Identification of an ATP-Binding Cassette Transporter Implicated in Aluminum Tolerance in Wild Soybean (Glycine soja)
Abstract
:1. Introduction
2. Results
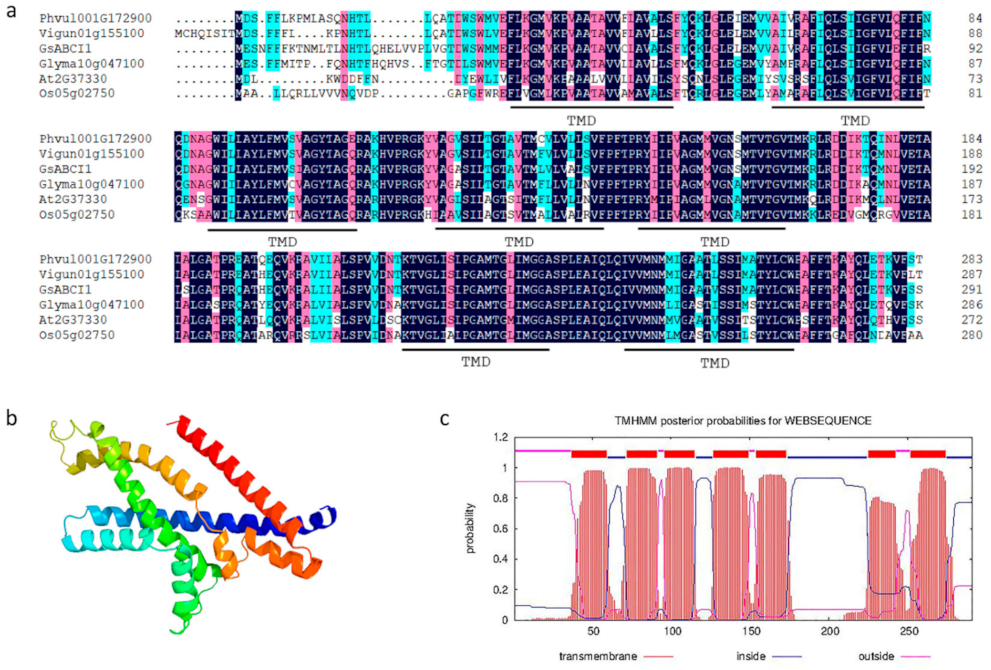
2.1. GsABCI1 Encodes an ATP-Binding Cassette Transporter Protein
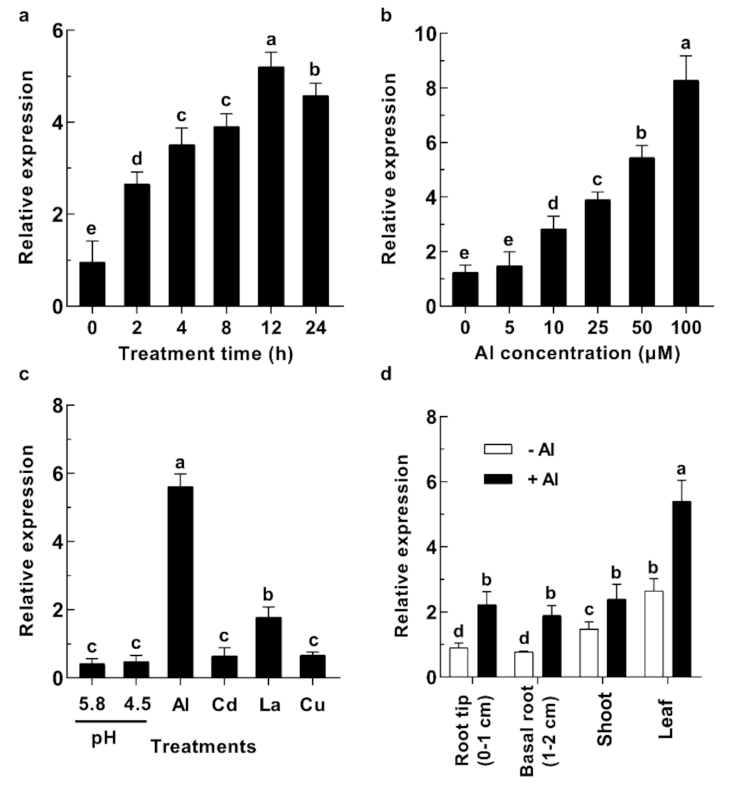
2.2. GsABCI1 Expression Responses to Aluminum Stress in Wild Soybean (Glycine soja)
2.3. Generation and Detection of GsABCI1 Transgenic Lines
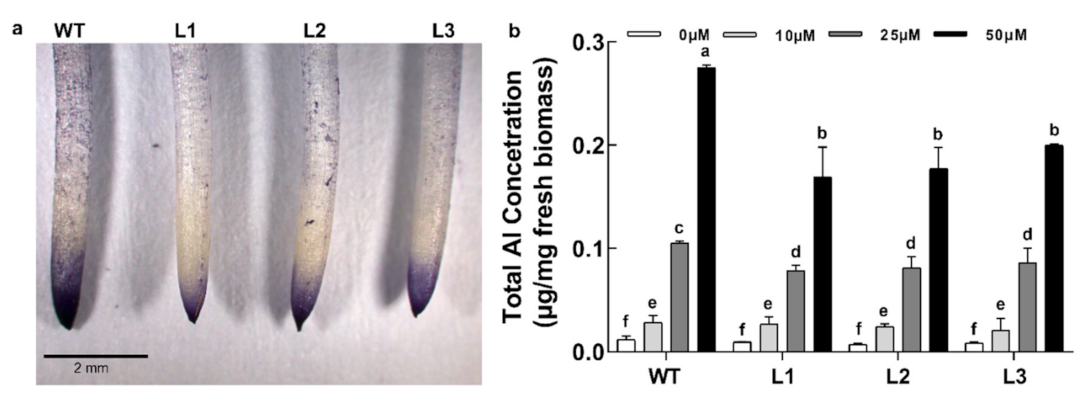
2.4. Overexpression of GsABCI1 Enhances Phenotypic Tolerance to Aluminum
2.5. GsABCI1 Involves the Translocation of Aluminum in Roots
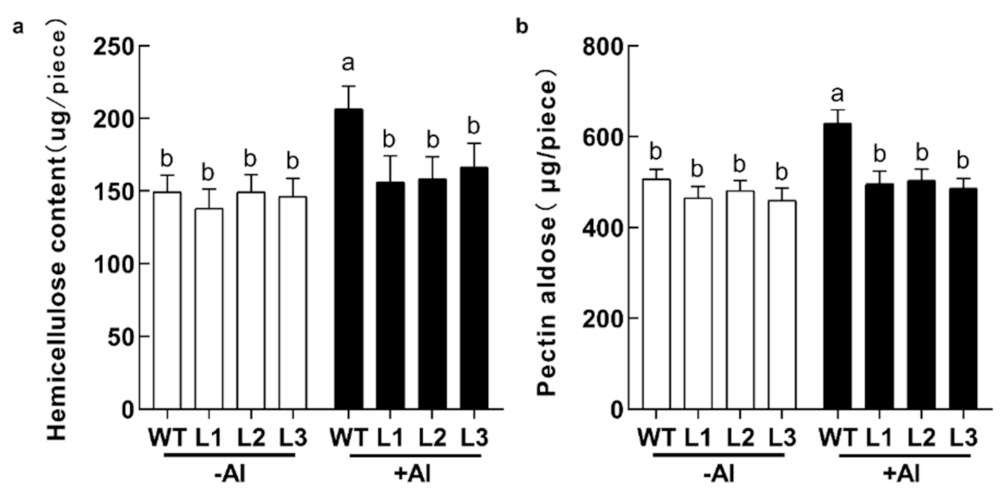
2.6. Root Component Changes of GsABCI1 Transgenic Lines under Aluminum Stress
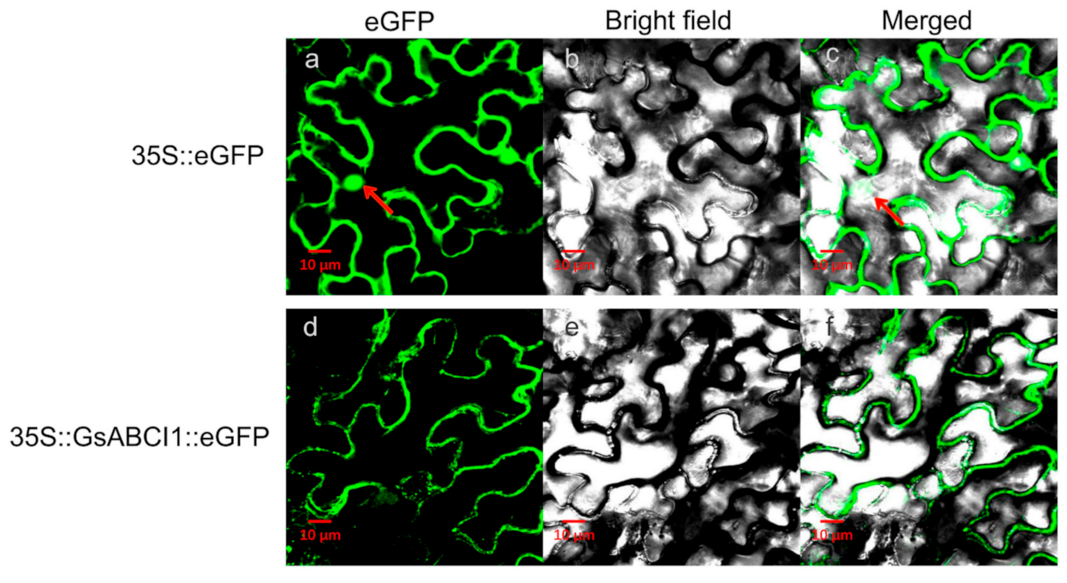
2.7. GsABCI1 Subcellular Localization in the Plasma Membrane
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growing Conditions
4.2. Isolation of the GsABCI1 Gene from Wild Soybean and Vector Construction
4.3. Generation of GsABCI1 Transgenic Lines
4.4. Detection of GsABCI1 Transgenic Lines
4.5. Phenotypes of GsABCI1 Transgenic Lines
4.6. Bioinformatics Analysis of GsABCI1
4.7. Subcellular Localization of GsABCI1
4.8. Expression Pattern of GsABCI1
4.9. Analysis of Al Accumulation Patterns
4.10. Root Component Analysis of GsABCI1 Transgenic Lines
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xia, J.; Yamaji, N.; Kasai, T.; Ma, J.F. Plasma membrane-localized transporter for aluminum in rice. Proc. Natl. Acad. Sci. USA 2010, 107, 18381–18385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Von Uexküll, H.R.; Mutert, E. Global extent, development and economic impact of acid soils. Plant Soil 1995, 171, 1–15. [Google Scholar] [CrossRef]
- Kochian, L.V.; Hoekenga, O.A.; Piñeros, M.A. How do crop plants tolerate acid soils? Mechanisms of aluminum tolerance and phosphorous efficiency. Annu. Rev. Plant Biol. 2004, 55, 459–493. [Google Scholar] [CrossRef] [PubMed]
- Eticha, D.; Zahn, M.; Bremer, M.; Yang, Z.; Rangel, A.F.; Rao, I.M.; Horst, W.J. Transcriptomic analysis reveals differential gene expression in response to aluminium in common bean (Phaseolus vulgaris) genotypes. Ann. Bot. 2010, 105, 1119–1128. [Google Scholar] [CrossRef] [Green Version]
- Barceló, J.; Charlotte, P. Fast root growth responses, root exudates, and internal detoxification as clues to the mechanisms of aluminium toxicity and resistance: A review. Environ. Exp. Bot. 2002, 48, 75–92. [Google Scholar] [CrossRef]
- Kochian, L.V. Cellular mechanisms of aluminum toxicity and resistance in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1995, 46, 237–260. [Google Scholar] [CrossRef]
- Ma, J.F.; Ryan, P.R.; Delhaize, E. Aluminium tolerance in plants and the complexing role of organic acids. Trends Plant Sci. 2001, 6, 273–278. [Google Scholar] [CrossRef]
- Kinraide, T.B. Identity of the rhizotoxic aluminum species. Plant Soil 1991, 134, 167–178. [Google Scholar] [CrossRef]
- Kollmeier, M.; Felle, H.H.; Horst, W.J. Genotypical Differences in Aluminum Resistance of Maize Are Expressed in the Distal Part of the Transition Zone. Is Reduced Basipetal Auxin Flow Involved in Inhibition of Root Elongation by Aluminum? Plant Physiol. 2000, 122, 945–956. [Google Scholar] [CrossRef] [Green Version]
- Sivaguru, M.; Balusška, F.; Volkmann, D.; Felle, H.H.; Horst, W.J. Impacts of Aluminum on the Cytoskeleton of the Maize Root Apex. Short-Term Effects on the Distal Part of the Transition Zone1. Plant Physiol. 1999, 119, 1073–1082. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, J.F.; Hiradate, S. Form of aluminium for uptake and translocation in buckwheat (Fagopyrum esculentum Moench). Planta 2000, 211, 355–360. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.F.; Taketa, S.; Yang, Z.M. Aluminum Tolerance Genes on the Short Arm of Chromosome 3R Are Linked to Organic Acid Release in Triticale. Plant Physiol. 2000, 122, 687–694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sade, H.; Meriga, B.; Surapu, V.; Gadi, J.; Sunita, M.S.L.; Suravajhala, P.; Kishor, P.B.K. Toxicity and tolerance of aluminum in plants: Tailoring plants to suit to acid soils. BioMetals 2016, 29, 187–210. [Google Scholar] [CrossRef] [PubMed]
- Magalhaes, J.V.; Liu, J.; Guimarães, C.T.; Lana, U.G.P.; Alves, V.M.C.; Wang, Y.-H.; E Schaffert, R.; A Hoekenga, O.; Pineros, M.; E Shaff, J.; et al. A gene in the multidrug and toxic compound extrusion (MATE) family confers aluminum tolerance in sorghum. Nat. Genet. 2007, 39, 1156–1161. [Google Scholar] [CrossRef]
- Horst, W.J.; Schmohl, N.; Kollmeier, M.; Ka, F.E.B.; Sivaguru, M. Does aluminium affect root growth of maize through interaction with the cell wall—Plasma membrane—Cytoskeleton continuum? Plant Soil 1999, 215, 163–174. [Google Scholar] [CrossRef]
- Li, X.F.; Ma, J.F.; Matsumoto, H. Pattern of Aluminum-Induced Secretion of Organic Acids Differs between Rye and Wheat. Plant Physiol. 2000, 123, 1537–1544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.L.; Zhu, X.F.; Peng, Y.X.; Zheng, C.; Li, G.X.; Liu, Y.; Shi, Y.Z.; Zheng, S.J. Cell Wall Hemicellulose Contributes Significantly to Aluminum Adsorption and Root Growth in Arabidopsis. Plant Physiol. 2011, 155, 1885–1892. [Google Scholar] [CrossRef] [Green Version]
- Takeda, K.; Kariuda, M.; Itoi, H. Blueing of sepal colour of Hydrangea macrophylla. Phytochemistry 1985, 24, 2251–2254. [Google Scholar] [CrossRef]
- Ma, J.F.; Hiradate, S.; Nomoto, K.; Iwashita, T.; Matsumoto, H. Internal Detoxification Mechanism of Al in Hydrangea (Identification of Al Form in the Leaves). Plant Physiol. 1997, 113, 1033–1039. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, S.J.; Ma, J.F.; Matsumoto, H. High aluminum resistance in buckwheat.I. Al-induced specific secretion of oxalic acid from root tips. Plant Physiol. 1998, 117, 745–751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simões, C.C.; Melo, J.O.; Magalhaes, J.V.; Guimarães, C.T. Genetic and molecularmechanisms of aluminum tolerance in plants. Genet. Mol. Res. 2012, 11, 1949–1957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, R.; Ma, J.; Kyo, M.; Iwashita, T. Compartmentation of aluminium in leaves of an Al-accumulator, Fagopyrum esculentum Moench. Planta 2002, 215, 394–398. [Google Scholar] [CrossRef] [PubMed]
- Locher, K.P. Mechanistic diversity in ATP-binding cassette (ABC) transporters. Nat. Struct. Mol. Biol. 2016, 23, 487–493. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, V.N.T.; Moon, S.; Jung, K.-H. Genome-wide expression analysis of rice ABC transporter family across spatio-temporal samples and in response to abiotic stresses. J. Plant Physiol. 2014, 171, 1276–1288. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.K.; Choi, J.; Rabbee, M.F.; Baek, K.H. In Silico Genome-Wide Analysis of the ATP-Binding Cassette Transporter Gene Family in Soybean (Glycine max L.) and Their Expression Profiling. BioMed Res. Int. 2019, 2019, 8150523. [Google Scholar] [PubMed] [Green Version]
- Garcia, O.; Bouige, P.; Forestier, C.; Dassa, E. Inventory and Comparative Analysis of Rice and Arabidopsis ATP-binding Cassette (ABC) Systems. J. Mol. Biol. 2004, 343, 249–265. [Google Scholar] [CrossRef]
- Arana, M.R.; Altenberg, G.A.; Altenberg, M.R.A.A.G. ATP-binding Cassette Exporters: Structure and Mechanism with a Focus on P-glycoprotein and MRP1. Curr. Med. Chem. 2019, 26, 1062–1078. [Google Scholar] [CrossRef] [PubMed]
- Verrier, P.J.; Bird, D.; Burla, B.; Dassa, E.; Forestier, C.; Geisler, M.; Klein, M.; Kolukisaoglu, U.; Lee, Y.; Martinoia, E.; et al. Plant ABC proteins--a unified nomenclature and updated inventory. Trends Plant Sci. 2008, 13, 151–159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watts, G.F. An ABC of HDL: A paradigm shift in managing lipid disorders. Mod. Med. 2004, 29, 31–42. [Google Scholar]
- Dong, J.; Pineros, M.; Li, X.; Yang, H.; Liu, Y.; Murphy, A.S.; Kochian, L.V.; Liu, D. An Arabidopsis ABC Transporter Mediates Phosphate Deficiency-Induced Remodeling of Root Architecture by Modulating Iron Homeostasis in Roots. Mol. Plant 2017, 10, 244–259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rea, P.A. Plant ATP-Binding Cassette Transporters. Annu. Rev. Plant Biol. 2007, 58, 347–375. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.C.; Yamaji, N.; Motoyama, R.; Nagamura, Y.; Ma, J.F. Up-Regulation of a Magnesium Transporter Gene OsMGT1 Is Required for Conferring Aluminum Tolerance in Rice. Plant Physiol. 2012, 159, 1624–1633. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.-F.; Yamaji, N.; Mitani, N.; Yano, M.; Nagamura, Y.; Ma, J.F. A Bacterial-Type ABC Transporter Is Involved in Aluminum Tolerance in Rice. Plant Cell 2009, 21, 655–667. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tahara, K.; Nishiguchi, M.; Frolov, A.; Mittasch, J.; Milkowski, C. Identification of UDP glucosyltransferases from the aluminum-resistant tree Eucalyptus camaldulensis forming β-glucogallin, the precursor of hydrolyzable tannins. Phytochemistry 2018, 152, 154–161. [Google Scholar] [CrossRef]
- Roselló, M.; Poschenrieder, C.; Gunse, B.; Barceló, J.; Llugany, M. Differential activation of genes related to aluminium tolerance in two contrasting rice cultivars. J. Inorg. Biochem. 2015, 152, 160–166. [Google Scholar] [CrossRef] [PubMed]
- Che, J.; Yamaji, N.; Yokosho, K.; Shen, R.F.; Ma, J.F. Two Genes Encoding a Bacterial-Type ATP-Binding Cassette Transporter are Implicated in Aluminum Tolerance in Buckwheat. Plant Cell Physiol. 2018, 59, 2502–2511. [Google Scholar] [CrossRef]
- Gao, J.; Liang, Y.; Li, J.; Wang, S.; Zhan, M.; Zheng, M.; Li, H.; Yang, Z. Identification of a bacterial-type ATP-binding cassette transporter implicated in aluminum tolerance in sweet sorghum (Sorghum bicolor L.). Plant Signal. Behav. 2021, 16, 1916211. [Google Scholar] [CrossRef] [PubMed]
- Larsen, P.B.; Geisler, M.; Jones, C.A.; Williams, K.M.; Cancel, J.D. ALS3 encodes a phloem-localized ABC transporter-like protein that is required for aluminum tolerance in Arabidopsis. Plant J. 2005, 41, 353–363. [Google Scholar] [CrossRef]
- Reyna-Llorens, I.; Corrales, I.; Poschenrieder, C.; Barcelo, J.; Cruz-Ortega, R. Both aluminum and ABA induce the expression of an ABC-like transporter gene (FeALS3) in the Al-tolerant species Fagopyrum esculentum. Environ. Exp. Bot. 2015, 111, 74–82. [Google Scholar] [CrossRef]
- Agrahari, R.K.; Kobayashi, Y.; Borgohain, P. Aluminum-Specific Upregulation of GmALS3 in the Shoots of Soybeans: A Potential Biomarker for Managing Soybean Production in Acidic Soil Regions. Agronomy 2020, 10, 1228. [Google Scholar] [CrossRef]
- Liu, W.; Feng, X.; Cao, F.; Wu, D.; Zhang, G.; Vincze, E.; Wang, Y.; Chen, Z.-H.; Wu, F. An ATP binding cassette transporter HvABCB25 confers aluminum detoxification in wild barley. J. Hazard. Mater. 2021, 401, 123371. [Google Scholar] [CrossRef]
- Niu, L.; Li, H.; Song, Z.; Dong, B.; Cao, H.; Liu, T.; Du, T.; Yang, W.; Amin, R.; Wang, L.; et al. The functional analysis of ABCG transporters in the adaptation of pigeon pea (Cajanus cajan) to abiotic stresses. PeerJ 2021, 9, e10688. [Google Scholar] [CrossRef]
- Lei, G.J.; Yokosho, K.; Yamaji, N.; Fujii-Kashino, M.; Ma, J.F. Functional characterization of two half-size ABC transporter genes in aluminium-accumulating buckwheat. New Phytol. 2017, 215, 1080–1089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, C.-F.; Yamaji, N.; Chen, Z.; Ma, J.F. A tonoplast-localized half-size ABC transporter is required for internal detoxification of aluminum in rice. Plant J. 2012, 69, 857–867. [Google Scholar] [CrossRef]
- Liu, Y.; Du, H.; Li, P.; Shen, Y.; Peng, H.; Liu, S.; Zhou, G.-A.; Zhang, H.; Liu, Z.; Shi, M.; et al. Pan-Genome of Wild and Cultivated Soybeans. Cell 2020, 182, 162–176. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; You, Q.; Duan, G.; Ren, J.; Chu, S.; Zhao, J.; Li, X.; Zhou, X.; Jiao, Y. Quantitative trait loci analysis of seed oil content and composition of wild and cultivated soybean. BMC Plant Biol. 2020, 20, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Magalhaes, J.V.; Shaff, J.; Kochian, L.V. Aluminum-activated citrate and malate transporters from the MATE and ALMT families function independently to confer Arabidopsis aluminum tolerance. Plant J. 2009, 57, 389–399. [Google Scholar] [CrossRef] [Green Version]
- Ma, J.F. Syndrome of Aluminum Toxicity and Diversity of Aluminum Resistance in Higher Plants. Adv. Appl. Microbiol. 2007, 264, 225–252. [Google Scholar]
- Ma, J.F. Role of Organic Acids in Detoxification of Aluminum in Higher Plants. Plant Cell Physiol. 2000, 41, 383–390. [Google Scholar] [CrossRef] [Green Version]
- Hoekenga, O.A.; Maron, L.G.; Piñeros, M.A.; Cançado, G.M.A.; Shaff, J.; Kobayashi, Y.; Ryan, P.R.; Dong, B.; Delhaize, E.; Sasaki, T.; et al. AtALMT1, which encodes a malate transporter, is identified as one of several genes critical for aluminum tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 2006, 103, 9738–9743. [Google Scholar] [CrossRef] [Green Version]
- Sasaki, T.; Yamamoto, Y.; Ezaki, B.; Katsuhara, M.; Ahn, S.J.; Ryan, P.R.; Delhaize, E.; Matsumoto, H. A wheat gene encoding an aluminum-activated malate transporter. Plant J. 2004, 37, 645–653. [Google Scholar] [CrossRef]
- Liang, C.; Pineros, M.; Tian, J.; Yao, Z.; Sun, L.; Liu, J.; Shaff, J.; Coluccio, A.; Kochian, L.; Liao, H. Low pH, Aluminum, and Phosphorus Coordinately Regulate Malate Exudation through GmALMT1 to Improve Soybean Adaptation to Acid Soils. Plant Physiol. 2013, 161, 1347–1361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lou, H.Q.; Fan, W.; Xu, J.M.; Gong, Y.L.; Jin, J.F.; Chen, W.W.; Liu, L.Y.; Hai, M.R.; Yang, J.L.; Zheng, S.J. An Oxalyl-CoA Synthetase Is Involved in Oxalate Degradation and Aluminum Tolerance. Plant Physiol. 2016, 172, 1679–1690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia-Oliveira, A.L.; Martins-Lopes, P.; Tolrá, R.; Poschenrieder, C.; Tarquis, M.; Guedes-Pinto, H.; Benito, C. Molecular characterization of the citrate transporter gene TaMATE1 and expression analysis of upstream genes involved in organic acid transport under Al stress in bread wheat (Triticum aestivum). Physiol. Plant 2014, 152, 441–452. [Google Scholar] [CrossRef] [PubMed]
- Yokosho, K.; Yamaji, N.; Fujii-Kashino, M.; Ma, J.F. Functional analysis of a MATE gene OsFRDL2 revealed its involvement in Al-induced secretion of citrate, but a lower contribution to Al tolerance in rice. Plant Cell Physiol. 2016, 57, 976–985. [Google Scholar] [CrossRef] [Green Version]
- Deng, W.; Luo, K.; Li, D.; Zheng, X.; Wei, X.; Smith, W.; Thammina, C.; Lu, L.; Li, Y.; Pei, Y. Overexpression of an Arabidopsis magnesium transport gene, AtMGT1, in Nicotiana benthamiana confers Al tolerance. J. Exp. Bot. 2006, 57, 4235–4243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, A.; Chen, X.; Wang, H. Genome-wide investigation and expression analysis suggest diverse roles and genetic redundancy of Pht1 family genes in response to Pi deficiency in tomato. BMC Plant Biol. 2014, 14, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iuchi, S.; Koyama, H.; Iuchi, A.; Kobayashi, Y.; Kitabayashi, S.; Ikka, T.; Hirayama, T.; Shinozaki, K.; Kobayashi, M.; Kobayashi, Y.; et al. Zinc finger protein STOP1 is critical for proton tolerance in Arabidopsis and coregulates a key gene in aluminum tolerance. Proc. Natl. Acad. Sci. USA 2007, 104, 9900–9905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, J.; Park, J.; Choi, H.; Burla, B.; Kretzschmar, T.; Lee, Y.; Martinoia, E. Plant ABC Transporters. Arab. Book 2011, 9, e0153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larsen, P.B.; Cancel, J.; Rounds, M.; Ochoa, V. Arabidopsis ALS1 encodes a root tip and stele localized half type ABC transporter required for root growth in an aluminum toxic environment. Planta 2007, 225, 1447–1458. [Google Scholar] [CrossRef]
- Li, C.X.; Yan, J.Y.; Ren, J.Y.; Sun, L.; Xu, C.; Li, G.X.; Ding, Z.J.; Zheng, S.J. A WRKY transcription factor confers aluminum tolerance via regulation of cell wall modifying genes. J. Integr. Plant Biol. 2019, 62, 1176–1192. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.-F.; Yamaji, N.; Ma, J.F. Knockout of a Bacterial-Type ATP-Binding Cassette Transporter Gene, AtSTAR1, Results in Increased Aluminum Sensitivity in Arabidopsis. Plant Physiol. 2010, 153, 1669–1677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jaskowiak, J.; Kwasniewska, J.; Milewska-Hendel, A.; Kurczynska, E.U.; Szurman-Zubrzycka, M.; Szarejko, I. Aluminum Alters the Histology and Pectin Cell Wall Composition of Barley Roots. Int. J. Mol. Sci. 2019, 20, 3039. [Google Scholar] [CrossRef] [Green Version]
- Sujkowska-Rybkowska, M.; Borucki, W. Pectins esterification in the apoplast of aluminum-treated pea root nodules. J. Plant Physiol. 2015, 184, 1–7. [Google Scholar] [CrossRef]
- Yang, J.L.; Li, Y.Y.; Zhang, Y.J.; Zhang, S.S.; Wu, Y.R.; Wu, P.; Zheng, S.J. Cell Wall Polysaccharides Are Specifically Involved in the Exclusion of Aluminum from the Rice Root Apex. Plant Physiol. 2008, 146, 602–611. [Google Scholar] [CrossRef]
- Guillon, F.; Moïse, A.; Quemener, B.; Bouchet, B.; Devaux, M.-F.; Alvarado, C.; Lahaye, M. Remodeling of pectin and hemicelluloses in tomato pericarp during fruit growth. Plant Sci. 2017, 257, 48–62. [Google Scholar] [CrossRef] [PubMed]
- Exley, C.; Burgess, E.; Day, J.P.; Jeffery, E.H.; Melethil, S.; Yokel, R.A. Aluminum toxicokinetics. J. Toxicol. Environ. Health 1996, 48, 569–584. [Google Scholar] [PubMed]
- Zeng, Q.-Y.; Yang, C.-Y.; Ma, Q.-B.; Li, X.-P.; Dong, W.-W.; Nian, H. Identification of wild soybean miRNAs and their target genes responsive to aluminum stress. BMC Plant Biol. 2012, 12, 182. [Google Scholar] [CrossRef] [Green Version]
- Hoagland, D.R.; Arnon, D.I. The water-culture method for growing plants without soil. Circ. Calif. Agric. Exp. Stn. 1950, 347, 32. [Google Scholar]
- Lu, J.; Suo, H.; Yi, R.; Ma, Q.; Nian, H. Glyma11g13220, a homolog of the vernalization pathway gene VERNALIZATION 1 from soybean [Glycine max (L.) Merr.], promotes flowering in Arabidopsis thaliana. BMC Plant Biol. 2015, 15, 232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frame, B.R.; Shou, H.; Chikwamba, R.K.; Zhang, Z.; Xiang, C.; Fonger, T.M.; Pegg, S.E.K.; Li, B.; Nettleton, D.S.; Pei, D.; et al. Agrobacterium tumefaciens-Mediated Transformation of Maize Embryos Using a Standard Binary Vector System. Plant Physiol. 2002, 129, 13–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- E Hood, E.; Helmer, G.L.; Fraley, R.T.; Chilton, M.D. The hypervirulence of Agrobacterium tumefaciens A281 is encoded in a region of pTiBo542 outside of T-DNA. J. Bacteriol. 1986, 168, 1291–1301. [Google Scholar] [CrossRef] [Green Version]
- Suo, H.; Ma, Q.; Ye, K.; Yang, C.; Tang, Y.; Hao, J.; Zhang, Z.J.; Chen, M.; Feng, Y.-Q.; Nian, H. Overexpression of AtDREB1A Causes a Severe Dwarf Phenotype by Decreasing Endogenous Gibberellin Levels in Soybean [Glycine max (L.) Merr.]. PLoS ONE 2012, 7, e45568. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [Green Version]
- Murashige, T.; Skoog, F. A Revised Medium for Rapid Growth and Bio Assays with Tobacco Tissue Cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Ma, Q.; Yi, R.; Li, L.; Liang, Z.; Zeng, T.; Zhang, Y.; Huang, H.; Zhang, X.; Yin, X.; Cai, Z.; et al. GsMATE encoding a multidrug and toxic compound extrusion transporter enhances aluminum tolerance in Arabidopsis thaliana. BMC Plant Biol. 2018, 18, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Zia, M.; Rizvi, Z.F.; And, R. Agrobacterium mediated transformation of soybean (Glycine Max L.): Some conditions standardization. Pak. J. Bot. 2010, 42, 2269–2279. [Google Scholar]
- Nezames, C.D.; Ochoa, V.; Larsen, P.B. Mutational loss of Arabidopsis SLOW WALKER2 results in reduced endogenous spermine concomitant with increased aluminum sensitivity. Funct. Plant Biol. 2013, 40, 67–78. [Google Scholar] [CrossRef]
- Cai, Z.; Cheng, Y.; Xian, P.; Ma, Q.; Wen, K.; Xia, Q.; Zhang, G.; Nian, H. Acid phosphatase gene GmHAD1 linked to low phosphorus tolerance in soybean, through fine mapping. Theor. Appl. Genet. 2018, 131, 1715–1728. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Xia, Z.; Cai, Z.; Li, L.; Cheng, Y.; Liu, J.; Nian, H. GmWRKY16 Enhances Drought and Salt Tolerance Through an ABA-Mediated Pathway in Arabidopsis thaliana . Front. Plant Sci. 2019, 9, 1979. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delhaize, E.; Craig, S.; Beaton, C.D.; Bennet, R.J.; Jagadish, V.C.; Randall, P.J. Aluminum Tolerance in Wheat (Triticum aestivum L.) (I. Uptake and Distribution of Aluminum in Root Apices). Plant Physiol. 1993, 103, 685–693. [Google Scholar] [CrossRef] [Green Version]
- Konishi, T.; Kotake, T.; Tsumuraya, Y. Chain elongation of pectic beta-(1-->4)-galactan by a partially purified galactosyltransferase from soybean (Glycine max Merr.) hypocotyls. Planta 2007, 226, 571–579. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wen, K.; Pan, H.; Li, X.; Huang, R.; Ma, Q.; Nian, H. Identification of an ATP-Binding Cassette Transporter Implicated in Aluminum Tolerance in Wild Soybean (Glycine soja). Int. J. Mol. Sci. 2021, 22, 13264. https://doi.org/10.3390/ijms222413264
Wen K, Pan H, Li X, Huang R, Ma Q, Nian H. Identification of an ATP-Binding Cassette Transporter Implicated in Aluminum Tolerance in Wild Soybean (Glycine soja). International Journal of Molecular Sciences. 2021; 22(24):13264. https://doi.org/10.3390/ijms222413264
Chicago/Turabian StyleWen, Ke, Huanting Pan, Xingang Li, Rong Huang, Qibin Ma, and Hai Nian. 2021. "Identification of an ATP-Binding Cassette Transporter Implicated in Aluminum Tolerance in Wild Soybean (Glycine soja)" International Journal of Molecular Sciences 22, no. 24: 13264. https://doi.org/10.3390/ijms222413264
APA StyleWen, K., Pan, H., Li, X., Huang, R., Ma, Q., & Nian, H. (2021). Identification of an ATP-Binding Cassette Transporter Implicated in Aluminum Tolerance in Wild Soybean (Glycine soja). International Journal of Molecular Sciences, 22(24), 13264. https://doi.org/10.3390/ijms222413264





