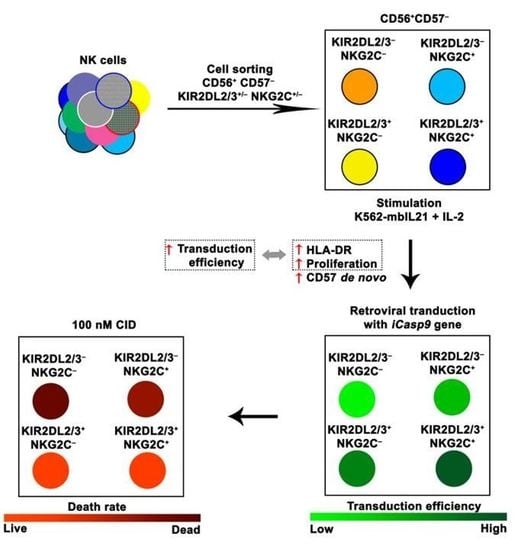
Increased Susceptibility of the CD57− NK Cells Expressing KIR2DL2/3 and NKG2C to iCasp9 Gene Retroviral Transduction and the Relationships with Proliferative Potential, Activation Degree, and Death Induction Response
Abstract
1. Introduction
2. Results
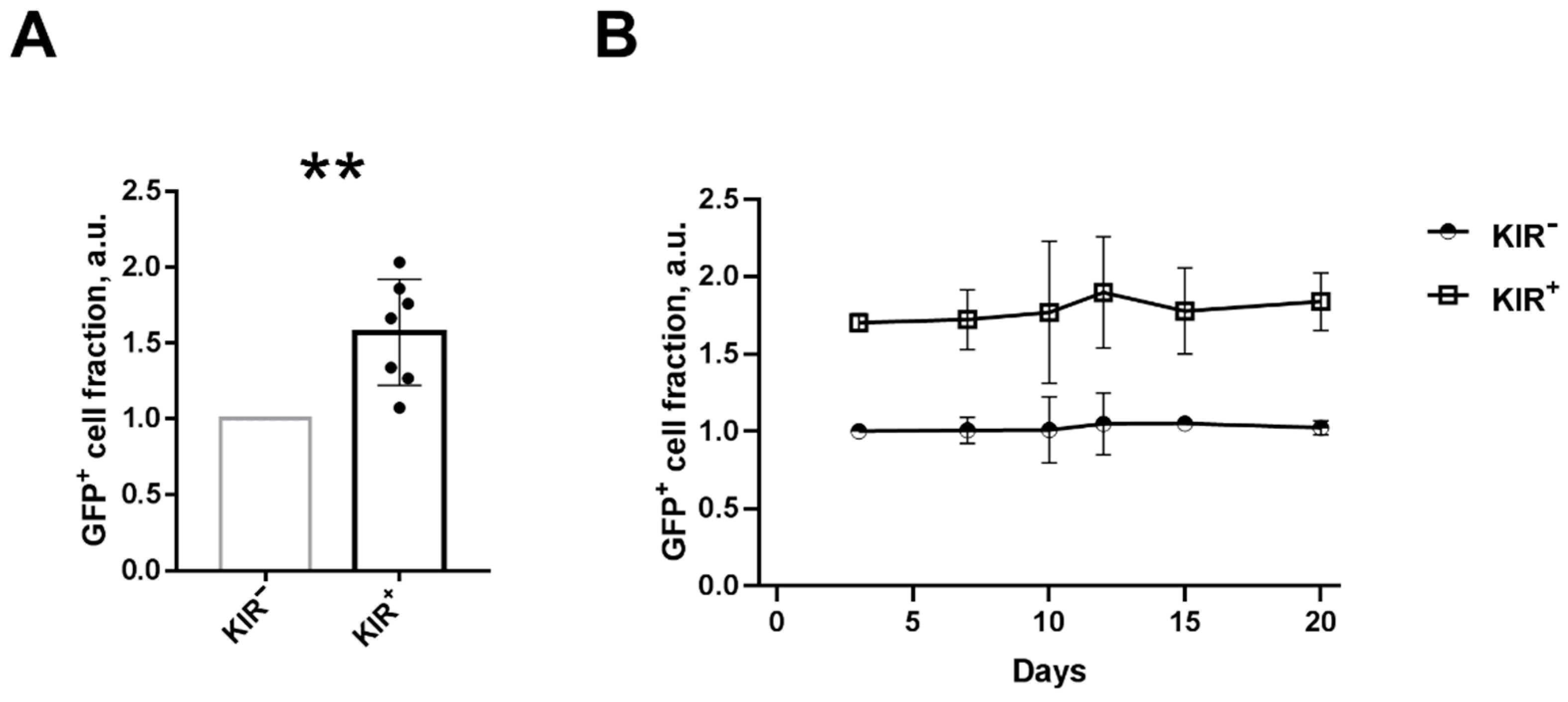
2.1. NK Cells with Surface KIR2DL2/3 Expression Show Greater Retroviral Transduction Efficiency Compared to KIR2DL2/3-Negative NK Cells
2.2. Frequencies of NK Cells Expressing KIR2DL2/3 and/or NKG2C Receptors Differ in CD56bright and CD56dimCD57− NK Cell Subsets
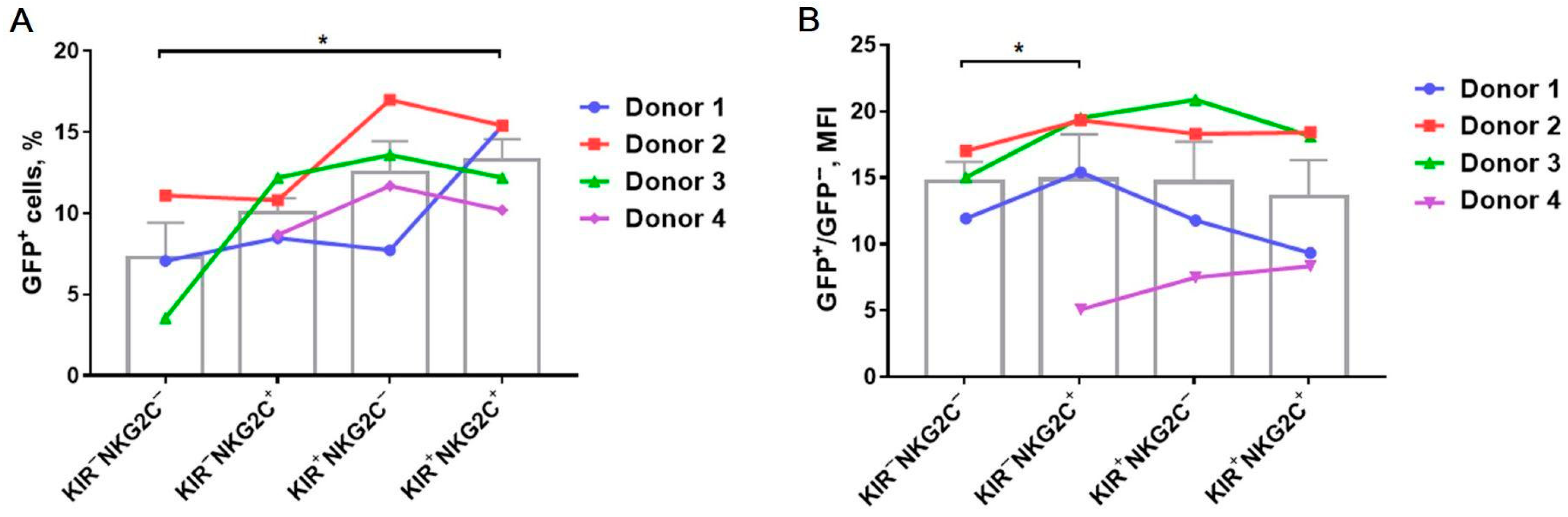
2.3. Transduction Efficiency Depends on KIR2DL2/3 and NKG2C Expression with the Higher Transduction Rate among the KIR2DL2/3+ and NKG2C+ NK Cells
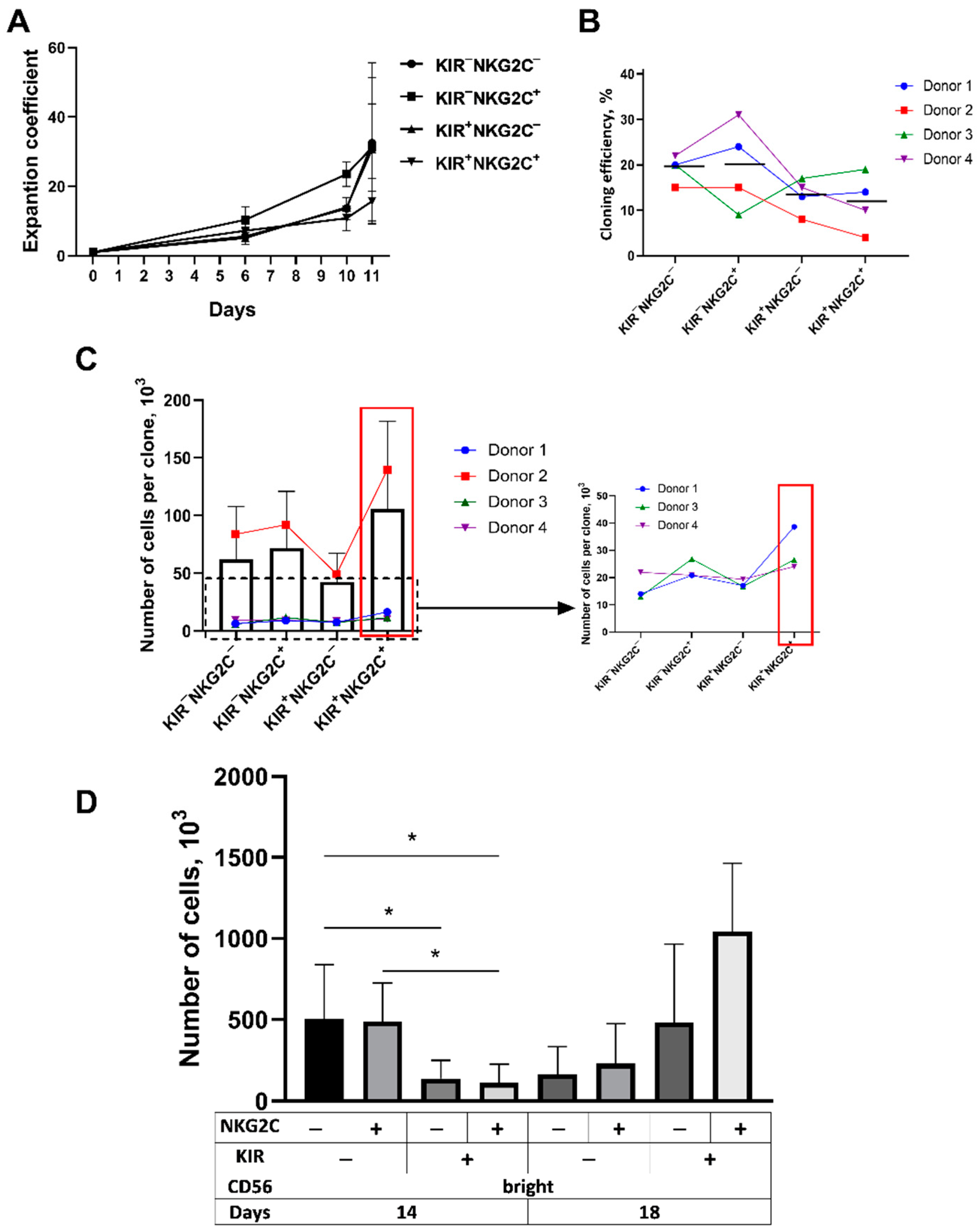
2.4. The Proliferative Potential of the CD57−KIR2DL2/3+ Subsets Is Better Realized at Later Stages of Cultivation
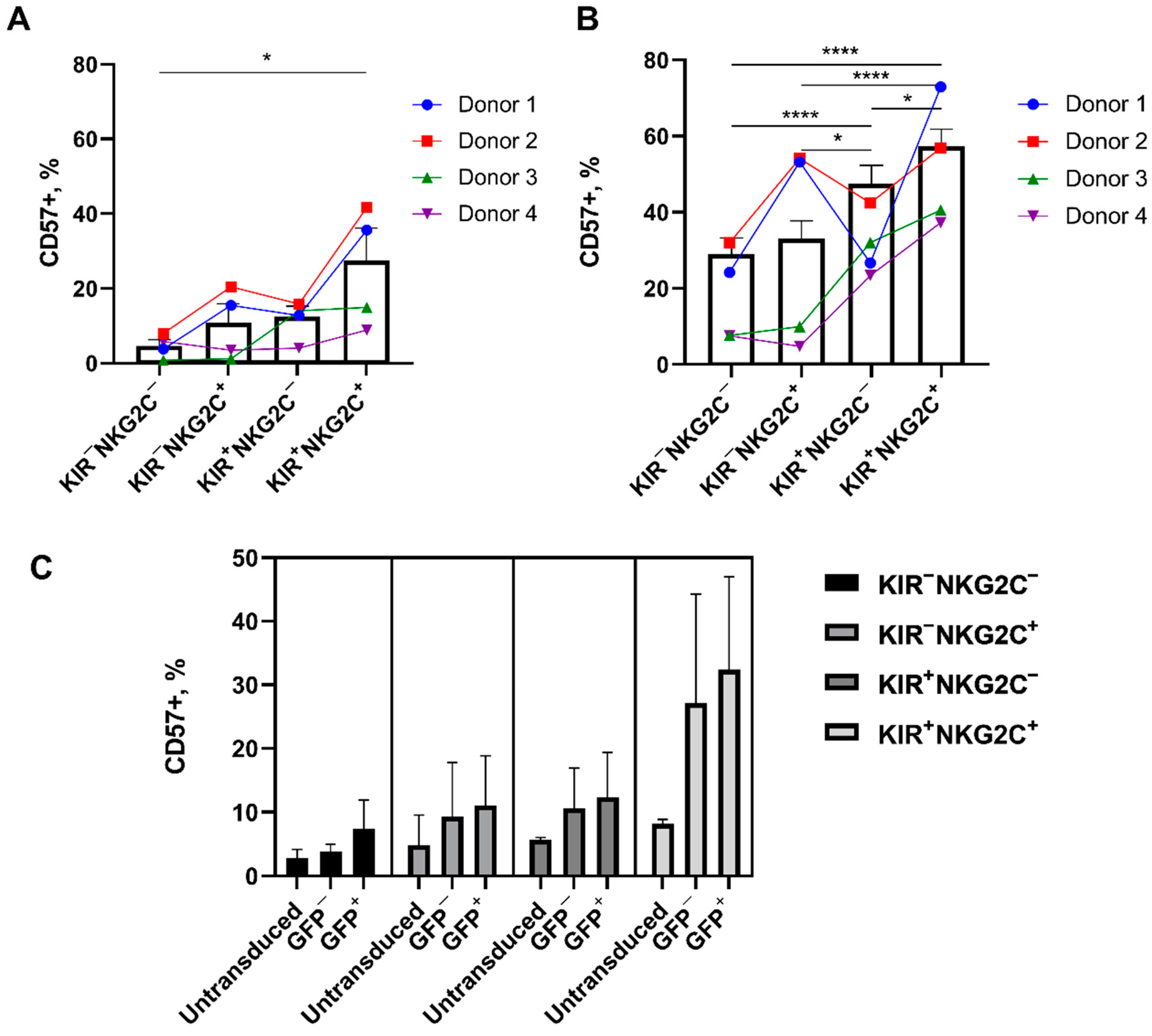
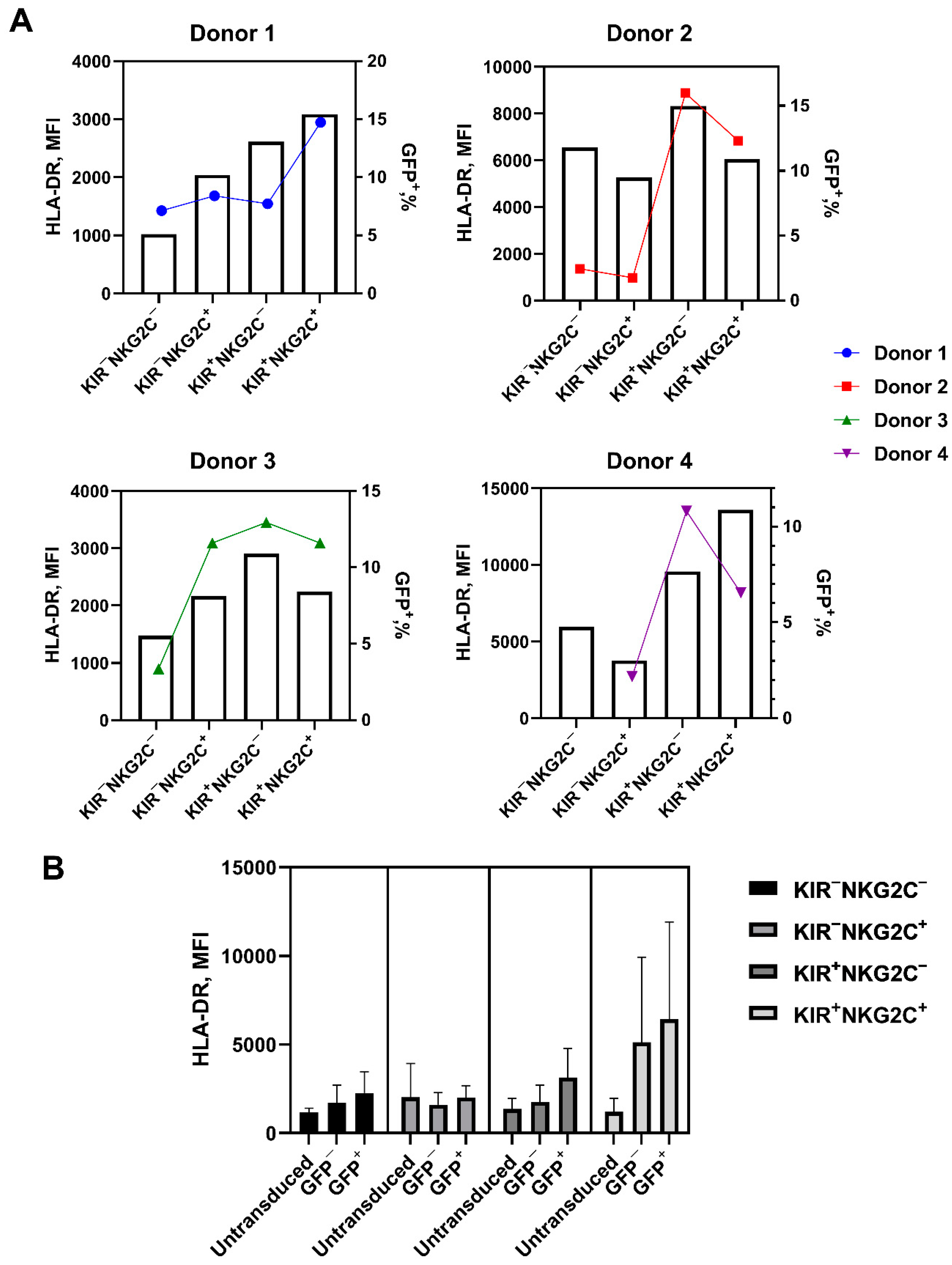
2.5. Retroviral Transduction Success Is Associated with the Surface Level of the HLA-DR Activation Marker and CD57 De Novo Expression
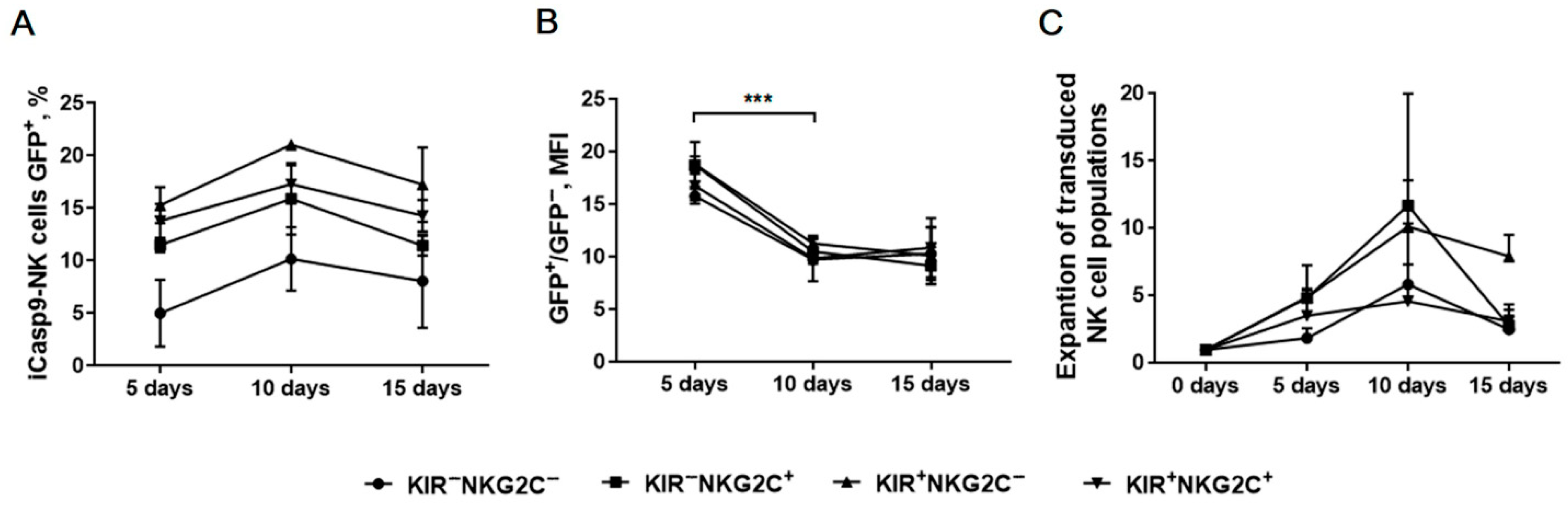
2.6. Verification of iCasp9 Transgene Stability in Modified NK Cells by Monitoring Reporter Gene Expression Level
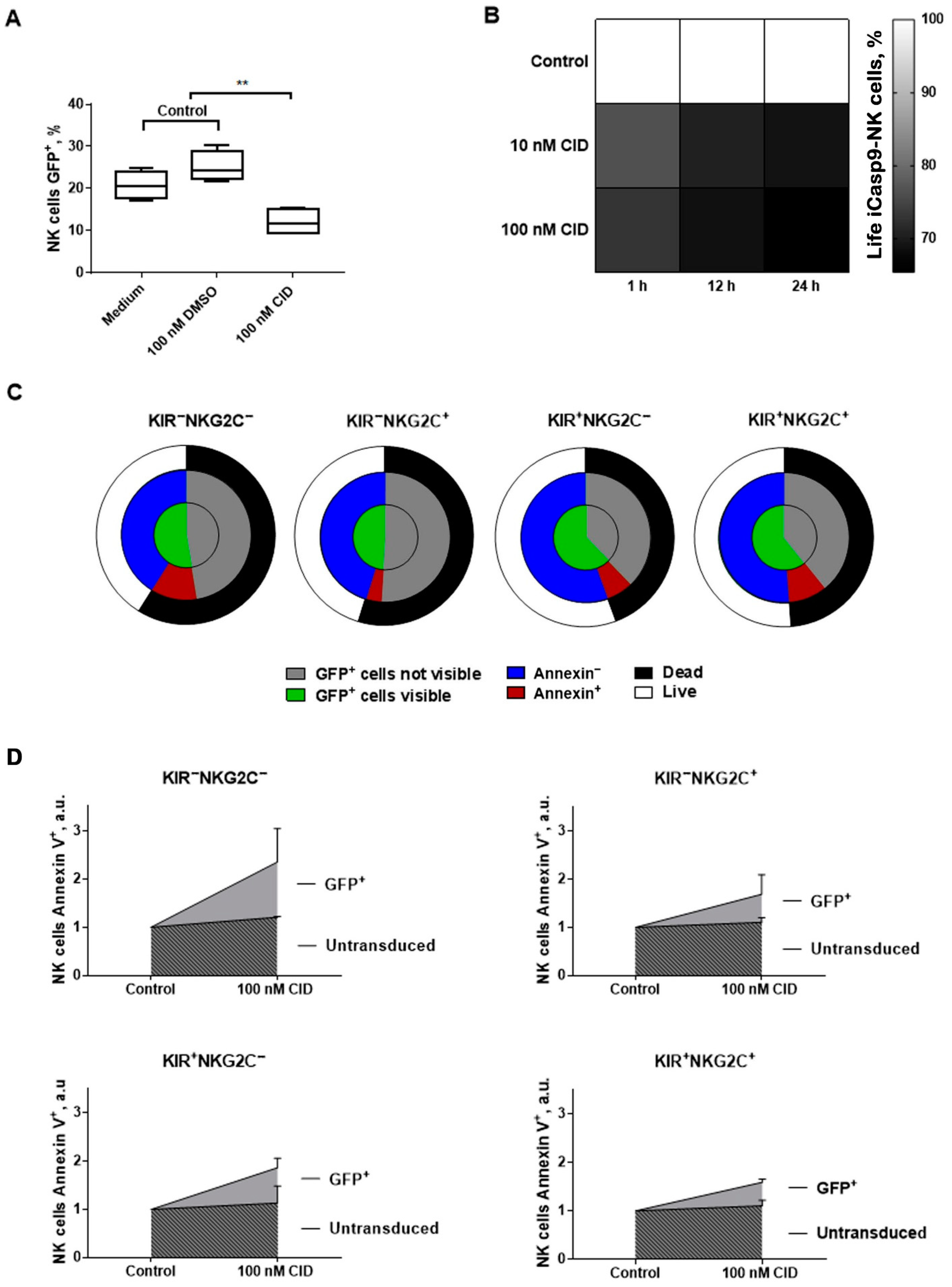
2.7. The Response of iCasp9-NK Cells to the Chemical Inductor of Dimerization in the CD57−KIR2DL2/3+/−NKG2C+/−Subsets
3. Discussion
4. Materials and Methods
4.1. Cell Lines
4.2. NK Cell Isolation
4.3. HCMV Serology Status
4.4. Generation of NK Cell Clones and Populations
4.5. Cell Staining and Flow Cytometry
4.6. Proliferative Capacity Measuring
4.7. Retroviral Transduction
4.8. Cell Death Induction in iCasp9-NK Cells by Chemical Inductor of Dimerization
4.9. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Morgan, M.A.; Büning, H.; Sauer, M.; Schambach, A. Use of cell and genome modification technologies to generate improved “off-the-shelf” CAR T and CAR NK cells. Front. Immunol. 2020, 11, 1965. [Google Scholar] [CrossRef] [PubMed]
- Tey, S.K. Adoptive T-cell therapy: Adverse events and safety switches. Clin. Transl. Immunol. 2014, 3, e17-7. [Google Scholar] [CrossRef] [PubMed]
- Matosevic, S. Viral and nonviral engineering of natural killer cells as emerging adoptive cancer immunotherapies. J. Immunol. Res. 2018, 2018, 4054815. [Google Scholar] [CrossRef]
- Lanier, L.L. Evolutionary struggles between NK cells and viruses. Nat. Rev. Immunol. 2008, 8, 259–268. [Google Scholar] [CrossRef] [PubMed]
- Van Erp, E.A.; Van Kampen, M.R.; Van Kasteren, P.B.; De Wit, J. Viral infection of human natural killer cells. Viruses 2019, 11, 243. [Google Scholar] [CrossRef] [PubMed]
- Streltsova, M.A.; Barsov, E.; Erokhina, S.A.; Kovalenko, E.I. Retroviral gene transfer into primary human NK cells activated by IL-2 and K562 feeder cells expressing membrane-bound IL-21. J. Immunol. Methods 2017, 450, 90–94. [Google Scholar] [CrossRef] [PubMed]
- Denman, C.J.; Senyukov, V.V.; Somanchi, S.S.; Phatarpekar, P.V.; Kopp, L.M.; Johnson, J.L.; Singh, H.; Hurton, L.; Maiti, S.N.; Huls, M.H.; et al. Membrane-bound IL-21 promotes sustained ex vivo proliferation of human natural killer cells. PLoS ONE 2012, 7, e30264. [Google Scholar] [CrossRef] [PubMed]
- Streltsova, M.A.; Erokhina, S.A.; Kanevskiy, L.M.; Lee, D.A.; Telford, W.G.; Sapozhnikov, A.M.; Kovalenko Id, E.I. Analysis of NK cell clones obtained using interleukin-2 and gene-modified K562 cells revealed the ability of «senescent» NK cells to lose CD57 expression and start expressing NKG2A. PLoS ONE 2018, 13, e0208469. [Google Scholar] [CrossRef]
- Streltsova, M.A.; Erokhina, S.A.; Kanevskiy, L.M.; Grechikhina, M.V.; Kobyzeva, P.A.; Lee, D.A.; Telford, W.G.; Sapozhnikov, A.M.; Kovalenko, E.I. Recurrent stimulation of natural killer cell clones with K562 expressing membrane-bound interleukin-21 affects their phenotype, interferon-γ production, and lifespan. Int. J. Mol. Sci. 2019, 20, 443. [Google Scholar] [CrossRef]
- Yang, C.; Siebert, J.R.; Burns, R.; Gerbec, Z.J.; Bonacci, B.; Rymaszewski, A.; Rau, M.; Riese, M.J.; Rao, S.; Carlson, K.S.; et al. Heterogeneity of human bone marrow and blood natural killer cells defined by single-cell transcriptome. Nat. Commun. 2019, 10, 1–16. [Google Scholar] [CrossRef]
- Lopez-Vergès, S.; Milush, J.M.; Pandey, S.; York, V.A.; Arakawa-Hoyt, J.; Pircher, H.; Norris, P.J.; Nixon, D.F.; Lanier, L.L. CD57 defines a functionally distinct population of mature NK cells in the human CD56dimCD16+ NK-cell subset. Blood 2010, 116, 3865–3874. [Google Scholar] [CrossRef] [PubMed]
- Chin, D.S.; Lim, C.S.Y.; Nordin, F.; Arifin, N.; Jun, T.G. Antibody-dependent cell-mediated cytotoxicity through natural killer (NK) cells: Unlocking NK cells for future immunotherapy. Curr. Pharm. Biotechnol. 2021, 22. [Google Scholar] [CrossRef]
- Tu, M.M.; Mahmoud, A.B.; Makrigiannis, A.P. Licensed and unlicensed NK cells: Differential roles in cancer and viral control. Front. Immunol. 2016, 7, 1–11. [Google Scholar] [CrossRef]
- Tesi, B.; Schlums, H.; Cichocki, F.; Bryceson, Y.T. Epigenetic regulation of adaptive NK cell diversification. Trends Immunol. 2016, 37, 451–461. [Google Scholar] [CrossRef] [PubMed]
- Beziat, V.; Liu, L.L.; Malmberg, J.; Ivarsson, M.A.; Sohlberg, E.; Björklund, A.T.; Reti, C.; Sverremark-Ekström, E.; Traherne, J.; Ljungman, P.; et al. NK cell responses to cytomegalovirus infection lead to stable imprints in the human KIR repertoire and involve activating KIRs. Blood 2013, 121, 2678–2689. [Google Scholar] [CrossRef] [PubMed]
- Kovalenko, E.I.; Streltsova, M.A.; Kanevskiy, L.M.; Erokhina, S.A.; Telford, W.G. Identification of human memory-like NK cells. Curr. Protoc. Cytom. 2017, 2017, 9. [Google Scholar] [CrossRef] [PubMed]
- Kobyzeva, P.A.; Streltsova, M.A.; Erokhina, S.A.; Kanevskiy, L.M.; Telford, W.G.; Sapozhnikov, A.M.; Kovalenko, E.I. CD56dimCD57−NKG2C+ NK cells retaining proliferative potential are possible precursors of CD57+NKG2C+ memory-like NK cells. J. Leukoc. Biol. 2020, 108, 1379–1395. [Google Scholar] [CrossRef] [PubMed]
- Jones, B.S.; Lamb, L.S.; Goldman, F.; Di Stasi, A. Improving the safety of cell therapy products by suicide gene transfer. Front. Pharmacol. 2014, 5, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Marin, V.; Cribioli, E.; Philip, B.; Tettamanti, S.; Pizzitola, I.; Biondi, A.; Biagi, E.; Pule, M. Comparison of different suicide-gene strategies for the safety improvement of genetically manipulated T cells. Hum. Gene Ther. Methods 2012, 23, 376–386. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Zhou, L.; Zhao, T.; Liu, X.; Zhang, P.; Liu, Y.; Zheng, X.; Li, Q. Caspase-9: Structure, mechanisms and clinical application. Oncotarget 2017, 8, 23996–24008. [Google Scholar] [CrossRef]
- Oelsner, S.; Waldmann, A.; Billmeier, A.; Röder, J.; Lindner, A.; Ullrich, E.; Marschalek, R.; Dotti, G.; Jung, G.; Große-Hovest, L.; et al. Genetically engineered CAR NK cells display selective cytotoxicity against FLT3-positive B-ALL and inhibit in vivo leukemia growth. Int. J. Cancer 2019, 145, 1935–1945. [Google Scholar] [CrossRef]
- Streltsova, M.A.; Ustiuzhanina, M.O.; Barsov, E.V.; Kust, S.A.; Velichinskii, R.A.; Kovalenko, E.I. Telomerase reverse transcriptase increases proliferation and lifespan of human NK cells without immortalization. Biomedicines 2021, 9, 662. [Google Scholar] [CrossRef]
- Costa-García, M.; Ataya, M.; Moraru, M.; Vilches, C.; López-Botet, M.; Muntasell, A. Human cytomegalovirus antigen presentation by HLA-DR+ NKG2C+ adaptive NK cells specifically activates polyfunctional effector memory CD4+ T lymphocytes. Front. Immunol. 2019, 10, 1–14. [Google Scholar] [CrossRef]
- Caligiuri, M.A.; Zmuidzinas, A.; Manley, T.J.; Levine, H.; Smith, K.A.; Ritz, J. Functional consequences of interleukin 2 receptor expression on resting human lymphocytes: Identification of a novel natural killer cell subset with high affinity receptors. J. Exp. Med. 1990, 171, 1509–1526. [Google Scholar] [CrossRef] [PubMed]
- Erokhina, S.A.; Streltsova, M.A.; Kanevskiy, L.M.; Telford, W.G.; Sapozhnikov, A.M.; Kovalenko, E.I. HLA-DR+ NK cells are mostly characterized by less mature phenotype and high functional activity. Immunol. Cell Biol. 2018, 96, 212–228. [Google Scholar] [CrossRef]
- Lange, C.; Blankenstein, T. Loss of retroviral gene expression in bone marrow reconstituted mice correlates with down-regulation of gene expression in long-term culture initiating cells. Gene Ther. 1997, 4, 303–308. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lupo, K.B.; Matosevic, S. Natural killer cells as allogeneic effectors in adoptive cancer immunotherapy. Cancers 2019, 11, 769. [Google Scholar] [CrossRef] [PubMed]
- Kordelas, L.; Steckel, N.K.; Horn, P.A.; Beelen, D.W.; Rebmann, V. The activating NKG2C receptor is significantly reduced in NK cells after allogeneic stem cell transplantation in patients with severe graft-versus-host disease. Int. J. Mol. Sci. 2016, 17, 1797. [Google Scholar] [CrossRef]
- Béziat, V.; Dalgard, O.; Asselah, T.; Halfon, P.; Bedossa, P.; Boudifa, A.; Hervier, B.; Theodorou, I.; Martinot, M.; Debré, P.; et al. CMV drives clonal expansion of NKG2C + NK cells expressing self-specific KIRs in chronic hepatitis patients. Eur. J. Immunol. 2012, 42, 447–457. [Google Scholar] [CrossRef]
- Bigley, A.B.; Baker, F.L.; Simpson, R.J. Cytomegalovirus: An unlikely ally in the fight against blood cancers? Clin. Exp. Immunol. 2018, 193, 265–274. [Google Scholar] [CrossRef]
- Di Vito, C.; Mikulak, J.; Mavilio, D. On the way to become a natural killer cell. Front. Immunol. 2019, 10, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Thomas, M.L.; Peterson, M.E.; Long, E.O. Cutting edge: NK cell licensing modulates adhesion to target cells. J. Immunol. 2013, 191, 3981–3985. [Google Scholar] [CrossRef] [PubMed]
- Liu, E.; Tong, Y.; Dotti, G.; Shaim, H.; Savoldo, B.; Mukherjee, M.; Orange, J.; Wan, X.; Lu, X.; Reynolds, A.; et al. Cord blood NK cells engineered to express IL-15 and a CD19- targeted CAR show long-term persistence and potent anti-tumor activity. Leukemia 2018, 32, 520–531. [Google Scholar] [CrossRef]
- Emery, D.W.; Yannaki, E.; Tubb, J.; Stamatoyannopoulos, G. A chromatin insulator protects retrovirus vectors from chromosomal position effects. Proc. Natl. Acad. Sci. USA 2000, 97, 9150–9155. [Google Scholar] [CrossRef]
- Straathof, K.C.; Pulè, M.A.; Yotnda, P.; Dotti, G.; Vanin, E.F.; Brenner, M.K.; Heslop, H.E.; Spencer, D.M.; Rooney, C.M. An inducible caspase 9 safety switch for T-cell therapy. Blood 2005, 105, 4247–4254. [Google Scholar] [CrossRef]
- Spencer, D.M.; Belshaw, P.J.; Chen, L.; Ho, S.N.; Randazzo, F.; Crabtree, G.R.; Schreiber, S.L. Functional analysis of Fas signaling in vivo using synthetic inducers of dimerization. Curr. Biol. 1996, 6, 839–847. [Google Scholar] [CrossRef][Green Version]
- MacCorkle, R.A.; Freeman, K.W.; Spencer, D.M. Synthetic activation of caspases: Artificial death switches. Proc. Natl. Acad. Sci. USA 1998, 95, 3655–3660. [Google Scholar] [CrossRef] [PubMed]
- Ramos, C.A.; Asgari, Z.; Liu, E.; Yvon, E.; Heslop, H.E.; Rooney, C.M.; Brenner, M.K.; Dotti, G. An inducible caspase 9 suicide gene to improve the safety of mesenchymal stromal cell therapies. Stem Cells 2010, 28, 1107–1115. [Google Scholar] [CrossRef]
- Thomis, D.C.; Marktel, S.; Bonini, C.; Traversari, C.; Gilman, M.; Bordignon, C.; Clackson, T. A Fas-based suicide switch in human T cells for the treatment of graft-versus-host disease. Blood 2001, 97, 1249–1257. [Google Scholar] [CrossRef]
- Barese, C.N.; Felizardo, T.C.; Sellers, S.E.; Keyvanfar, K.; Stasi, D.; Metzger, M.E.; Krouse, A.E.; Donahue, R.E.; Spencer, D.M. Regulated apoptosis of genetically-modified hematopoietic stem and progenitor cells via an inducible caspase-9 suicide gene in rhesus macaques. Stem Cells 2015, 33, 91–100. [Google Scholar] [CrossRef]
- Galluzzi, L.; Vitale, I.; Aaronson, S.A.; Abrams, J.M.; Adam, D.; Agostinis, P.; Alnemri, E.S.; Altucci, L.; Amelio, I.; Andrews, D.W.; et al. Molecular mechanisms of cell death: Recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 2018, 25, 486–541. [Google Scholar] [CrossRef] [PubMed]
- Felices, M.; Lenvik, T.R.; Ankarlo, D.E.M.; Foley, B.; Curstinger, J.; Luo, X.; Blazar, B.R.; Anderson, S.K.; Miller, J.S. Functional NK cell repertoires are maintained through IL-2Rα and FasL. J. Immunol. 2014, 192, 3889–3897. [Google Scholar] [CrossRef] [PubMed]
- Viant, C.; Guia, S.; Hennessy, R.J.; Rautela, J.; Pham, K.; Bernat, C.; Goh, W.; Jiao, Y.; Delconte, R.; Roger, M.; et al. Cell cycle progression dictates the requirement for BCL2 in natural killer cell survival. J. Exp. Med. 2017, 214, 491–510. [Google Scholar] [CrossRef] [PubMed]
- Sandrin, V.; Boson, B.; Salmon, P.; Gay, W.; Nègre, D.; Le Grand, R.; Trono, D.; Cosset, F.L. Lentiviral vectors pseudotyped with a modified RD114 envelope glycoprotein show increased stability in sera and augmented transduction of primary lymphocytes and CD34+ cells derived from human and nonhuman primates. Blood 2002, 100, 823–832. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Palamarchuk, A.I.; Alekseeva, N.A.; Streltsova, M.A.; Ustiuzhanina, M.O.; Kobyzeva, P.A.; Kust, S.A.; Grechikhina, M.V.; Boyko, A.A.; Shustova, O.A.; Sapozhnikov, A.M.; et al. Increased Susceptibility of the CD57− NK Cells Expressing KIR2DL2/3 and NKG2C to iCasp9 Gene Retroviral Transduction and the Relationships with Proliferative Potential, Activation Degree, and Death Induction Response. Int. J. Mol. Sci. 2021, 22, 13326. https://doi.org/10.3390/ijms222413326
Palamarchuk AI, Alekseeva NA, Streltsova MA, Ustiuzhanina MO, Kobyzeva PA, Kust SA, Grechikhina MV, Boyko AA, Shustova OA, Sapozhnikov AM, et al. Increased Susceptibility of the CD57− NK Cells Expressing KIR2DL2/3 and NKG2C to iCasp9 Gene Retroviral Transduction and the Relationships with Proliferative Potential, Activation Degree, and Death Induction Response. International Journal of Molecular Sciences. 2021; 22(24):13326. https://doi.org/10.3390/ijms222413326
Chicago/Turabian StylePalamarchuk, Anastasia I., Nadezhda A. Alekseeva, Maria A. Streltsova, Maria O. Ustiuzhanina, Polina A. Kobyzeva, Sofya A. Kust, Maria V. Grechikhina, Anna A. Boyko, Olga A. Shustova, Alexander M. Sapozhnikov, and et al. 2021. "Increased Susceptibility of the CD57− NK Cells Expressing KIR2DL2/3 and NKG2C to iCasp9 Gene Retroviral Transduction and the Relationships with Proliferative Potential, Activation Degree, and Death Induction Response" International Journal of Molecular Sciences 22, no. 24: 13326. https://doi.org/10.3390/ijms222413326
APA StylePalamarchuk, A. I., Alekseeva, N. A., Streltsova, M. A., Ustiuzhanina, M. O., Kobyzeva, P. A., Kust, S. A., Grechikhina, M. V., Boyko, A. A., Shustova, O. A., Sapozhnikov, A. M., & Kovalenko, E. I. (2021). Increased Susceptibility of the CD57− NK Cells Expressing KIR2DL2/3 and NKG2C to iCasp9 Gene Retroviral Transduction and the Relationships with Proliferative Potential, Activation Degree, and Death Induction Response. International Journal of Molecular Sciences, 22(24), 13326. https://doi.org/10.3390/ijms222413326