNA in Early Embryos and Embryonic Stem Cells. Cell Rep. 2019, 29, 2461–2472. [Google Scholar] [CrossRef]
- Lubas, M.; Andersen, P.R.; Schein, A.; Dziembowski, A.; Kudla, G.; Jensen, T.H. The human nuclear exosome targeting complex is loaded onto newly synthesized RNA to direct early ribonucleolysis. Cell Rep. 2015, 10, 178–192. [Google Scholar] [CrossRef] [PubMed]
- Meola, N.; Domanski, M.; Karadoulama, E.; Chen, Y.; Gentil, C.; Pultz, D.; Vitting-Seerup, K.; Lykke-Andersen, S.; Andersen, J.S.; Sandelin, A.; et al. Identification of a Nuclear Exosome Decay Pathway for Processed Transcripts. Mol. Cell 2016, 64, 520–533. [Google Scholar] [CrossRef] [PubMed]
- Silla, T.; Schmid, M.; Dou, Y.; Garland, W.; Milek, M.; Imami, K.; Johnsen, D.; Polak, P.; Andersen, J.S.; Selbach, M.; et al. The human ZC3H3 and RBM26/27 proteins are critical for PAXT-mediated nuclear RNA decay. Nucleic Acids Res. 2020, 48, 2518–2530. [Google Scholar] [CrossRef]
- Ogami, K.; Richard, P.; Chen, Y.; Hoque, M.; Li, W.; Moresco, J.J.; Yates, J.R., 3rd; Tian, B.; Manley, J.L. An Mtr4/ZFC3H1 complex facilitates turnover of unstable nuclear RNAs to prevent their cytoplasmic transport and global translational repression. Genes Dev. 2017, 31, 1257–1271. [Google Scholar] [CrossRef]
- Andersen, P.R.; Domanski, M.; Kristiansen, M.S.; Storvall, H.; Ntini, E.; Verheggen, C.; Schein, A.; Bunkenborg, J.; Poser, I.; Hallais, M.; et al. The human cap-binding complex is functionally connected to the nuclear RNA exosome. Nat. Struct. Mol. Biol. 2013, 20, 1367–1376. [Google Scholar] [CrossRef]
- Winczura, K.; Schmid, M.; Iasillo, C.; Molloy, K.R.; Harder, L.M.; Andersen, J.S.; LaCava, J.; Jensen, T.H. Characterizing ZC3H18, a Multi-domain Protein at the Interface of RNA Production and Destruction Decisions. Cell Rep. 2018, 22, 44–58. [Google Scholar] [CrossRef]
- Wu, G.; Schmid, M.; Rib, L.; Polak, P.; Meola, N.; Sandelin, A.; Jensen, T.H. A Two-Layered Targeting Mechanism Underlies Nuclear RNA Sorting by the Human Exosome. Cell Rep. 2020, 30, 2387–2401. [Google Scholar] [CrossRef]
- Rambout, X.; Maquat, L.E. The nuclear cap-binding complex as choreographer of gene transcription and pre-mRNA processing. Genes Dev. 2020, 34, 1113–1127. [Google Scholar] [CrossRef] [PubMed]
- Giacometti, S.; Benbahouche, N.E.H.; Domanski, M.; Robert, M.C.; Meola, N.; Lubas, M.; Bukenborg, J.; Andersen, J.S.; Schulze, W.M.; Verheggen, C.; et al. Mutually Exclusive CBC-Containing Complexes Contribute to RNA Fate. Cell Rep. 2017, 18, 2635–2650. [Google Scholar] [CrossRef] [PubMed]
- Marzluff, W.F.; Koreski, K.P. Birth and Death of Histone mRNAs. Trends Genet. TIG 2017, 33, 745–759. [Google Scholar] [CrossRef] [PubMed]
- Iasillo, C.; Schmid, M.; Yahia, Y.; Maqbool, M.A.; Descostes, N.; Karadoulama, E.; Bertrand, E.; Andrau, J.C.; Jensen, T.H. ARS2 is a general suppressor of pervasive transcription. Nucleic Acids Res. 2017, 45, 10229–10241. [Google Scholar] [CrossRef] [PubMed]
- Ogami, K.; Manley, J.L. Mtr4/ZFC3H1 protects polysomes through nuclear RNA surveillance. Cell Cycle 2017, 16, 1999–2000. [Google Scholar] [CrossRef]
- Fan, J.; Kuai, B.; Wu, G.; Wu, X.; Chi, B.; Wang, L.; Wang, K.; Shi, Z.; Zhang, H.; Chen, S.; et al. Exosome cofactor hMTR4 competes with export adaptor ALYREF to ensure balanced nuclear RNA pools for degradation and export. EMBO J. 2017, 36, 2870–2886. [Google Scholar] [CrossRef] [PubMed]
- Silla, T.; Karadoulama, E.; Mąkosa, D.; Lubas, M.; Jensen, T.H. The RNA Exosome Adaptor ZFC3H1 Functionally Competes with Nuclear Export Activity to Retain Target Transcripts. Cell Rep. 2018, 23, 2199–2210. [Google Scholar] [CrossRef]
- Wang, Y.; Fan, J.; Wang, J.; Zhu, Y.; Xu, L.; Tong, D.; Cheng, H. ZFC3H1 prevents RNA trafficking into nuclear speckles through condensation. Nucleic Acids Res. 2021, 49, 10630–10643. [Google Scholar] [CrossRef] [PubMed]
- Richard, P.; Ogami, K.; Chen, Y.; Feng, S.; Moresco, J.J.; Yates, J.R., 3rd; Manley, J.L. NRDE-2, the human homolog of fission yeast Nrl1, prevents DNA damage accumulation in human cells. RNA Biol. 2018, 15, 868–876. [Google Scholar] [CrossRef]
- Wang, J.; Chen, J.; Wu, G.; Zhang, H.; Du, X.; Chen, S.; Zhang, L.; Wang, K.; Fan, J.; Gao, S.; et al. NRDE2 negatively regulates exosome functions by inhibiting MTR4 recruitment and exosome interaction. Genes Dev. 2019, 33, 536–549. [Google Scholar] [CrossRef] [PubMed]
- Palazzo, A.F.; Koonin, E.V. Functional Long Non-coding RNAs Evolve from Junk Transcripts. Cell 2020, 183, 1151–1161. [Google Scholar] [CrossRef] [PubMed]
- Ponting, C.P. Biological function in the twilight zone of sequence conservation. BMC Biol. 2017, 15, 71. [Google Scholar] [CrossRef] [PubMed]
- Seila, A.C.; Calabrese, J.M.; Levine, S.S.; Yeo, G.W.; Rahl, P.B.; Flynn, R.A.; Young, R.A.; Sharp, P.A. Divergent transcription from active promoters. Science 2008, 322, 1849–1851. [Google Scholar] [CrossRef]
- Iglesias, N.; Moazed, D. Silencing repetitive DNA. eLife 2017, 6, e29503. [Google Scholar] [CrossRef]
- Lukeš, J.; Archibald, J.M.; Keeling, P.J.; Doolittle, W.F.; Gray, M.W. How a neutral evolutionary ratchet can build cellular complexity. IUBMB Life 2011, 63, 528–537. [Google Scholar] [CrossRef]
- Gould, S.J.; Lewontin, R.C. The spandrels of San Marco and the Panglossian paradigm: A critique of the adaptationist programme. Proc. R. Soc. Lond. Ser. B Biol. Sci. 1979, 205, 581–598. [Google Scholar] [CrossRef]
- Chen, S.; Krinsky, B.H.; Long, M. New genes as drivers of phenotypic evolution. Nat. Rev. Genet. 2013, 14, 645–660. [Google Scholar] [CrossRef]
- Long, M.; VanKuren, N.W.; Chen, S.; Vibranovski, M.D. New gene evolution: Little did we know. Annu. Rev. Genet. 2013, 47, 307–333. [Google Scholar] [CrossRef] [PubMed]
- Lai, F.; Orom, U.A.; Cesaroni, M.; Beringer, M.; Taatjes, D.J.; Blobel, G.A.; Shiekhattar, R. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature 2013, 494, 497–501. [Google Scholar] [CrossRef]
- Sigova, A.A.; Abraham, B.J.; Ji, X.; Molinie, B.; Hannett, N.M.; Guo, Y.E.; Jangi, M.; Giallourakis, C.C.; Sharp, P.A.; Young, R.A. Transcription factor trapping by RNA in gene regulatory elements. Science 2015, 350, 978–981. [Google Scholar] [CrossRef]
- Engreitz, J.M.; Haines, J.E.; Perez, E.M.; Munson, G.; Chen, J.; Kane, M.; McDonel, P.E.; Guttman, M.; Lander, E.S. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 2016, 539, 452–455. [Google Scholar] [CrossRef] [PubMed]
- Kaida, D.; Berg, M.G.; Younis, I.; Kasim, M.; Singh, L.N.; Wan, L.; Dreyfuss, G. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature 2010, 468, 664–668. [Google Scholar] [CrossRef] [PubMed]
- Almada, A.E.; Wu, X.; Kriz, A.J.; Burge, C.B.; Sharp, P.A. Promoter directionality is controlled by U1 snRNP and polyadenylation signals. Nature 2013, 499, 360–363. [Google Scholar] [CrossRef]
- Ntini, E.; Järvelin, A.I.; Bornholdt, J.; Chen, Y.; Boyd, M.; Jørgensen, M.; Andersson, R.; Hoof, I.; Schein, A.; Andersen, P.R.; et al. Polyadenylation site-induced decay of upstream transcripts enforces promoter directionality. Nat. Struct. Mol. Biol. 2013, 20, 923–928. [Google Scholar] [CrossRef]
- Chiu, A.C.; Suzuki, H.I.; Wu, X.; Mahat, D.B.; Kriz, A.J.; Sharp, P.A. Transcriptional Pause Sites Delineate Stable Nucleosome-Associated Premature Polyadenylation Suppressed by U1 snRNP. Mol. Cell 2018, 69, 648–663.e647. [Google Scholar] [CrossRef]
- Yin, Y.; Lu, J.Y.; Zhang, X.; Shao, W.; Xu, Y.; Li, P.; Hong, Y.; Cui, L.; Shan, G.; Tian, B.; et al. U1 snRNP regulates chromatin retention of noncoding RNAs. Nature 2020, 580, 147–150. [Google Scholar] [CrossRef]
- Kilchert, C.; Wittmann, S.; Vasiljeva, L. The regulation and functions of the nuclear RNA exosome complex. Nat. Reviews. Mol. Cell Biol. 2016, 17, 227–239. [Google Scholar] [CrossRef]
- Basu, U.; Meng, F.L.; Keim, C.; Grinstein, V.; Pefanis, E.; Eccleston, J.; Zhang, T.; Myers, D.; Wasserman, C.R.; Wesemann, D.R.; et al. The RNA exosome targets the AID cytidine deaminase to both strands of transcribed duplex DNA substrates. Cell 2011, 144, 353–363. [Google Scholar] [CrossRef] [PubMed]
- Pefanis, E.; Wang, J.; Rothschild, G.; Lim, J.; Chao, J.; Rabadan, R.; Economides, A.N.; Basu, U. Noncoding RNA transcription targets AID to divergently transcribed loci in B cells. Nature 2014, 514, 389–393. [Google Scholar] [CrossRef] [PubMed]
- Pefanis, E.; Wang, J.; Rothschild, G.; Lim, J.; Kazadi, D.; Sun, J.; Federation, A.; Chao, J.; Elliott, O.; Liu, Z.P.; et al. RNA exosome-regulated long non-coding RNA transcription controls super-enhancer activity. Cell 2015, 161, 774–789. [Google Scholar] [CrossRef]
- Lim, J.; Giri, P.K.; Kazadi, D.; Laffleur, B.; Zhang, W.; Grinstein, V.; Pefanis, E.; Brown, L.M.; Ladewig, E.; Martin, O.; et al. Nuclear Proximity of Mtr4 to RNA Exosome Restricts DNA Mutational Asymmetry. Cell 2017, 169, 523–537. [Google Scholar] [CrossRef]
- Rothschild, G.; Zhang, W.; Lim, J.; Giri, P.K.; Laffleur, B.; Chen, Y.; Fang, M.; Chen, Y.; Nair, L.; Liu, Z.P.; et al. Noncoding RNA transcription alters chromosomal topology to promote isotype-specific class switch recombination. Sci. Immunol. 2020, 5. [Google Scholar] [CrossRef] [PubMed]
- Laffleur, B.; Lim, J.; Zhang, W.; Chen, Y.; Pefanis, E.; Bizarro, J.; Batista, C.R.; Wu, L.; Economides, A.N.; Wang, J.; et al. Noncoding RNA processing by DIS3 regulates chromosomal architecture and somatic hypermutation in B cells. Nat. Genet. 2021, 53, 230–242. [Google Scholar] [CrossRef]
- Richard, P.; Feng, S.; Manley, J.L. A SUMO-dependent interaction between Senataxin and the exosome, disrupted in the neurodegenerative disease AOA2, targets the exosome to sites of transcription-induced DNA damage. Genes Dev. 2013, 27, 2227–2232. [Google Scholar] [CrossRef] [PubMed]
- Kazadi, D.; Lim, J.; Rothschild, G.; Grinstein, V.; Laffleur, B.; Becherel, O.; Lavin, M.J.; Basu, U. Effects of senataxin and RNA exosome on B-cell chromosomal integrity. Heliyon 2020, 6, e03442. [Google Scholar] [CrossRef] [PubMed]
- Domingo-Prim, J.; Endara-Coll, M.; Bonath, F.; Jimeno, S.; Prados-Carvajal, R.; Friedländer, M.R.; Huertas, P.; Visa, N. EXOSC10 is required for RPA assembly and controlled DNA end resection at DNA double-strand breaks. Nat. Commun. 2019, 10, 2135. [Google Scholar] [CrossRef]
- Singh, I.; Contreras, A.; Cordero, J.; Rubio, K.; Dobersch, S.; Günther, S.; Jeratsch, S.; Mehta, A.; Krüger, M.; Graumann, J.; et al. MiCEE is a ncRNA-protein complex that mediates epigenetic silencing and nucleolar organization. Nat. Genet. 2018, 50, 990–1001. [Google Scholar] [CrossRef] [PubMed]
- Garland, W.; Comet, I.; Wu, M.; Radzisheuskaya, A.; Rib, L.; Vitting-Seerup, K.; Lloret-Llinares, M.; Sandelin, A.; Helin, K.; Jensen, T.H. A Functional Link between Nuclear RNA Decay and Transcriptional Control Mediated by the Polycomb Repressive Complex 2. Cell Rep. 2019, 29, 1800–1811.e1806. [Google Scholar] [CrossRef]
- Kawabe, Y.; Mori, K.; Yamashita, T.; Gotoh, S.; Ikeda, M. The RNA exosome complex degrades expanded hexanucleotide repeat RNA in C9orf72 FTLD/ALS. EMBO J. 2020, 39, e102700. [Google Scholar] [CrossRef] [PubMed]
- Bergeron, D.; Pal, G.; Beaulieu, Y.B.; Chabot, B.; Bachand, F. Regulated Intron Retention and Nuclear Pre-mRNA Decay Contribute to PABPN1 Autoregulation. Mol. Cell. Biol. 2015, 35, 2503–2517. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Pedersen, J.S.; Kwon, S.C.; Belair, C.D.; Kim, Y.K.; Yeom, K.H.; Yang, W.Y.; Haussler, D.; Blelloch, R.; Kim, V.N. Posttranscriptional crossregulation between Drosha and DGCR8. Cell 2009, 136, 75–84. [Google Scholar] [CrossRef]
- Fraga de Andrade, I.; Mehta, C.; Bresnick, E.H. Post-transcriptional control of cellular differentiation by the RNA exosome complex. Nucleic Acids Res. 2020, 48, 11913–11928. [Google Scholar] [CrossRef] [PubMed]
- Mistry, D.S.; Chen, Y.; Sen, G.L. Progenitor function in self-renewing human epidermis is maintained by the exosome. Cell Stem Cell 2012, 11, 127–135. [Google Scholar] [CrossRef] [PubMed]
- McIver, S.C.; Kang, Y.A.; DeVilbiss, A.W.; O’Driscoll, C.A.; Ouellette, J.N.; Pope, N.J.; Camprecios, G.; Chang, C.J.; Yang, D.; Bouhassira, E.E.; et al. The exosome complex establishes a barricade to erythroid maturation. Blood 2014, 124, 2285–2297. [Google Scholar] [CrossRef]
- McIver, S.C.; Katsumura, K.R.; Davids, E.; Liu, P.; Kang, Y.A.; Yang, D.; Bresnick, E.H. Exosome complex orchestrates developmental signaling to balance proliferation and differentiation during erythropoiesis. eLife 2016, 5, e17877. [Google Scholar] [CrossRef] [PubMed]
- Lloret-Llinares, M.; Karadoulama, E.; Chen, Y.; Wojenski, L.A.; Villafano, G.J.; Bornholdt, J.; Andersson, R.; Core, L.; Sandelin, A.; Jensen, T.H. The RNA exosome contributes to gene expression regulation during stem cell differentiation. Nucleic Acids Res. 2018, 46, 11502–11513. [Google Scholar] [CrossRef] [PubMed]
- Belair, C.; Sim, S.; Kim, K.Y.; Tanaka, Y.; Park, I.H.; Wolin, S.L. The RNA exosome nuclease complex regulates human embryonic stem cell differentiation. J. Cell Biol. 2019, 218, 2564–2582. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Dou, X.; Chen, C.; Chen, C.; Liu, C.; Xu, M.M.; Zhao, S.; Shen, B.; Gao, Y.; Han, D.; et al. N (6)-methyladenosine of chromosome-associated regulatory RNA regulates chromatin state and transcription. Science 2020, 367, 580–586. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, Y.; Toth, J.I.; Petroski, M.D.; Zhang, Z.; Zhao, J.C. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat. Cell Biol. 2014, 16, 191–198. [Google Scholar] [CrossRef]
- Batista, P.J.; Molinie, B.; Wang, J.; Qu, K.; Zhang, J.; Li, L.; Bouley, D.M.; Lujan, E.; Haddad, B.; Daneshvar, K.; et al. m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell 2014, 15, 707–719. [Google Scholar] [CrossRef] [PubMed]
- Geula, S.; Moshitch-Moshkovitz, S.; Dominissini, D.; Mansour, A.A.; Kol, N.; Salmon-Divon, M.; Hershkovitz, V.; Peer, E.; Mor, N.; Manor, Y.S.; et al. Stem cells. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science 2015, 347, 1002–1006. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Hao, Y.J.; Zhang, Y.; Li, M.M.; Wang, M.; Han, W.; Wu, Y.; Lv, Y.; Hao, J.; Wang, L.; et al. m(6)A RNA methylation is regulated by microRNAs and promotes reprogramming to pluripotency. Cell Stem Cell 2015, 16, 289–301. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Xie, W.; Pickering, B.F.; Chu, K.L.; Savino, A.M.; Yang, X.; Luo, H.; Nguyen, D.T.; Mo, S.; Barin, E.; et al. N(6)-Methyladenosine on mRNA facilitates a phase-separated nuclear body that suppresses myeloid leukemic differentiation. Cancer Cell 2021, 39, 958–972. [Google Scholar] [CrossRef] [PubMed]
- Mullani, N.; Porozhan, Y.; Mangelinck, A.; Rachez, C.; Costallat, M.; Batsché, E.; Goodhardt, M.; Cenci, G.; Mann, C.; Muchardt, C. Reduced RNA turnover as a driver of cellular senescence. Life Sci. Alliance 2021, 4. [Google Scholar] [CrossRef] [PubMed]
- De Amorim, J.; Slavotinek, A.; Fasken, M.B.; Corbett, A.H.; Morton, D.J. Modeling Pathogenic Variants in the RNA Exosome. RNA Dis. 2020, 7, e1166. [Google Scholar]
- Morton, D.J.; Kuiper, E.G.; Jones, S.K.; Leung, S.W.; Corbett, A.H.; Fasken, M.B. The RNA exosome and RNA exosome-linked disease. RNA 2018, 24, 127–142. [Google Scholar] [CrossRef] [PubMed]
- Fasken, M.B.; Morton, D.J.; Kuiper, E.G.; Jones, S.K.; Leung, S.W.; Corbett, A.H. The RNA Exosome and Human Disease. Methods Mol. Biol. 2020, 2062, 3–33. [Google Scholar] [CrossRef]
- Wan, J.; Yourshaw, M.; Mamsa, H.; Rudnik-Schöneborn, S.; Menezes, M.P.; Hong, J.E.; Leong, D.W.; Senderek, J.; Salman, M.S.; Chitayat, D.; et al. Mutations in the RNA exosome component gene EXOSC3 cause pontocerebellar hypoplasia and spinal motor neuron degeneration. Nat. Genet. 2012, 44, 704–708. [Google Scholar] [CrossRef] [PubMed]
- Boczonadi, V.; Müller, J.S.; Pyle, A.; Munkley, J.; Dor, T.; Quartararo, J.; Ferrero, I.; Karcagi, V.; Giunta, M.; Polvikoski, T.; et al. EXOSC8 mutations alter mRNA metabolism and cause hypomyelination with spinal muscular atrophy and cerebellar hypoplasia. Nat. Commun. 2014, 5, 4287. [Google Scholar] [CrossRef]
- Burns, D.T.; Donkervoort, S.; Müller, J.S.; Knierim, E.; Bharucha-Goebel, D.; Faqeih, E.A.; Bell, S.K.; AlFaifi, A.Y.; Monies, D.; Millan, F.; et al. Variants in EXOSC9 Disrupt the RNA Exosome and Result in Cerebellar Atrophy with Spinal Motor Neuronopathy. Am. J. Hum. Genet. 2018, 102, 858–873. [Google Scholar] [CrossRef] [PubMed]
- Slavotinek, A.; Misceo, D.; Htun, S.; Mathisen, L.; Frengen, E.; Foreman, M.; Hurtig, J.E.; Enyenihi, L.; Sterrett, M.C.; Leung, S.W.; et al. Biallelic variants in the RNA exosome gene EXOSC5 are associated with developmental delays, short stature, cerebellar hypoplasia and motor weakness. Hum. Mol. Genet. 2020, 29, 2218–2239. [Google Scholar] [CrossRef]
- Somashekar, P.H.; Kaur, P.; Stephen, J.; Guleria, V.S.; Kadavigere, R.; Girisha, K.M.; Bielas, S.; Upadhyai, P.; Shukla, A. Bi-allelic missense variant, p.Ser35Leu in EXOSC1 is associated with pontocerebellar hypoplasia. Clin. Genet. 2021, 99, 594–600. [Google Scholar] [CrossRef] [PubMed]
- Van Dijk, T.; Baas, F.; Barth, P.G.; Poll-The, B.T. What’s new in pontocerebellar hypoplasia? An update on genes and subtypes. Orphanet J. Rare Dis. 2018, 13, 92. [Google Scholar] [CrossRef] [PubMed]
- Di Donato, N.; Neuhann, T.; Kahlert, A.K.; Klink, B.; Hackmann, K.; Neuhann, I.; Novotna, B.; Schallner, J.; Krause, C.; Glass, I.A.; et al. Mutations in EXOSC2 are associated with a novel syndrome characterised by retinitis pigmentosa, progressive hearing loss, premature ageing, short stature, mild intellectual disability and distinctive gestalt. J. Med. Genet. 2016, 53, 419–425. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Georgiou, M.; Atkinson, R.; Collin, J.; Al-Aama, J.; Nagaraja-Grellscheid, S.; Johnson, C.; Ali, R.; Armstrong, L.; Mozaffari-Jovin, S.; et al. Pre-mRNA Processing Factors and Retinitis Pigmentosa: RNA Splicing and Beyond. Front. Cell Dev. Biol. 2021, 9, 700276. [Google Scholar] [CrossRef]
- Brais, B.; Bouchard, J.P.; Xie, Y.G.; Rochefort, D.L.; Chrétien, N.; Tomé, F.M.; Lafrenière, R.G.; Rommens, J.M.; Uyama, E.; Nohira, O.; et al. Short GCG expansions in the PABP2 gene cause oculopharyngeal muscular dystrophy. Nat. Genet. 1998, 18, 164–167. [Google Scholar] [CrossRef] [PubMed]
- Giunta, M.; Edvardson, S.; Xu, Y.; Schuelke, M.; Gomez-Duran, A.; Boczonadi, V.; Elpeleg, O.; Müller, J.S.; Horvath, R. Altered RNA metabolism due to a homozygous RBM7 mutation in a patient with spinal motor neuropathy. Hum. Mol. Genet. 2016, 25, 2985–2996. [Google Scholar] [CrossRef] [PubMed]
- Gable, D.L.; Gaysinskaya, V.; Atik, C.C.; Talbot, C.C., Jr.; Kang, B.; Stanley, S.E.; Pugh, E.W.; Amat-Codina, N.; Schenk, K.M.; Arcasoy, M.O.; et al. ZCCHC8, the nuclear exosome targeting component, is mutated in familial pulmonary fibrosis and is required for telomerase RNA maturation. Genes Dev. 2019, 33, 1381–1396. [Google Scholar] [CrossRef]
- Fukushima, K.; Satoh, T.; Sugihara, F.; Sato, Y.; Okamoto, T.; Mitsui, Y.; Yoshio, S.; Li, S.; Nojima, S.; Motooka, D.; et al. Dysregulated Expression of the Nuclear Exosome Targeting Complex Component Rbm7 in Nonhematopoietic Cells Licenses the Development of Fibrosis. Immunity 2020, 52, 542–556. [Google Scholar] [CrossRef] [PubMed]
- Fabre, A.; Charroux, B.; Martinez-Vinson, C.; Roquelaure, B.; Odul, E.; Sayar, E.; Smith, H.; Colomb, V.; Andre, N.; Hugot, J.P.; et al. SKIV2L mutations cause syndromic diarrhea, or trichohepatoenteric syndrome. Am. J. Hum. Genet. 2012, 90, 689–692. [Google Scholar] [CrossRef] [PubMed]
- Hartley, J.L.; Zachos, N.C.; Dawood, B.; Donowitz, M.; Forman, J.; Pollitt, R.J.; Morgan, N.V.; Tee, L.; Gissen, P.; Kahr, W.H.; et al. Mutations in TTC37 cause trichohepatoenteric syndrome (phenotypic diarrhea of infancy). Gastroenterology 2010, 138, 2388–2398. [Google Scholar] [CrossRef]
- Meyers, R.M.; Bryan, J.G.; McFarland, J.M.; Weir, B.A.; Sizemore, A.E.; Xu, H.; Dharia, N.V.; Montgomery, P.G.; Cowley, G.S.; Pantel, S.; et al. Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat. Genet. 2017, 49, 1779–1784. [Google Scholar] [CrossRef] [PubMed]
- Dempster, J.M.; Boyle, I.; Vazquez, F.; Root, D.; Boehm, J.S.; Hahn, W.C.; Tsherniak, A.; McFarland, J.M. Chronos: A CRISPR cell population dynamics model. bioRxiv 2021. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef]
- Kang, J.U. Chromosome 8q as the most frequent target for amplification in early gastric carcinoma. Oncol. Lett. 2014, 7, 1139–1143. [Google Scholar] [CrossRef] [PubMed]
- Neel, B.G.; Jhanwar, S.C.; Chaganti, R.S.; Hayward, W.S. Two human c-onc genes are located on the long arm of chromosome 8. Proc. Natl. Acad. Sci. USA 1982, 79, 7842–7846. [Google Scholar] [CrossRef] [PubMed]
- Baykara, O.; Bakir, B.; Buyru, N.; Kaynak, K.; Dalay, N. Amplification of chromosome 8 genes in lung cancer. J. Cancer 2015, 6, 270–275. [Google Scholar] [CrossRef] [PubMed]
- Chapman, M.A.; Lawrence, M.S.; Keats, J.J.; Cibulskis, K.; Sougnez, C.; Schinzel, A.C.; Harview, C.L.; Brunet, J.P.; Ahmann, G.J.; Adli, M.; et al. Initial genome sequencing and analysis of multiple myeloma. Nature 2011, 471, 467–472. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Ley, T.J.; Larson, D.E.; Miller, C.A.; Koboldt, D.C.; Welch, J.S.; Ritchey, J.K.; Young, M.A.; Lamprecht, T.; McLellan, M.D.; et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature 2012, 481, 506–510. [Google Scholar] [CrossRef]
- Lionetti, M.; Barbieri, M.; Todoerti, K.; Agnelli, L.; Fabris, S.; Tonon, G.; Segalla, S.; Cifola, I.; Pinatel, E.; Tassone, P.; et al. A compendium of DIS3 mutations and associated transcriptional signatures in plasma cell dyscrasias. Oncotarget 2015, 6, 26129–26141. [Google Scholar] [CrossRef][Green Version]
- Pertesi, M.; Vallée, M.; Wei, X.; Revuelta, M.V.; Galia, P.; Demangel, D.; Oliver, J.; Foll, M.; Chen, S.; Perrial, E.; et al. Exome sequencing identifies germline variants in DIS3 in familial multiple myeloma. Leukemia 2019, 33, 2324–2330. [Google Scholar] [CrossRef] [PubMed]
- Milbury, K.L.; Paul, B.; Lari, A.; Fowler, C.; Montpetit, B.; Stirling, P.C. Exonuclease domain mutants of yeast DIS3 display genome instability. Nucleus 2019, 10, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Cui, K.; Liu, C.; Li, X.; Zhang, Q.; Li, Y. Comprehensive characterization of the rRNA metabolism-related genes in human cancer. Oncogene 2020, 39, 786–800. [Google Scholar] [CrossRef]
- Pan, Y.; Tong, J.H.M.; Kang, W.; Lung, R.W.M.; Chak, W.P.; Chung, L.Y.; Wu, F.; Li, H.; Yu, J.; Chan, A.W.H.; et al. EXOSC4 functions as a potential oncogene in development and progression of colorectal cancer. Mol. Carcinog. 2018, 57, 1780–1791. [Google Scholar] [CrossRef] [PubMed]
- Pan, H.; Pan, J.; Song, S.; Ji, L.; Lv, H.; Yang, Z. EXOSC5 as a Novel Prognostic Marker Promotes Proliferation of Colorectal Cancer via Activating the ERK and AKT Pathways. Front. Oncol. 2019, 9, 643. [Google Scholar] [CrossRef]
- Zhang, W.; Zhu, J.; He, X.; Liu, X.; Li, J.; Li, W.; Yang, P.; Wang, J.; Hu, K.; Zhang, X.; et al. Exosome complex genes mediate RNA degradation and predict survival in mantle cell lymphoma. Oncol. Lett. 2019, 18, 5119–5128. [Google Scholar] [CrossRef] [PubMed]
- Kp, M.; Kumar, A.; Biswas, D.; Moiyadi, A.; Shetty, P.; Gupta, T.; Epari, S.; Shirsat, N.; Srivastava, S. The proteomic analysis shows enrichment of RNA surveillance pathways in adult SHH and extensive metabolic reprogramming in Group 3 medulloblastomas. Brain Tumor Pathol. 2021, 38, 96–108. [Google Scholar] [CrossRef] [PubMed]
- Yoshino, S.; Matsui, Y.; Fukui, Y.; Seki, M.; Yamaguchi, K.; Kanamori, A.; Saitoh, Y.; Shimamura, T.; Suzuki, Y.; Furukawa, Y.; et al. EXOSC9 depletion attenuates P-body formation, stress resistance, and tumorigenicity of cancer cells. Sci. Rep. 2020, 10, 9275. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Kim, J.; Jiang, L.; Feng, B.; Ying, Y.; Ji, K.Y.; Tang, Q.; Chen, W.; Mai, T.; Dou, W.; et al. MTR4 drives liver tumorigenesis by promoting cancer metabolic switch through alternative splicing. Nat. Commun. 2020, 11, 708. [Google Scholar] [CrossRef] [PubMed]
- Xi, P.W.; Zhang, X.; Zhu, L.; Dai, X.Y.; Cheng, L.; Hu, Y.; Shi, L.; Wei, J.F.; Ding, Q. Oncogenic action of the exosome cofactor RBM7 by stabilization of CDK1 mRNA in breast cancer. NPJ Breast Cancer 2020, 6, 58. [Google Scholar] [CrossRef]
- Fish, L.; Navickas, A.; Culbertson, B.; Xu, Y.; Nguyen, H.C.B.; Zhang, S.; Hochman, M.; Okimoto, R.; Dill, B.D.; Molina, H.; et al. Nuclear TARBP2 Drives Oncogenic Dysregulation of RNA Splicing and Decay. Mol. Cell 2019, 75, 967–981.e969. [Google Scholar] [CrossRef] [PubMed]
- Banani, S.F.; Lee, H.O.; Hyman, A.A.; Rosen, M.K. Biomolecular condensates: Organizers of cellular biochemistry. Nat. Reviews. Mol. Cell Biol. 2017, 18, 285–298. [Google Scholar] [CrossRef] [PubMed]
- Sharp, P.A.; Chakraborty, A.K.; Henninger, J.E.; Young, R.A. RNA in formation and regulation of transcriptional condensates. RNA 2021, rna-078997. [Google Scholar] [CrossRef] [PubMed]
- Gao, A.; Shrinivas, K.; Lepeudry, P.; Suzuki, H.I.; Sharp, P.A.; Chakraborty, A.K. Evolution of weak cooperative interactions for biological specificity. Proc. Natl. Acad. Sci. USA 2018, 115, E11053–E11060. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ogami, K.; Suzuki, H.I. Nuclear RNA Exosome and Pervasive Transcription: Dual Sculptors of Genome Function. Int. J. Mol. Sci. 2021, 22, 13401. https://doi.org/10.3390/ijms222413401
Ogami K, Suzuki HI. Nuclear RNA Exosome and Pervasive Transcription: Dual Sculptors of Genome Function. International Journal of Molecular Sciences. 2021; 22(24):13401. https://doi.org/10.3390/ijms222413401
Chicago/Turabian StyleOgami, Koichi, and Hiroshi I. Suzuki. 2021. "Nuclear RNA Exosome and Pervasive Transcription: Dual Sculptors of Genome Function" International Journal of Molecular Sciences 22, no. 24: 13401. https://doi.org/10.3390/ijms222413401
APA StyleOgami, K., & Suzuki, H. I. (2021). Nuclear RNA Exosome and Pervasive Transcription: Dual Sculptors of Genome Function. International Journal of Molecular Sciences, 22(24), 13401. https://doi.org/10.3390/ijms222413401